Abstract
Endothelial monocyte-activating polypeptide II (EMAP II) is a proinflammatory cytokine and a chemoattractant for monocytes. We show here that, in the mouse embryo, EMAP II mRNA was most abundant at sites of tissue remodeling where many apoptotic cells could be detected by terminal deoxynucleotidyltransferase-mediated dUTP end labeling. Removal of dead cells is known to require macrophages, and these were found to colocalize with areas of EMAP II mRNA expression and programmed cell death. In cultured cells, post-translational processing of pro-EMAP II protein to the mature released EMAP II form (23 kDa) occurred coincidentally with apoptosis. Cleavage of pro-EMAP II could be abrogated in cultured cells by using a peptide-based inhibitor, which competes with the ASTD cleavage site of pro-EMAP II. Our results suggest that the coordinate program of cell death includes activation of a caspase-like activity that initiates the processing of a cytokine responsible for macrophage attraction to the sites of apoptosis.
On the basis of their ability to induce tissue factor (the initiator of coagulation) on endothelial cells in vitro, three factors were isolated from the supernatants of cultured Methylcholanthrene A (Meth A)-transformed fibrosarcoma cells. These were termed endothelial monocyte-activating polypeptides I–III (EMAP I–III). EMAP I/Meth A-factor (1) and EMAP II (2) were previously unknown cytokines, whereas EMAP III was found to be identical to the previously described vascular endothelial growth factor (3). EMAP II has emerged as a potential proinflammatory mediator with the ability to stimulate the chemotactic migration of mononuclear phagocytes and polymorphonuclear leukocytes and to induce the expression of tissue factor on endothelial cells (2). In vivo, the injection of EMAP II into the footpad of mice induces an inflammatory swelling response, which is characterized by a cell infiltrate and edema in the subcutaneous tissues (2).
The cloning of the EMAP II-cDNA revealed that the protein is encoded by a 1.0-kb transcript, which, after translation, results in a precursor protein with a predicted molecular mass of 34 kDa, which lacks a conventional secretion signal peptide (4). The cleavage of the pro-EMAP II protein results in the formation of a “mature” 23-kDa fragment and other as yet uncharacterized fragments. It is the mature 23 kDa form that is released from the cells and that has been described to convey the biological activity (2). As both the human and the murine EMAP II precursor proteins are cleaved after an aspartate residue, the EMAP II cleavage site shares similarities to the cleavage sites, which are hydrolyzed by cysteinyl–aspartate-specific proteases (caspases). A well known member of this family is the interleukin-1β- (IL-1β) converting enzyme (ICE; caspase-1), which activates pro-IL-1β (5, 6). Cleavage of IL-1β after an aspartate residue leads to the released proinflammatory form, which resembles the situation for EMAP II. Furthermore, caspases play a pivotal role in the execution phase of apoptosis, also known as programmed cell death (7, 8). Apoptosis can be distinguished from necrotic cell death by a series of morphological and cellular features, amongst which the most striking is DNA fragmentation (9). The morphological features of programmed cell death were first observed during embryogenesis, in which large numbers of apoptotic cells are phagocytosed at sites of extensive tissue remodeling (10).
In this study, we have analyzed the expression pattern of the EMAP II gene. Our results demonstrate that EMAP II mRNA is expressed predominantly at sites of tissue remodeling during embryogenesis of the mouse. Therefore, we further examined whether the expression and release of the processed EMAP II protein were regulated by apoptosis. After the induction of programmed cell death in myeloid precursor cells (32D), we observed release of the mature EMAP II protein, while the transcription of the EMAP II gene remained unaffected. This release of mature EMAP II protein from apoptotic cells was blocked by the tetra-peptide compound benzyloxycarbonyl-Ala-Ser-Thr-Asp-fluoromethylketone (Z-ASTD-FMK), which acts as a competitive inhibitor for the cleavage site of pro-EMAP II.
EXPERIMENTAL PROCEDURES
Materials.
All reagents were purchased from Sigma (Deisenhofen, Germany) unless otherwise indicated. Tumor necrosis factor (TNF) was a gift from Jochen Salfeld (BASF Bioresearch, Worcester, MA). Z-ASTD-FMK was obtained from Enzyme Systems (Livermore, CA), benzyloxycarbonyl-Asp-Glu-Val-Asp-chloromethylketone (Z-DEVD-CMK) and benzyloxycarbonyl-Tyr-Val-Ala-Asp-chloromethylketone (Z-YVAD-CMK) were purchased from Bachem.
Cell Culture and Induction of Apoptosis in Vitro.
Meth A fibrosarcoma cells (11) were a gift from J. Lloyd Old (Ludwig Cancer Institute, New York). They were cultivated in RPMI medium 1640/5% FCS, and apoptosis was induced by treating the confluent cells for 12 h with a combination of 0.5 nM TNF and 3 μg/ml cycloheximide. Treatment of confluent cells for 12 h with the calcium-ionophore A23187 (5 μg/ml) was used to induce necrosis. 32D myeloid precursor cells (American Type Culture Collection) were cultivated and induced to undergo apoptosis by IL-3 deprivation as described (12). Where indicated, the inhibitors Z-ASTD-FMK, Z-DEVD-CMK, or Z-YVAD-CMK were added to 32D cells after 150 min of IL-3 withdrawal. Supernatants were harvested 21 h later and subjected to Western blot analysis.
