Abstract
In Trypanosoma brucei, transcription by RNA polymerase II and 5′ capping of messenger RNA are uncoupled: a capped spliced leader is trans spliced to every RNA. This decoupling makes it possible to have protein-coding gene transcription driven by RNA polymerase I. Indeed, indirect evidence suggests that the genes for the major surface glycoproteins, variant surface glycoproteins (VSGs) in bloodstream-form trypanosomes, are transcribed by RNA polymerase I. In a single trypanosome, only one VSG expression site is maximally transcribed at any one time, and it has been speculated that transcription takes place at a unique site within the nucleus, perhaps in the nucleolus. We tested this by using fluorescence in situ hybridization. With probes that cover about 50 kb of the active 221 expression site, we detected nuclear transcripts of this site in a single fluorescent spot, which did not colocalize with the nucleolus. Analysis of marker gene-tagged active expression site DNA by fluorescent DNA in situ hybridization confirmed the absence of association with the nucleolus. Even an active expression site in which the promoter had been replaced by an rDNA promoter did not colocalize with the nulceolus. As expected, marker genes inserted in the rDNA array predominantly colocalize with the nucleolus, whereas the tubulin gene arrays do not. We conclude that transcription of the active VSG expression site does not take place in the nucleolus.
Keywords: antigenic variation/in situ hybridization/nucleolus/RNA polymerase I
Trypanosoma brucei, an extracellular parasite of mammals, uses antigenic variation of its coat to escape complete destruction by the immune system of the host (reviewed in refs. 1–5). The coat consists of a single protein, the variant surface glycoprotein (VSG). There are hundreds of VSG genes (VSGs) spread throughout the genome, and only one of these is expressed at any given time from one of approximately 20 VSG expression sites (ESs) located at the ends of chromosomes. The VSGs present in expression sites can be replaced by others by recombination mechanisms. Each ES is a polycistronic transcription unit controlled by a single promoter, located 40 to 60 kb upstream of the telomeric VSG (see Fig. 1A and refs. 6–8). A set of expression site-associated genes (ESAGs), which may meet specific metabolic requirements such as transferrin uptake (2), is cotranscribed with the VSG (9, 10).
Figure 1.
(A) Representation of the 221 VSG expression site. The VSG, pseudo-VSG (Ψ gene), and ESAGs are represented by boxes. ESAGs are numbered according to the nomenclature of Pays et al. (8) for the AnTat 1.3A ES and were determined by analysis of steady-state RNA (7) and complete or partial sequence analysis. The positions of genes in the 221 ES corresponding to ESAGs 2, 4, and 5 from the AnTat 1.3A ES have not been determined. This ES is a 60-kb transcription unit, under control of a single promoter (flag). The recombinant inserts used to form probe mix 4 are depicted as black bars underneath the ES. Numbers refer to recombinant clones pTg221.11, pTg221.14, pTg221.4, and pTg221.8 described in Kooter et al. (7). A detailed description of the probes is given in Materials and Methods. (B) Map of the 3174 transformant of T. brucei variant 221a (36). The single-copy marker gene cassettes (H, hyg; N, neo) are represented by black rectangles flanked by hatched boxes (which correspond to the processing signals). Other symbols are the same as in A. (C) Differences in the genomic integration of the constructs RPhygro and r4 in the rDNA. The top drawing represents the RPhygro transfectant, which contains a hyg cassette (H); the middle drawing shows part of an rDNA array (wild-type); the bottom drawing represents the r4 transfectant, which contains a neo cassette (N). The open box labeled 18S corresponds to the 18S rRNA gene. The constructs are bordered by the restriction enzyme at the integration site (a, AvaI; c, ClaI). In the middle drawing, the black bars represent the homology region used in the constructs. The flags represent transcription initiation (open flag: endogenous rDNA promoter; filled flag: duplicated rDNA promoter).
Because it has approximately 20 ESs, the trypanosome needs mechanisms to activate and inactivate an ES and to prevent more than one ES from being active at any one time. Until recently, it seemed probable that activation/inactivation was controlled by a form of telomeric silencing (2, 11–16). Our work has failed to confirm this, however, and has indicated that there must be some form of crosstalk between ESs (17). A plausible form of crosstalk would be competition between ESs for a single nuclear site (17, 18).
Ever since Kooter and Borst (19) found that the transcription of ESs is insensitive to high concentrations of α-amanitin, a characteristic property of RNA polymerase (Pol) I, evidence has been accumulating that ESs are transcribed by Pol I (reviewed in refs. 20 and 21) rather than by a modified form of Pol II (22–24). In contrast to the situation in animal cells, Pol I of T. brucei can efficiently mediate the synthesis of mRNA (25–27), because trypanosome mRNAs get their caps by trans splicing from an independently synthesized and capped precursor RNA (reviewed in refs. 28–30). Three other arguments also favor Pol I as the polymerase transcribing the VSGs: transcription of these genes (like the rRNA genes) is completely insensitive to concentrations of Sarkosyl that abolish the transcription of other protein-coding genes (31); the ES remains fully active and can still be switched off and on when the VSG ES promoter is replaced by a ribosomal promoter (14); and VSG/rRNA chimeric promoters are functional (32).