Northern Blot Analysis.
Poly(A)+RNA (10 μg) from 32D cells was fractionated by denaturing agarose gel electrophoresis and transferred onto Hybond N+ membranes (Amersham-Buchler, Braunschweig, Germany). For hybridization, a full-length cDNA for mouse EMAP II (1.0 kb) or the ribosomal protein L28 (13) was used as a probe. Detection and quantification of the hybridization signals was performed by using the Fuji phosphoimager BAS-2500.
In Situ Hybridization.
In situ hybridization of mouse embryos was performed as described (14). The full-length cDNA of mEMAP II in pBluescript II SK+ (Stratagene) was used to generate 35S-labeled sense and antisense probes by using T3 or T7 RNA polymerases (Stratagene). Hybridization was performed overnight at 42°C and washing at 37°C. The slides were then coated with photographic emulsion (Kodak NTB-2) and counterstained with toluidine blue before photography.
In Situ Detection of Apoptosis.
DNA strand breaks were detected on frozen sections by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) by using a fluorescein detection system according to Gavrieli et al. (15). Sections were mounted in a solution of Mowiol 488 (Calbiochem) and analyzed with a Leitz fluorescence microscope.
F4/80 Immunohistochemistry.
Frozen sections of unfixed murine embryos were treated for 30 min with PBS/10% goat serum to block nonspecific antibody binding. The macrophage-specific rat anti-mouse F4/80 antibody (Dianova, Hamburg, Germany; diluted 1:100 in PBS) (16) was applied to sections for 16 h at 37°C. After washing, bound antibody was detected with a Cy3-conjugated goat anti-rat IgG (Jackson ImmunoResearch).
DNA Fragmentation.
After induction of apoptosis, 1 × 106 cells were harvested, washed with PBS, and incubated in lysis buffer (10 mM Tris⋅HCl, pH 7.5/10 mM EDTA/0.2% Triton X-100) for 15 min at 4°C. After centrifugation for 10 min at 13,000 rpm, the supernatant was first extracted with phenol (pH 8.0) and then re-extracted with phenol/chloroform/isoamylalcohol (25:24:1, pH 8.0). The DNA fragments were precipitated and dissolved in 10 mM Tris (pH 7.5)/1 mM EDTA/RNase A (20 μg/ml). After RNA digestion, the samples were analyzed on a 1.5% agarose gel.
Western Blot Analysis.
SDS/PAGE and immunoblotting were performed as described by Towbin (17). Briefly, the cell supernatants were harvested and protein was precipitated by addition of trichloroacetic acid. The samples were denatured and separated on 15% SDS/PAGE gels. After transfer onto nitrocellulose (Schleicher & Schüll, Dassel, Germany) and blocking in PBS/3% BSA, the membranes were incubated in the EMAP II antiserum (SA 2846; diluted 1:500 in PBS/0.1% Tween 20) for 2 h at room temperature, washed, and incubated in a peroxidase-coupled goat anti-rabbit IgG (Jackson; diluted 1:3,500) for 1 h at room temperature. Detection was performed with the ECL kit (Amersham-Buchler). The rabbit antiserum SA 2846 was generated (Eurogentec, Seraing, Belgium) against recombinant mature EMAP II (23 kDa), and specifically recognized EMAP II. For detection of poly(ADP ribose) polymerase (PARP), cells were boiled in Laemmli buffer and the lysates were electrophoresed on 8% SDS/PAGE gels. After transfer onto nitrocellulose, the membranes were blocked with PBS/5% nonfat dry milk and incubated in an anti-PARP antibody (C2.10; Enzyme Systems Products, Livermore, CA; diluted 1:10,000). After washing, the membranes were incubated in a peroxidase-coupled goat anti-mouse IgG (Jackson; diluted 1:3,500) and developed by enhanced chemiluminescence.
RESULTS
EMAP II Expression During Mouse Embryogenesis.
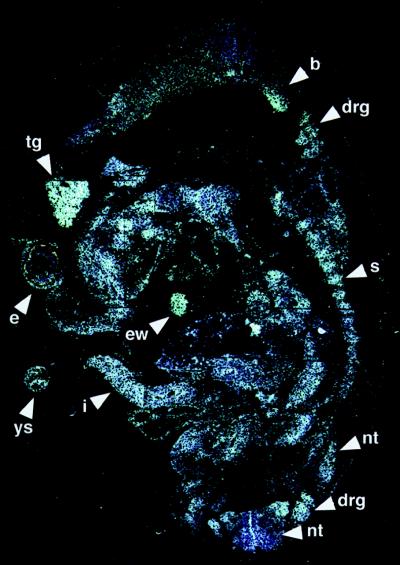
In situ hybridization studies showed that EMAP II mRNA was expressed throughout the 9.5 days post coitum (dpc) mouse embryo (data not shown), with the strongest hybridization signals observed in the neural tube and mesencephalon, optic vesicle, limb buds, lung bud, and the mandibular and hyoid arches. In the 11.5-dpc embryo, there was an overall low level of hybridization signals for EMAP II mRNA (Fig. 1). Prominent signals were detected within the residue of the yolk sac, the intestine within the umbilical cord, the endocardial wall, and in the sclerotomal component of the somites. In the eye, hybridization signals for EMAP II could be detected in the retina and the lens vesicle epithelium as well as in the developing eyelid and the ectoderm covering the eye. In addition, the signal intensity was high in neuronal tissues such as the brain, trigeminal ganglion (V), dorsal root ganglia, and the neural tube (all Fig. 1). In 13.5–17.5-dpc embryos, EMAP II mRNA was abundant in developing neuronal tissues such as the brain, spinal cord, dorsal root ganglia, and ganglia of different cranial nerves. Moreover, prominent signals for EMAP II were detected in areas with ongoing ossification. These included the mesenchymal condensations of the developing frontal bone and chondrification centers of the ribs at 13.5 dpc, the ossification centers of the palatal shelf of the maxilla and mandible on days 15.5 dpc and 17.5 dpc, as well as other forming bones. Moreover, the thymus and the intestinal tract showed a strong EMAP II expression on 13.5–17.5 dpc. On 15.5 and 17.5 dpc, we found a strong hybridization signal for EMAP II mRNA in the developing incisors (especially the dental papilla) and molar teeth. In general, hybridization signals were most abundant in tissues undergoing extensive remodeling and differentiation, such as ganglia, developing bones, and teeth.
Figure 1.
In situ hybridization analysis of EMAP II expression in an 11.5-dpc mouse embryo. Shown here is a parasaggital section that was hybridized overnight with 35S-UTP antisense riboprobe for the detection of mouse EMAP II mRNA. Note the high expression of EMAP II mRNA in the residuum of the yolk sac (ys), in the eye (e), the trigeminal ganglion (tg), the intestine within the umbilical cord (i), the endocardial wall (ew), the sclerotome of the somites (s), the dorsal root ganglia (drg), the brain (b), and the neural tube (nt). A sense probe was used as a control and showed no signal (data not shown).
High Levels of EMAP II Expression Correlate with Sites of Extensive Remodeling and Apoptosis in Embryos of Different Developmental Stages.
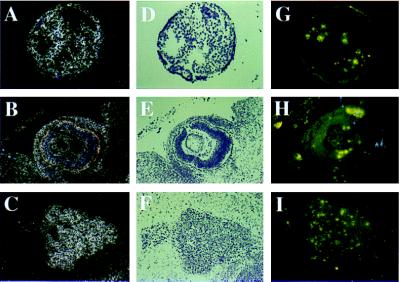
The observation that EMAP II mRNA was elevated in tissues undergoing extensive remodeling raised the possibility that programmed cell death is involved in the regulation of EMAP II expression. To determine whether EMAP II mRNA expression colocalizes with high numbers of apoptotic cells, DNA fragmentation was visualized by TUNEL staining on sections adjacent to those used for in situ hybridization. Large numbers of TUNEL-positive nuclei were found in areas of the embryo in which the strongest hybridization signals for EMAP II mRNA were detected. In the 9.5-dpc embryo, apoptosis was detectable at sites of EMAP II expression in the neural tube, the hyoid and the mandibular arches, optic vesicles, and limb buds (data not shown). In the 11.5-dpc embryo, TUNEL labeled nuclei were observed in the residuum of the yolk sac (Fig. 2G), the eye and the developing eyelid (Fig. 2H), the somites, and the liver primordium. Moreover, neuronal tissues such as the brain, trigeminal ganglion (V) (Fig. 2I), and dorsal root ganglia showed large numbers of TUNEL-positive nuclei. In the 13.5-, 15.5-, and 17.5-dpc embryo, a similar correlation between EMAP II expression and TUNEL-positive nuclei was observed, most noticeably in neuronal tissues such as the brain, trigeminal ganglion (V), spinal cord, and dorsal root ganglia but also in thymus, gut, liver, limbs, ossification centers, and the epithelium of the nasal sinus (data not shown).
Figure 2.
EMAP II expression and apoptosis in an 11.5-dpc mouse embryo. Serial sections were either hybridized with 35S-UTP antisense riboprobe to detect EMAP II transcripts (in dark field A–C and in bright field D–F) or stained for DNA fragmentation by using the TUNEL assay (G–I). Shown here are sections of the yolk sac (A, D, and G), the eye (B, E, and H), and the trigeminal ganglion (C, F, and I). EMAP II sense probe (for the in situ hybridization) and omission of the terminal transferase (for the TUNEL assay) were used as controls and showed no signals (data not shown).
Detection of Macrophages at Sites of High EMAP II mRNA Expression During Mouse Embryonic Development.