If a VSG ES were transcribed at a single site in the nucleus and if transcription were carried out by Pol I, which is normally restricted to transcription of the major rRNA genes in the nucleolus, then the site of VSG ES transcription may be the nucleolus as well. We have used dual-fluorescence in situ hybridization to test this hypothesis.
MATERIALS AND METHODS
Trypanosome Culture.
The trypanosomes used belong to strain 427 of T. brucei brucei (33). Procyclic-form trypanosomes were grown in semidefined medium at 28°C as described (34). Bloodstream-form trypanosomes were cultured in vitro in HMI-9 medium (35). The 221a variant (MiTat 1.2a) of T. brucei was used, which expresses the VSG 221 from the 221 ES (33). Transformants of variant 221a used included 3174, which contains a resistance gene for neomycin and hygromycin between ESAG1 and the 221 VSG (see Fig. 1B; ref. 36); and RPhygro, which contains a gene for resistance to hygromycin in the ribosomal array. Transformants of procyclic trypanosomes derived from bloodstream-form variant 221a included r4, which has a resistance gene for neomycin in the ribosomal array (31) and RPhygro. RPhygro and r4 constructs differ in the size of the ribosomal promoter used as the target and in the processing signals for the marker genes tubulin, in the case of RPhygro and PARP (procyclic acidic repetitive protein), in the case of r4 (see Fig. 1C).
The pro.Anv.pTSA.CAT.HYG.NM8.NsiI (proCAT) transformant of EATRO 1125 stock of T. brucei (procyclic form) was used to localize the active PARP A locus (37).
Cell Fixation and Preparation of the Microscope Slides.
Midlogarithmic-phase culture-form trypanosomes were harvested, washed in PBS (0.15 M NaCl, 10 mM sodium phosphate, pH 7.2), and resuspended in PBS prior to fixation. In vitro-cultured bloodstream-form trypanosomes were handled in the same way, but the buffer used was phosphate/saline/glucose (60 mM Na2HPO4/3 mM NaH2PO4/44 mM NaCl/55 mM glucose, pH 8.0). The cells were then diluted 1:2 in 2× fixation solution (1× fixation solution: 4% formaldehyde and 5% acetic acid in PBS) and incubated at room temperature for 20 min on a rotating wheel. Fixed cells were centrifuged for 10 min at 3,000 × g and the fixation medium was replaced by 70% ethanol, followed by two additional washes in 70% ethanol to remove all traces of formaldehyde. At this stage, cells could be stored at 4°C for several weeks without apparent loss of signal quality in in situ hybridization. Microscope slides were prepared by dropping 20 μl of the fixed-cell suspension on glass slides precleaned with ethanol/ether, 1:1 (vol/vol). Slides were allowed to air dry, and then were baked at 80°C for 10 min to improve cell adherence to the glass.
DNA Probes.
The probes derived from the VSG ES used in the in situ hybridization shown in Fig. 3 are shown in Fig. 1. pTg221.8 is an 11.3-kb BglII fragment inserted into pAT153 (19); pTg221.4 is an 8.5-kb EcoRI fragment inserted into pAT153; pTg221.14 is a 15-kb BamHI fragment inserted into pAT153; and pTg221.11 is a 21.5-kb BglII fragment inserted into pAT153 (7). The clone pTg221.11 is not derived from the 221 ES, but from a homologous ES, and does not contain any 50-bp repeat sequences. The ES probe mix 4 contained the clones pTg221.8, pTg221.4, pTg221.14, and pTg221.11. The 50-bp repeat probe is a 1-kb fragment of the 221 ES 50-bp repeat array inserted into pBlueScriptSK. The rDNA probe pR2 is a 1.4-kb BglII/HindIII fragment of the 18S gene inserted into pGEM3. Plasmid sequences alone were used as a negative control, and did not hybridize to fixed trypanosome nuclei (data not shown). The neo probe is an XbaI/SmaI fragment of the pNeo (Pharmacia), inserted into pGEM3 (pGEM3.neo). The hyg probe corresponds to the hygromycin resistance gene inserted into pUC18 (pHA57). The tubulin probe is a 2.9-kb EcoRI/HindIII fragment of one αβ repeat unit, inserted into pGEM4. The chloramphenicol acetyltransferase (CAT) probe was the pCAT basic plasmid (Promega). Probes neo+hyg and cat+hyg contained both neo and hyg probes and cat and hyg probes, respectively, in equal amounts.
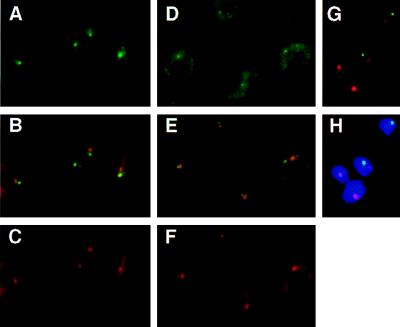
Figure 3.