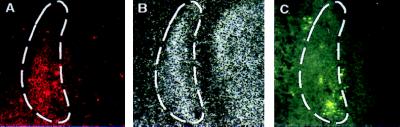
Accumulation of macrophages has been observed in tissues where programmed cell death is a feature of embryonic development (18, 19). As EMAP II is a potent chemoattractant for monocytes, we hypothesized that sites in which EMAP II and apoptotic events colocalize should have numerous macrophages. Sections adjacent to those used for detection of EMAP II mRNA and TUNEL labeling were stained with the macrophage-specific F4/80-antibody. From 11.5 dpc onwards, we observed colocalization of F4/80 positive cells in embryonic regions that showed strong hybridization signals for EMAP II mRNA as well as many apoptotic cells. F4/80 staining was prominent in the mesenchymal condensations of the developing frontal bone in the 13.5-dpc embryo (Fig. 3A), a region positive for both EMAP II mRNA (Fig. 3B) and TUNEL labeling (Fig. 3C).
Figure 3.
Comparison of EMAP II expression, apoptosis, and mononuclear phagocyte infiltration in the developing os frontale of a 13.5-dpc mouse embryo. Serial sections were either analyzed for the presence of mononuclear phagocytes by staining with the F4/80-antibody (A), for the presence of EMAP II mRNA by hybridization with 35S-UTP antisense probe for murine EMAP II (B), or for DNA fragmentation by using the TUNEL assay (C). The area of colocalization is labeled (dashed line).
Apoptosis Leads to the Release of the Mature EMAP II Protein but Not to an Increase in Transcript Accumulation.
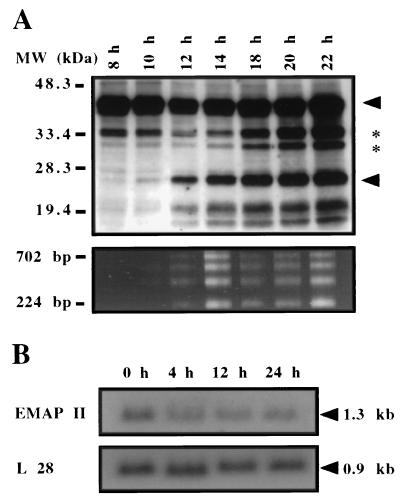
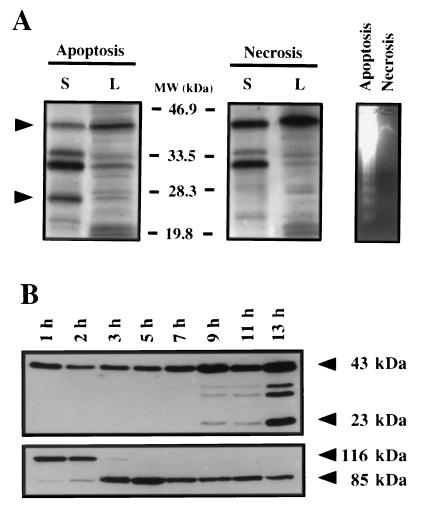
EMAP II is synthesized as a precursor protein, cleaved to the mature form, and released from the cell (4). We investigated the effect of the induction of apoptosis on the post-translational processing of EMAP II. For this purpose, we chose an established model of apoptosis, the myeloid precursor cell line 32D. These cells are dependent on IL-3, and withdrawal of this cytokine induces apoptosis (12). Fig. 4A shows a time course experiment in which 32D cells were cultured in IL-3-free medium for differing lengths of time. At the indicated time points, the supernatants were subjected to Western blot analysis for EMAP II protein (Fig. 4A Upper), and DNA of the cells was tested for fragmentation (Fig. 4A Lower). Six bands were recognized in the Western blot by a polyclonal antiserum directed against EMAP II. Two bands with molecular masses below 20 kDa are considered to be unspecific because they also were recognized by the pre-immune serum (data not shown). The band with an apparent size of 43 kDa (arrow) corresponds to the size of in vitro translated pro-EMAP II (data not shown) but migrated more slowly than predicted from the amino acid sequence. In addition, we observed three further bands with apparent molecular masses of 34 kDa, 31 kDa (asterisks), and 23 kDa (arrow). Because the 23-kDa band corresponds to the originally described mature and active form of EMAP II protein (2), we focused our attention on this band. An increase in the amount of mature EMAP II in comparison to control samples was observed as early as 10 h after withdrawal of IL-3 and was maximal at 22 h (Fig. 4A Upper). Concomitant with the increase of mature EMAP II protein, DNA fragmentation was observed from 12 h on (Fig. 4A Lower), demonstrating a close correlation between the amount of extracellular mature EMAP II and apoptosis. In addition to 32D cells, other cells including primary bovine aortic endothelial cells and murine embryonic fibroblasts showed release of mature EMAP II upon induction of apoptosis (U.E.K., unpublished observation).
Figure 4.
The influence of apoptosis on the post-translational processing of the EMAP II protein and on the expression of the EMAP II mRNA in 32D cells. (A Upper) Western blot of 32D cell supernatants after withdrawal of IL-3. At the indicated time points, the supernatants were submitted to Western blot analysis with the antiserum SA 2846. Arrows indicate the position of pro-EMAP II (upper arrowhead) and mature EMAP II (lower arrowhead). Two intermediate bands are indicated by asterisks. Two bands of apparent molecular masses <20 kDa are due to antibody artifacts. (A Lower) In a parallel experiment, the time course of apoptosis was determined by DNA fragmentation. (B) Northern blot analysis of EMAP II mRNA expression after induction of apoptosis. After withdrawal of IL-3, poly(A)+RNA was extracted at the indicated time points and analyzed by Northern blot by using the murine EMAP II cDNA as a probe (Upper). The blot was rehybridized with the cDNA for the ribosomal protein L28 to ensure equal loading of RNA (Lower).
In 32D cells, the post-translational processing of EMAP II appeared to be regulated by the induction of programmed cell death. Therefore, we tested the hypothesis that induction of apoptosis also regulates EMAP II transcription in vitro. However, when apoptosis was induced in 32D cells by withdrawal of IL-3, no obvious changes in EMAP II mRNA levels were observed by Northern blot analysis (Fig. 4B). Quantification of the hybridization signal of EMAP II mRNA in comparison to the mRNA coding for the ribosomal protein L28 (13) indicated that the EMAP II mRNA levels of this cell type were not influenced by programmed cell death, although a slight decline in the EMAP II mRNA levels was measured 24 h after the induction of apoptosis (Fig. 4B).
Release of Processed EMAP II Is Linked to Programmed but Not to Necrotic Cell Death.
To determine whether EMAP II release is specifically induced by apoptosis and is not merely related to loss of cell membrane integrity, supernatants of cells subjected to either apoptotic or necrotic stimuli were analyzed for mature EMAP II. Apoptosis was induced in Meth A cells by treatment with TNF and cycloheximide whereas necrosis was induced by the calcium-ionophore A23187. The different modes of cell death were verified by a DNA fragmentation assay in which necrotic cells, in contrast to apoptotic cells, show no distinct fragmentation pattern (Fig. 5A Right). Western blot analysis demonstrated that the mature EMAP II is clearly detectable in supernatants (S) but not in the lysates (L) of apoptotic cells (Fig. 5A Left). In contrast, the mature form of EMAP II can neither be observed in the supernatants nor in the lysates of necrotic Meth A cells. This result demonstrates that the cleavage and release of mature EMAP II are specifically linked to apoptosis and do not occur as a result of necrosis.
Figure 5.
(A) Release of the mature EMAP II protein is induced by apoptosis but not by necrosis. For the induction of apoptosis, Meth A cells were treated for 12 h with TNF and cycloheximide. Necrosis was induced by treatment for 12 h with the calcium-ionophore A23187. DNA fragmentation was used to determine apoptosis of the treated cells and to distinguish them from necrotic cells (Right). Supernatants (S) and total cell lysates (L) of apoptotic and necrotic cells were subjected to Western blot analysis by using an anti-EMAP II antiserum. Arrows indicate the position of pro-EMAP II (upper arrowhead) and processed EMAP II (lower arrowhead). (B) Western blot analysis of EMAP II processing and PARP cleavage after induction of apoptosis. Meth A cells were treated with TNF and cycloheximide for the times indicated, and supernatants were analyzed for EMAP II (Upper) and cell lysates for PARP (Lower). Arrowheads indicate the position of pro-EMAP II and mature EMAP II (Upper) and PARP or the PARP fragment (Lower).
As the release of mature EMAP II is associated with programmed cell death, we wanted to know whether the kinetics of pro-EMAP II processing is temporally related to the cleavage of PARP, an event observed in virtually every form of apoptosis (20). The 116-kDa PARP protein is specifically cleaved by caspase-3 to produce an 85-kDa fragment (8, 21). Apoptosis was induced in Meth A cells, and at the indicated time points, cell lysates were assessed for PARP cleavage, while the supernatants were analyzed for the presence of mature EMAP II. Western blot analysis revealed that proteolysis of PARP starts 2 h after the induction of apoptosis, with complete cleavage detectable after 5 h (Fig. 5B). In comparison, release of the mature EMAP II from Meth A cells starts 9 h after induction of apoptosis, with increased levels detectable after 13 h.
Inhibition of EMAP II Maturation and Release.
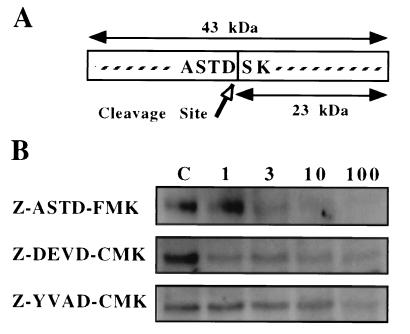
Several possible mechanisms for the post-translational regulation of EMAP II can be proposed including the differential expression or activation of the proteases responsible for the maturation of EMAP II in apoptotic vs. nonapoptotic cells. As the cleavage site of pro-EMAP II (ASTD/S) contains an aspartate residue at the P1 position (Fig. 6A), it is possible that a putative EMAP II-converting enzyme belongs to the caspase family of proteases. To test this hypothesis, we investigated the effect of the inhibitor Z-ASTD-FMK on the processing of the EMAP II precursor protein. The peptide sequence of this inhibitor represents the cleavage site of murine pro-EMAP II, whereas the fluoromethylketone residue irreversibly binds to and blocks the central cysteine residue of caspases. Similar tetra-peptide inhibitors, Z-DEVD-CMK and Z-YVAD-CMK, have been described to inhibit the activity of caspase-1 (22) and caspase-3 (21), respectively. Apoptosis was induced in 32D cells in either the presence or absence of these inhibitors, and supernatants were assayed for the presence of mature EMAP II. With increasing concentrations of Z-ASTD-FMK (Fig. 6B Top), the amount of released mature EMAP II was clearly diminished in comparison to controls (supernatants from apoptotic 32D cells without inhibitor, lane C). When a maximal concentration of 100 μM of inhibitor was used, the 23-kDa band was no longer detectable. Addition of the inhibitor did not affect the induction of apoptosis in 32D cells as determined by DNA fragmentation (data not shown). The caspase-3 inhibitor Z-DEVD-CMK had a weak inhibitory effect on the cleavage of pro-EMAP II (Fig. 6B Middle). Z-YVAD-CMK only slightly diminished the processing of EMAP II if added at the highest concentration of 100 μM (Fig. 6B Bottom). These results indicate that the active site of the hypothetical EMAP II converting enzyme shares a structural similarity to members of the caspase family and that the activity of this protease is not required for apoptosis in 32D cells.