(A–C) Subnuclear localization of the nuclear transcripts derived from the active VSG ES. T. brucei variant 221a cells hybridized with probe mix 4 (see Fig. 1A) and the ribosomal probe R2. Cells were treated with DNase before hybridization. (A) Detection of probe mix 4 (Fig. 1A and Materials and Methods) with FITC-conjugated antibodies. The single intense spot, in the nucleus, shows the localization of the nascent transcripts derived from the active site. The more diffuse staining in the cytoplasm corresponds to ES mRNAs. (B) Simultaneous detection of the active ES RNA (green) and the nucleolus (red). Overlap between the two signals appears in yellow. (C) Detection of the ribosomal probe R2 (Materials and Methods), with Texas Red-conjugated antibodies. The circle in the nucleus shows the signal derived from the nuclear rRNA, forming the nucleolus (together with the rDNA and the processing machinery). The cytoplasmic signal comes from the rRNA in ribosomes. (D–H) Subnuclear localization of marker genes integrated in the active VSG ES in the 3174 transformant of T. brucei variant 221a (ref. 36; Fig. 1B). (D) Detection of probe neo+hyg (active VSG ES) with FITC-conjugated antibodies. The probe hybridized to the marker genes appears as a small fluorescent signal in the nucleus; a cytoplasmic signal attributable to mRNA is also visible. (E) Simultaneous detection of the active ES (green) and the rDNA in the nucleolus (red). The overlap of signals appears as yellow (or slightly orange). (F) Detection of the signal derived from the ribosomal probe R2. (G and H) Simultaneous detection of the active ES (green) and the rDNA (red) in RNase-treated cells; H also shows the DAPI-stained nucleus).
DNA was nick-translated by established procedures using biotin-16-dUTP (Boehringer-Mannheim) and digoxigenin-11-dUTP (Boehringer-Mannheim) substituting for 60% of the dTTP, and unincorporated nucleotides were removed by G50 column-filtration and ethanol precipitation. DNase I in the nick-translation was titrated to obtain an average probe length of approximately 200 to 500 bp. Probes were dissolved to 10 ng of DNA per μl of hybridization mixture consisting of 60% deionized formamide, 2× SSC (1× SSC is 0.15 M NaCl/0.015 M sodium citrate, pH 7.0), and 50 mM sodium phosphate (pH 7.2) with 500 ng/μl denatured salmon sperm DNA and 500 ng/μl yeast RNA.
Probe R2 was labeled with biotin-16-dUTP, and all other probes were labeled with digoxigenin-11-dUTP.
In Situ Hybridization.
Before hybridization, the cells were subjected to partial hydrolysis and proteolysis by using 0.1% pepsin (Sigma) in 0.01 M HCl for 5 min at 37°C. The conditions described were found to be optimal for RNA retention, accessibility for probes, antibody recognition of hybrids after in situ hybridization, and preservation of the morphology throughout the in situ hybridization procedure. For mix 4 hybridization, cells were treated with DNase I (Promega, 0.2 unit of RQI DNase per μl/40 mM Tris⋅HCl [pH 7.9]/10 mM NaCl/6 mM MgCl2/0.1 mM CaCl2) after pepsin treatment and before hybridization for 1 h at 37°C in a humid chamber. For all other hybridizations, cells either were not nuclease treated or were subjected to an RNase treatment (200 μg of RNase A per ml in 2× SSC) for 1 h at 37°C, as indicated. Both incubations were followed by three washes in 2× SSC for 5 min each. Finally, cells were dehydrated with ethanol and air-dried. Prehybridization was not necessary. Hybridization mixture (7.5 μl; 37.5 ng of each probe in a double hybridization) was applied to the slides, and probe and target nucleic acids were denatured simultaneously under an 18 × 18 mm coverslip for 5 min on an 80°C plate. Hybridizations were performed at 37°C in a humid chamber for 16 h. After hybridization, slides were rinsed three times for 20 min each in 50% formamide/2× SSC, pH 7.0, at 37°C in a shaking water bath. Finally, slides were washed two times for 5 min each at room temperature in Tris/saline (0.1 M Tris⋅HCl/0.15 M NaCl, pH 7.4).
Immunocytochemical Detection.
For the detection of the biotinylated probes (R2 probe only), slides were incubated for 45 min at 37°C with streptavidin-Texas Red (Vector Laboratories) diluted 1:100, then for 30 min at 37°C with biotinylated goat anti-streptavidin (Vector) diluted 1:100, and finally for 30 min at 37°C with streptavidin-Texas Red diluted 1:100. All solutions were diluted with Tris/saline containing 0.5% blocking reagent (Boehringer Mannheim), and incubations were carried out in a humid chamber and followed by a 5-min wash at room temperature to remove coverslips, and three additional 5-min washes at room temperature in Tris/saline. For detection of the digoxigenin-labeled probes, slides were incubated for 45 min at 37°C with 1:200-diluted fluorescein isothiocyanate (FITC)-conjugated (Sigma)mouse anti-digoxigenin and then for 30 min at 37°C with (1:500-diluted FITC)-conjugated rabbit anti-mouse digoxigenin. For simultaneous detection of biotin- and digoxigenin-labeled probes, the first and second layers of antibodies were mixed in blocking solution. After the last wash, slides were dehydrated through an ethanol series, air dried, and mounted in antifading solution {10:1 glycerol:0.2 M Tris⋅Cl, pH 7.5/2% diazabicyclo[2.2.2]octane (DABCO)/0.02% NaN3} containing 75 ng of 4′,6-diamidino-2-phenylindole (DAPI) per ml.
Microscopy.
A Zeiss Axiovert 100 TV microscope equipped with a ×100 objective (numerical aperture 1.3), single band-pass filters for Texas Red, FITC, and DAPI fluorescence, and a double band-pass filter for simultaneous detection of red and green fluorescence was used for visual analysis. For photographic purposes, digital images were acquired with a Photometrics charge-coupled device Series 200 camera with a KAF 1400 chip. Weak signals from small targets were intensified to allow their visualization in the images. Images of RNase-treated trypanosomes are difficult to interpret for readers, because without the outline of the trypanosome delineated by cytoplasmic RNA hybridization, only (weak) fluorescent spots are visible, as shown in Figs. 3G and 4E. As nuclear localization studies with and without RNase pretreatment gave the same results, most of the pictures in Figs. 3–5 show DNA⋅RNA hybridization. However, all the data presented in Table 1 are based on RNase A-treated cells.
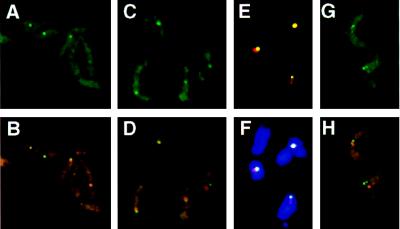
Figure 4.
(A–F) Localization of marker genes integrated in the ribosomal RNA gene array in two different cell lines r4 (A and B) and RPhygro (C–F) ilustrated in Fig. 1C. Only E and F show RNase-treated cells. In both strains the nuclear signal derived from the marker gene (probe neo, in the case of r4, and probe hyg, in the case of RPhygro, both detected with FITC-conjugated antibodies) appears as a single spot (A and C). r4 cells give a clearer picture because of the size of the target fragment (9 kb, compared with only 4 kb in RPhygro). The overlay of the neo and hyg signals (green) with the nucleolus (red) is shown in B and D–F, respectively (F also shows the overlay with DAPI-stained cells). (G and H) Nuclear localization of a Pol II transcription unit (tubulin) in T. brucei variant 221a (no RNase treatment). The probe for tubulin is described in Materials and Methods. (G) Detection of the signal derived from tubulin, using FITC-conjugated antibodies. The tubulin gene arrays appear as two dots in the nucleus, and the cytoplasmic signal corresponds to mRNA. (H) Simultaneous detection of the tubulin (green) and nucleolar (red) signals. For colocalization of both signals, one of the two tubulin signals (per cell) was counted.
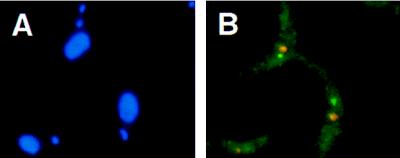
Figure 5.
Nuclear localization of the PARP A locus in the proCAT transformant of T. brucei stock EATRO 1125 (37) (see Materials and Methods; no RNase treatment). (A) DAPI-stained trypanosomes. Two stained structures are visible: the nuclear DNA (large circle) and the kinetoplast DNA (small circle). (B) Simultaneous detection of the signals derived from the PARP A locus (green; probe cat+hyg, described in Materials and Methods) and from the nucleolus (red).
Table 1.
Nuclear localization in Trypanosoma brucei
| Trypanosome clone | Markers inserted | Probe | Nucleic acid analyzed | Nuclease treatment | Overlap with nucleolus, % overlap (range) | Scoreable cells % | Cells analyzed |
|---|---|---|---|---|---|---|---|
| 221a | None | Mix 4 | RNA | DNase | 20 (18–22) | >80 | >450 |
| 3174 | neo+hyg (VSG ES) | neo+hyg | DNA | RNase | 25 (18–32) | 60 | >450 |
| RP2 | hyg (VSG ES) | hyg | DNA | RNase | 23 (20–31) | 60 | >450 |
| RPhygro | hyg (rDNA) | hgy | DNA | RNase | 75 (65–85) | 80 | >450 |
| r4 | neo (rDNA) | neo | NDA | RNase | 100 | 60 | 350 |
| 221a | None | Tubulin | DNA | RNase | 36 (32–40) | >90 | 450 |
| proCAT | cat+hyg (PARP A locus) | cat+hyg | DNA | RNase | 26 (23–30) | 60 | 450 |
RESULTS
Preserving Nuclear Substructure.