Figure 6.
Inhibition of EMAP II release by Z-ASTD-FMK. (A) Schematic drawing of the deduced EMAP II cleavage site. (B) Apoptosis was induced in 32D cells by IL-3 withdrawal in the presence or absence (lane C) of different concentrations (1, 3, 10, and 100 μM) of the inhibitors Z-ASTD-FMK (Top), Z-DEVD-CMK (Middle), or Z-YVAD-CMK (Bottom). Release of processed EMAP II was determined by Western blot analysis 24 h later. Only the mature 23-kDa EMAP II band is shown.
DISCUSSION
We have examined EMAP II mRNA expression during mouse development in vivo by in situ hybridization. Although EMAP II expression was widespread during early mouse embryogenesis (9.5–11.5 dpc), the strongest hybridization signals were detected in neuronal tissues, developing appendages and teeth, as well as in centers of ossification throughout these and later stages of embryonic development. All of these sites are associated with extensive tissue remodeling and also with large numbers of cells undergoing programmed cell death (reviewed in ref. 10). In all of the embryos analyzed, strong hybridization signals for EMAP II mRNA in neuronal tissues were observed in local correlation with a large number of TUNEL-positive cells. This finding is consistent with previous reports showing that during embryonic development apoptosis occurs in different types of neurons, in both the central and peripheral nervous systems (reviewed in ref. 23). It has been hypothesized that these large numbers of apoptotic cells correspond to the overproduction of neuronal cells and their incorrect projection to their target sites (24). The developing eye is another example where a colocalization of EMAP II mRNA and TUNEL staining was observed (Fig. 2). In this organ, the process of tissue remodeling during development is accompanied by apoptosis in the retina, lens, and optic nerve (25, 26). In addition, from 9.5 to 13.5 dpc, strong hybridization signals for EMAP II mRNA correlated with large numbers of TUNEL-positive cells in the limb buds, where apoptosis in the interdigital spaces is essential for the morphogenesis of the digits (27, 28).
One hypothesis that may explain the colocalization of areas with strong hybridization signals for EMAP II mRNA with sites of large numbers of TUNEL-positive cells is that EMAP II transcription is induced by apoptosis. However, Northern blot analysis of apoptotic cells in vitro revealed no change in EMAP II mRNA levels compared with nonapoptotic cells, which does not support the hypothesis of a direct effect of apoptosis on the transcription of the EMAP II gene (although it cannot be excluded that this event exclusively occurs in vivo). Alternatively, EMAP II mRNA may be up-regulated at sites of tissue remodeling by some unknown mechanism, which might be associated with increased cellular proliferation. Tumor cells, from which EMAP II was originally isolated, are characterized as embryonic-like undifferentiated cells in which both apoptosis and proliferation are increased (reviewed in ref. 29).
In the developing embryo, tissue remodeling is associated with the accumulation of apoptotic cells, which colocalize with F4/80-antigen positive monocytes/macrophages, particularly in the brain, the eye, and the limb buds (16, 19, 28). These macrophages are responsible for clearing sites of apoptotic cell-debris by phagocytosis, a function which has been described for macrophages in the mouse limb bud during the morphogenesis of the interdigital spaces and in the Wolffian and Müllerian ducts (28, 30). Although several molecules involved in the recognition of apoptotic cells by phagocytes have been reported (31, 32), the factors that lead to the accumulation of macrophages at these sites are unknown. Because EMAP II is a potent chemoattractant for monocytes (2), the expression and release of mature EMAP II may account for the attraction of phagocytotic cells to sites where large numbers of apoptotic cells are present.
The EMAP II protein is produced as a precursor that, following enzymatic cleavage, is released from cells as a processed 23 kDa form (2). There are as yet no available anti-EMAP II antibodies, which are specifically capable of recognizing the mature EMAP II in immunohistochemistry. Therefore, we decided to use an in vitro system to determine whether the release of mature EMAP II is temporally associated with the induction of programmed cell death. After the induction of apoptosis in Meth A and 32D cells, Western blot analysis of the cell supernatants revealed four EMAP II-specific bands. The mature EMAP II protein, initially described as a polypeptide of ≈22 kDa (2), corresponds to the 23-kDa band, whereas the other proteins of 31 and 34 kDa possibly represent intermediate cleavage products of the 43-kDa pro-EMAP II. Because the biological activity of EMAP II is conveyed by the 23-kDa form (2, 4), we further investigated the regulation of the enzymatic processing of pro-EMAP II and focused on the release of the mature EMAP II.