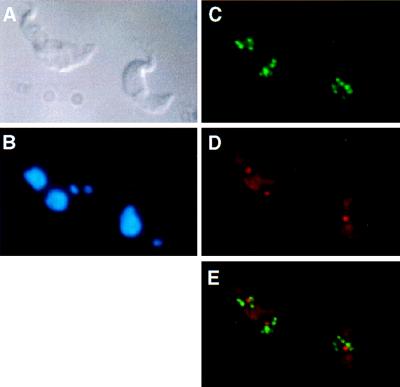
In initial experiments we used noncross-linking fixatives (38), resulting in high sensitivity but also distortion and flattening of the nucleus, making the subnuclear localization of sequences questionable. Therefore, we turned to fixatives containing cross-linking agents that appear to preserve the cell (Fig. 2A) and its nuclear morphology (Fig. 2B; see also Fig. 5A). A spherical region of low DNA density or condensation can be seen in some cases. This region corresponds to the transcription domain of rRNA genes in the nucleus (see below), i.e., the nucleolus. Because of the small diameter of the nuclei (2 μm) and the limited z-plane resolution of the confocal laser scanning microscopy (0.7 μm; ref. 39), we opted for conventional fluorescence microscopy for analysis of colocalization. The images are two-dimensional projections of three-dimensional objects. Consequently, fluorescense in situ hybridization signals resulting from DNA or RNA may accidentally colocalize by superimposition. We define colocalization here as total or partial overlap of signals.
Figure 2.
T. brucei variant 221a trypanosomes hybridised with the 50-bp repeat probe and the ribosomal probe R2. (A) Differential interference contrast image. (B) DAPI staining. Two stained structures are visible, the nuclear DNA (large circle) and the kinetoplast DNA (small circle). (C) Detection of the 50-bp repeat probe showing the distribution of VSG expression sites. (D) Detection of the ribosomal probe R2. Both the nucleolus (intense circle) and the cytoplasmic rRNA are visible. (E) Simultaneous detection of the 50-bp repeat probe (green) and probe R2 (red).
Localization of the Inactive Expression Sites in the Nucleus.
Previous work by Chung et al. (40) has shown that the long telomeric repeats of the approximately 100 minichromosomes and 25 larger chromosomes of T. brucei are clustered in 10–20 spots that tend to be localized in the periphery of the nucleus. To test whether the inactive expression sites also are clustered, we used probes hybridizing to the ES promoter or to the arrays of 50-bp repeats located upstream of each ES (Fig. 1A; refs. 41 and 42). No specific nuclear localization or clustering of expression sites was detected (Fig. 2 C and E). Although in some cases the fluorescent signals appeared to be in the periphery of the nucleus, in the majority of the cells analyzed these signals were randomly distributed in two dimensions within the nucleus.
Nuclear Localization of VSG Expression Site Nuclear Transcripts.
Studies on RNA synthesis in isolated nuclei indicate a high density of engaged RNA polymerase exists on the DNA template (7). We detected nuclear RNA transcripts at the active ES in cells treated with DNase I before hybridization (this procedure turned out to be more successful in eliminating crossreaction with silent ESs than doing the hybridization in trypanosomes that had not been denatured). The set of probes used (mix 4; Fig. 1A and Materials and Methods) covers a region of about 50 kb but does not extend into the VSG gene, as the highly abundant VSG mRNA results in a strong cytoplasmic signal that obscures the nuclear signal. Despite the large region spanned by mix 4, the green hybridization signal in Fig. 3A appears as a single intense fluorescent spot in the nucleus. The cytoplasmic signal reflects the mRNA derived from the ESAGs in the expression site. Both the nuclear and cytoplasmic signals were sensitive to RNase A (data not shown).
The ribosomal probe R2 (red) hybridizes to the 18S rRNA gene and RNA (see Materials and Methods), giving a fluorescent nuclear signal corresponding to the nucleolus, and also giving a cytoplasmic signal (Fig. 3C). Simultaneous analysis of both the ES and nucleolar signals (Fig. 3B) shows that the active ES colocalizes with the nucleolus in only 20% of the cases (Table 1). We attribute this colocalization to accidental two-dimensional overlap in the x–y plane of two signals from different z planes (43). The same result was obtained in a minimum of three independent experiments with 50–150 trypanosomes per analysis, as is the case for all of the observations reported in this section. Results obtained by two independent observers were similar. These results show that nuclear transcripts derived from the active VSG ES do not localize at the nucleolus.
Nuclear Localization of Marker Genes Integrated in the Active ES.
Trypanosome RNAs are rapidly processed (44), but to disprove that probe mix 4 nevertheless detects a processing site rather than nascent RNA, we analyzed the active ES at the DNA level in trypanosomes with an ES marked by unique marker gene sequences (Fig. 1B; ref. 36). To ensure that the marked expression site was the active one, cells were grown under drug selection. The probe used covers only 2 kb, and the percentage of cells that stain positive is approximately 60% (Table 1). The green signal derived from the neo+hyg probe appears as a small fluorescent spot in the nucleus (Fig. 3, D and G), and this spot overlapped only in 25% of the cases with the red signal of the ribosomal probe R2 (Fig. 3, E and G; Table 1). All quantitative results with the neo+hyg probe presented in Table 1 were obtained with RNase-treated cells. However, the same colocalization results were obtained without RNase. Because RNase treatment removes the visible outline of the trypanosome as the result of cytoplasmic RNA hybridization, the resulting spots in the dark (Fig. 3G) or in the DAPI-stained nucleus (Figure 3H) may be difficult to interpret for readers. Hence, most of the images presented in Figs. 3–5 are from cells not treated with RNase. These results are consistent with those obtained with probe mix 4 (nuclear RNA detection).
Nuclear Localization of an Active ES Driven by an rDNA Promoter.
Rudenko et al. (14) have constructed a trypanosome variant in which the promoter of the 221 ES has been replaced with an rDNA promoter. This replacement does not affect ES control (14). We analyzed this trypanosome variant by fluorescence in situ hybridization, and found that the active ES driven by the rDNA promoter does not colocalize with the nucleolus either (RP2; Table 1).
Localization of Genes Targeted to the rDNA or to a Pol II Transcription Unit.
To test whether marker genes inserted into an rDNA array localize at the nucleolus as expected, we analyzed two additional transfectants, each containing a marker gene in an rDNA array: RPhygro and r4 (Fig. 1C). In the r4 trypanosomes only 60% of the nuclei gave a spot with the neo probe (green), but there was 100% overlap between the neo and the nucleolar signals as previously reported by Rudenko et al. (31) for this transformant (Fig. 4B; Table 1). In the RPhygro trypanosomes, we found an overlap with the nucleolus in 75% of the cases, and ≈80% of the cells stained positive (Fig. 4 D–F; Table 1). Similar results were obtained with bloodstream-form and with procyclic-form trypanosomes. We attribute the difference in the results obtained with the two marker genes to the use of different ribosomal promoter fragments as targeting sequences in the two constructs. Nevertheless, these results show that a marker gene integrated in the rDNA is predominantly located at the nucleolus, as expected.
As another control, we used a probe for the tubulin transcription unit, which is transcribed by Pol II. Fig. 4 G and H show photographs of T. brucei variant 221a hybridized with a probe for tubulin (green) and the ribosomal probe R2 (red). Tubulin genes are present in the trypanosome genome as two multicopy clusters (45, 46). These clusters appear as two discrete spots in the nucleus (Fig. 4G). As the two tubulin clusters often are not in the same focal plane, the two signals can differ in intensity in photographs. More than 90% of the cells stained positive, and the signals derived from tubulin and from the nucleolus colocalized in 36% of the cases (Fig. 4G; Table 1). Because tubulin gives two signals, and the percentage of overlap obtained between one of the signals and the nucleolus is approximately twice that found for the single signal of the active ES, we attribute the colocalization to accidental overlap of signals in the preparations (in two dimensions).
Nuclear Localization of a Tagged PARP A Locus.
When a trypanosome is taken up by a tsetse fly, it replaces its VSG coat by an invariant coat consisting of procyclin or PARP. The PARP genes are transcribed by an RNA polymerase insensitive to α-amanitin (47–49), and it seems probable that this is the same Pol I-like polymerase that transcribes the VSG gene ES (20, 21).
Previous work has indicated that the PARP loci might be transcribed in the nucleolus (50), in contrast with our observations on the active VSG ES. However, a major difference between our results and those of Chung (50) is in the fraction of trypanosomes giving a fluorescent signal in the nucleus; more than 50% fluoresced in all of our analyses, and only 10–20% fluoresced in Chung’s analyses. To resolve this discrepancy, we have looked at the localization of a PARP transcription unit by using a procyclic line of trypanosomes (proCAT) in which the PARP A locus had been tagged with two single-copy genes, a hygromycin resistance marker and a CAT gene (37). By using equal amounts of the hyg and the CAT probe, we obtained a specific fluorescent nuclear signal in about 60% of the cells; in 74% of these cells the signal did not overlap with the nucleolus(Fig. 5). We conclude that the marked PARP A locus is not transcribed in the nucleolus.
DISCUSSION
Our results show that the actively transcribed VSG ES and a tagged PARP A locus are not located in the nucleolus, notwithstanding considerable indirect evidence that these transcription units are transcribed by an RNA polymerase with the characteristics of Pol I, an enzyme that is normally confined to the nucleolus, at least in mammalian cells (51–53).
Incidental overlap of any non-nucleolar signal with the large nucleolus in the trypanosome nucleus is high, as is also shown by our results with the tubulin gene probe: the average diameter of the nucleus is about 2 μm and that of the nucleolus is almost one-third of that. The value of approximately 20% overlap with the nucleolus, which we found for both the active ES and for one of the tubulin clusters, is within the theoretical range of accidental colocalization (43).