The release of mature EMAP II after the induction of apoptosis resembles the model described for the processing and release of IL-1β (6, 33). The IL-1β precursor is cleaved after an aspartate residue by caspase-1, resulting in the generation of the active processed form, which is subsequently released from the cell (6). Similarly, the cleavage site of pro-EMAP II (ASTD/S) carries an aspartate at the P1-position and a small amino acid at the P1′-position, but, apart from this, there is no significant homology to substrate sequences recognized by caspase-1. Fluoro- or chloromethylketone-based inhibitors, containing the minimal recognition sequence of the cleavage site for caspase-1 (YVAD) or other caspases, block the proteolytic activity of the corresponding enzymes (6, 22). A similarly structured inhibitor containing the EMAP II cleavage site (Z-ASTD-FMK) abrogated the release of mature EMAP II (Fig. 6) without affecting the induction of apoptosis or the other EMAP II proteins (data not shown). Together with the finding that the 23-kDa EMAP II is released after the induction of apoptosis but not necrosis, this supports the hypothesis that the proteolytic generation of mature EMAP II is a specific process, which involves a caspase-like enzyme. This enzyme is not identical to caspase-1 because Z-YVAD-CMK did not inhibit its activity. Moreover, the recombinant caspases-1, -2, -6, -8, -11, and -12 did not process in vitro translated EMAP II (Marc van de Craen, personal communication). Caspase-3 showed a weak ability to cleave pro-EMAP II, which may be explained by the cross-reactivity of this caspase with a related substrate. This hypothesis is supported by the finding that inhibition of EMAP II processing was more pronounced with the caspase-3 inhibitor Z-DEVD-CMK in comparison with the effect observed with the caspase-1 inhibitor Z-YVAD-CMK. However, the EMAP II-converting enzyme is unlikely to be identical to caspase-3 because pro-EMAP II cleavage occurred much later than the processing of the known caspase-3 substrate PARP. These results indicate that the EMAP II-cleaving enzyme is distinct from the caspases known so far, although its substrate specificity seems to be more closely related to that of caspase-3 than of caspase-1.
In conclusion, we have shown that EMAP II is specifically processed and released upon the induction of apoptosis in cultured cells. This release can be blocked by a specific tetrapeptide-based inhibitor (Z-ASTD-FMK). Although EMAP II mRNA expression was not induced by apoptosis in vitro, we found colocalization of high levels of EMAP II mRNA with large numbers of macrophages, specifically at sites which are characterized by extensive remodeling and apoptosis in the mouse embryo. We therefore propose that the activation of a caspase-like activity leads to the processing of pro-EMAP II, resulting in the release of a cytokine, which attracts macrophages to sites of apoptosis.
Acknowledgments
We are grateful to Dr. Simon Bamforth for critical reading of the manuscript and to Hiltrud Hölzinger for excellent photographic assistance. We also would like to thank Marc van de Craen and Peter Vandenabeele for the permission to communicate their unpublished data. This study was in part supported by a grant from the Deutsche Forschungsgemeinschaft (C5/SFB 547).
ABBREVIATIONS
- EMAP II
endothelial monocyte-activating polypeptide II
- IL
interleukin
- Meth A
methylcholanthrene A
- TNF
tumor necrosis factor
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- Z-ASTD-FMK
benzyloxycarbonyl-Ala-Ser-Thr-Asp-fluoromethylketone
- Z-DEVD-CMK
benzyloxycarbonyl-Asp-Glu-Val-Asp-chloromethylketone
- Z-YVAD-CMK
benzyloxycarbonyl-Tyr-Val-Ala-Asp-chloromethylketone
- PARP
poly(ADP ribose) polymerase
- dpc
days post coitum
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
To whom reprint requests should be addressed. e-mail: MClauss@kerckhoff.mpg.de.