The combination of a large nucleolus and weak signals from single-copy genes may also be responsible for the conclusion of Chung (50) that a marker gene under control of a PARP promoter is transcribed in the nucleolus. As PARP genes and VSG genes are transcribed by an RNA polymerase with the same characteristics, possibly Pol I, it seemed unlikely that the active PARP locus would be, and that the active VSG ES would not be, in the nucleolus. We now find that a transcribed marker gene in the PARP A locus does not colocalize with the nucleolus, the same result that we obtained for the active VSG ES.
Although our results show that the active VSG ES is not in the nucleolus, this does not imply that it could not be transcribed by Pol I. It would be of interest to test whether antibodies against Pol I colocalize with nuclear ES transcripts. The gene for the largest subunit of T. brucei Pol I has been cloned (23, 54), but so far polyclonal antisera (kindly provided by A. W. C. A. Cornelissen, University of Utrecht, The Netherlands) raised against a fusion protein, which were polymerase class-specific on Western blots , did not react with nuclei of fixed trypanosomes.
Transcription outside the nucleolus also does not mean that the ES could not be always in the same unique position attached to a special nuclear substructure. Such a unique location accommodating only a single active VSG ES would help to explain why only one VSG ES can be active at a time (17, 18). At present we cannot verify this, as we lack reference points in the nucleus. However, the alternative possibility that all inactive expression sites are bundled together in a single subcompartment is ruled out by our observation that expression sites are distributed throughout the nucleus. Attachment to the nuclear envelope cannot be excluded, as confocal laser scanning microscopy analysis of trypanosome nuclei is inconclusive because of their small size.
Formation of a nucleolus is directed by rDNA (51, 55) but requires transcription by Pol I because rDNA transcribed by Pol II does not result in normal nucleoli in yeast (56). A high rate of transcription by Pol I from an rDNA promoter is not sufficient, however, to obtain nucleolar localization in trypanosomes. Nor is an active VSG ES in which the endogenous promoter is replaced by an rDNA promoter (14) transcribed in the nucleolus (Table 1). The convenient ability of the trypanosome to generate mRNA from genes transcribed by Pol I (25, 26) should allow a further dissection of the requirements for nucleolar and nucleoplasmic localization of such transcription units.
Acknowledgments
We thank Dr. Magali Berberof, Dr. Mike Cross, Dr. Dennis Dooijes, Herlinde Gerrits, Dr. Rainer Mußman, Rudo Kieft, Dr. Gloria Rudenko, and Fred van Leeuwen for critical reading of this manuscript. We also thank Dr. Susan Gasser (Institut Suisse de Recherches Experimentales sur le Cancer; Lausanne, Switzerland), Dr. Keith Gull (University of Manchester, U.K.), Dr. Barbara Sollner-Webb (The Johns Hopkins University, Baltimore), and Dr. Lex van der Ploeg (Merk Research Lab., Rahway, NJ) for helpful comments. We thank Joop Wiegant (Leiden University, The Netherlands) for his introduction to DNA in situ hybridizations. We thank Lauran Oomen (The Netherlands Cancer Institute, Amsterdam) for photographic technical assistance, and Drs. Luc Vanhamme and Magali Berberof (Free University of Brussels, Belgium) for the transformant of stock EATRO 1125 with the marked PARP A locus. This work was supported by a grant from the Gulbenkian Ph.D. Program in Biology and Medicine (Portugal) to I.C. and by The Netherlands Foundation for Chemical Research (SON), with financial aid from the Netherlands Organisation for Scientific Research (NWO) to P.B.