References
- 1. Clauss M, Murray J C, Vianna M, de Waal R, Thurston G, Nawroth P, Gerlach H, Gerlach M, Bach R, Familletti P C, et al. J Biol Chem. 1990;265:7078–7083. [PubMed] [Google Scholar]
- 2.Kao J, Ryan J, Brett G, Chen J, Shen H, Fan Y-G, Godman G, Familletti P C, Wang F, Pan Y-C E, et al. J Biol Chem. 1992;267:20239–20247. [PubMed] [Google Scholar]
- 3.Clauss M, Gerlach M, Gerlach H, Brett J, Wang F, Familletti P C, Pan Y-C, Olander J V, Connolly D T, Stern D. J Exp Med. 1990;172:1535–1545. doi: 10.1084/jem.172.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kao J, Houck K, Fan Y, Haehnel I, Libutti S K, Kayton M L, Grikscheit T, Chabot J, Nowygrod R, Greenberg S, et al. J Biol Chem. 1994;269:25106–25119. [PubMed] [Google Scholar]
- 5.Black R A, Kronheim S R, Cantrell M, Deeley M C, March C J, Prickett K S, Wignall J, Conlon P J, Cosman D, Hopp T P, et al. J Biol Chem. 1988;263:9437–9442. [PubMed] [Google Scholar]
- 6.Thornberry N A, Bull H G, Calaycay J R, Chapman K T, Howard A D, Kostura M J, Miller D K, Molineaux S M, Weidner J R, Aunins J, et al. Nature (London) 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 7.Nicholson D W, Thornberry N A. Trends Biochem Sci. 1997;22:229–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 8.Van de Craen M, Vandenabeele P, Declercq W, Van den Brande I, Van Loo G, Molemans F, Schotte P, Van Criekinge W, Beyaert R, Fiers W. FEBS Lett. 1997;403:61–69. doi: 10.1016/s0014-5793(97)00026-4. [DOI] [PubMed] [Google Scholar]
- 9.Wyllie A H, Kerr J F K, Currie A R. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 10.Glücksmann A. Biol Rev. 1951;26:59–86. doi: 10.1111/j.1469-185x.1951.tb00774.x. [DOI] [PubMed] [Google Scholar]
- 11.Carswell E A, Old L J, Kassel R L, Green S, Fiore N, Williamson B. Proc Natl Acad Sci USA. 1975;72:3666–3670. doi: 10.1073/pnas.72.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baffy G, Miyashita T, Williamson J R, Reed J C. J Biol Chem. 1993;268:6511–6519. [PubMed] [Google Scholar]
- 13.Wenger R H, Rolfs A, Marti H H, Bauer C, Gassmann M. J Biol Chem. 1995;270:27865–27870. doi: 10.1074/jbc.270.46.27865. [DOI] [PubMed] [Google Scholar]
- 14.Breier G, Albrecht U, Sterrer S, Risau W. Development (Cambridge, UK) 1992;114:521–532. doi: 10.1242/dev.114.2.521. [DOI] [PubMed] [Google Scholar]
- 15.Gavrieli Y, Sherman Y, Ben-Sasson S A. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hume D A, Perry V H, Gordon S. J Cell Biol. 1983;97:253–257. doi: 10.1083/jcb.97.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rotello R J, Fernandez P A, Yuan J. Development (Cambridge, UK) 1994;120:1421–1431. doi: 10.1242/dev.120.6.1421. [DOI] [PubMed] [Google Scholar]
- 19.Perry V H, Hume D A, Gordon S. Neuroscience. 1985;15:313–326. doi: 10.1016/0306-4522(85)90215-5. [DOI] [PubMed] [Google Scholar]
- 20.Kaufmann S, Desnoyers S, Ottaviano Y, Davidson N E, Poirier G G. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 21.Nicholson D W, Ali A, Thornberry N A, Vaillancourt J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A, et al. Nature (London) 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 22.Enari M, Talanian R V, Wong W W, Nagata S. Nature (London) 1996;380:723–726. doi: 10.1038/380723a0. [DOI] [PubMed] [Google Scholar]
- 23.Oppenheim R W. Annu Rev Neurosc. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- 24.Clarke P G, Egloff M. Anat Embryol. 1988;179:103–108. doi: 10.1007/BF00304692. [DOI] [PubMed] [Google Scholar]
- 25.Pan H, Griep H E. Genes Dev. 1995;9:2157–2169. doi: 10.1101/gad.9.17.2157. [DOI] [PubMed] [Google Scholar]
- 26.Moujahid A, Navascues J, Marin-Teva J L, Cuadros M A. Anat Embryol. 1996;193:131–144. doi: 10.1007/BF00214704. [DOI] [PubMed] [Google Scholar]
- 27.Winter R M, Tickle C. Eur J Hum Genet. 1993;1:96–104. doi: 10.1159/000472392. [DOI] [PubMed] [Google Scholar]
- 28.Hopkinson-Woolley J, Hughes D, Gordon S, Martin P. J Cell Sci. 1994;107:1159–1167. doi: 10.1242/jcs.107.5.1159. [DOI] [PubMed] [Google Scholar]
- 29.Evan G. Int J Cancer. 1997;71:709–711. doi: 10.1002/(sici)1097-0215(19970529)71:5<709::aid-ijc2>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 30.De Felici M, Heasman J, Wylie C C, McLaren A. Cell Diff. 1986;338:119–129. doi: 10.1016/0045-6039(86)90006-0. [DOI] [PubMed] [Google Scholar]
- 31.Luciani M-F, Chimini G. EMBO J. 1996;15:226–235. [PMC free article] [PubMed] [Google Scholar]
- 32.Devitt A, Moffatt O D, Raykundalia C, Caprat J D, Simmons D L, Gregory C D. Nature (London) 1998;392:505–509. doi: 10.1038/33169. [DOI] [PubMed] [Google Scholar]
- 33.Cerretti D P, Kozlosky C J, Mosley B, Nelson N, Van Ness K, Greenstreet T A, March C J, Kronheim S R, Druck T, Cannizzaro L A, et al. Science. 1992;256:97–100. doi: 10.1126/science.1373520. [DOI] [PubMed] [Google Scholar]