ABBREVIATIONS
- VSG
variant surface glycoprotein
- ES
expression site
- ESAG
expression site-associated gene
- Pol
RNA polymerase
- FITC
fluorescein isothiocyanate
- PARP
procyclic acidic repetitive protein
- DAPI
4′,6-diamidino-2-phenylindole
- CAT
chloramphenicol acetyltransferase
References
- 1. Barry J D. Parasitol Today. 1997;13:212–218. doi: 10.1016/s0169-4758(97)01039-9. [DOI] [PubMed] [Google Scholar]
- 2.Borst P, Rudenko G, Blundell P A, van Leeuwen F, Cross M A, McCulloch R, Gerrits H, Chaves I. Behring Inst Mitt. 1997;99:1–15. [PubMed] [Google Scholar]
- 3.Cross G A. BioEssays. 1996;18:283–291. doi: 10.1002/bies.950180406. [DOI] [PubMed] [Google Scholar]
- 4.Donelson J E. J Biol Chem. 1995;270:7783–7786. doi: 10.1074/jbc.270.14.7783. [DOI] [PubMed] [Google Scholar]
- 5.Vanhamme L, Pays E. Microbiol Rev. 1995;59:223–240. doi: 10.1128/mr.59.2.223-240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson P J, Kooter J M, Borst P. Cell. 1987;51:273–281. doi: 10.1016/0092-8674(87)90154-1. [DOI] [PubMed] [Google Scholar]
- 7.Kooter J M, van der Spek H J, Wagter R, d’Oliveira C E, van der Hoeven F, Johnson P J, Borst P. Cell. 1987;51:261–272. doi: 10.1016/0092-8674(87)90153-x. [DOI] [PubMed] [Google Scholar]
- 8.Pays E, Tebabi P, Pays A, Coquelet H, Revelard P, Salmon D, Steinert M. Cell. 1989;57:835–845. doi: 10.1016/0092-8674(89)90798-8. [DOI] [PubMed] [Google Scholar]
- 9.Alexandre S, Guyaux M, Murphy N B, Coquelet H, Pays A, Steinert M, Pays E. Mol Cell Biol. 1988;8:2367–2378. doi: 10.1128/mcb.8.6.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cully D F, Gibbs C P, Cross G A. Mol Biochem Parasitol. 1986;21:189–197. doi: 10.1016/0166-6851(86)90022-8. [DOI] [PubMed] [Google Scholar]
- 11.Cornelissen A W, Johnson P J, Kooter J M, Van der Ploeg L H, Borst P. Cell. 1985;41:825–832. doi: 10.1016/s0092-8674(85)80063-5. [DOI] [PubMed] [Google Scholar]
- 12.Steverding D, Stierhof Y D, Chaudhri M, Ligtenberg M, Schell D, Beck-Sickinger A G, Overath P. Eur J Cell Biol. 1994;64:78–87. [PubMed] [Google Scholar]
- 13.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 14.Rudenko G, Blundell P A, Dirks-Mulder A, Kieft R, Borst P. Cell. 1995;83:547–553. doi: 10.1016/0092-8674(95)90094-2. [DOI] [PubMed] [Google Scholar]
- 15.Horn D, Cross G A. Cell. 1995;83:555–561. doi: 10.1016/0092-8674(95)90095-0. [DOI] [PubMed] [Google Scholar]
- 16.Horn D, Cross G A. Mol Biochem Parasitol. 1997;84:189–201. doi: 10.1016/s0166-6851(96)02794-6. [DOI] [PubMed] [Google Scholar]
- 17.Borst P, Bitter W, Blundell P A, Chaves I, Cross M A, Gerrits H, van Leeuwen F, McCulloch R, Taylor M C, Rudenko G. Mol Biochem Parasitol. 1998;91:67–76. doi: 10.1016/s0166-6851(97)00184-9. [DOI] [PubMed] [Google Scholar]
- 18.Rudenko G, Cross M A, Borst P. Trends Microbiol. 1998;6:113–117. doi: 10.1016/s0966-842x(97)01200-6. [DOI] [PubMed] [Google Scholar]
- 19.Kooter J M, Borst P. Nucleic Acids Res. 1984;12:9457–9472. doi: 10.1093/nar/12.24.9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung H M, Lee M G, Van der Ploeg L H. Parasitol Today. 1992;8:414–418. doi: 10.1016/0169-4758(92)90194-7. [DOI] [PubMed] [Google Scholar]
- 21.Lee M G, Van der Ploeg L H. Annu Rev Microbiol. 1997;51:463–489. doi: 10.1146/annurev.micro.51.1.463. [DOI] [PubMed] [Google Scholar]
- 22.Evers R, Hammer A, Kock J, Jess W, Borst P, Memet S, Cornelissen A W. Cell. 1989;56:585–597. doi: 10.1016/0092-8674(89)90581-3. [DOI] [PubMed] [Google Scholar]
- 23.Smith J L, Levin J R, Agabian N. J Biol Chem. 1989;264:18091–18099. [PubMed] [Google Scholar]
- 24.Grondal E J, Evers R, Kosubek K, Cornelissen A W. EMBO J. 1989;8:3383–3389. doi: 10.1002/j.1460-2075.1989.tb08502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudenko G, Chung H M, Pham V P, Van der Ploeg L H. EMBO J. 1991;10:3387–3397. doi: 10.1002/j.1460-2075.1991.tb04903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zomerdijk J C, Kieft R, Borst P. Nature (London) 1991;353:772–775. doi: 10.1038/353772a0. [DOI] [PubMed] [Google Scholar]
- 27.Zomerdijk J C, Kieft R, Shiels P G, Borst P. Nucleic Acids Res. 1991;19:5153–5158. doi: 10.1093/nar/19.19.5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borst P. Annu Rev Biochem. 1986;55:701–732. doi: 10.1146/annurev.bi.55.070186.003413. [DOI] [PubMed] [Google Scholar]
- 29.Van der Ploeg L H. Cell. 1986;47:479–480. doi: 10.1016/0092-8674(86)90608-2. [DOI] [PubMed] [Google Scholar]
- 30.Nilsen T W. Mol Biochem Parasitol. 1995;73:1–6. doi: 10.1016/0166-6851(94)00107-x. [DOI] [PubMed] [Google Scholar]
- 31.Rudenko G, Lee M G, Van der Ploeg L H. Nucleic Acids Res. 1992;20:303–306. doi: 10.1093/nar/20.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanhamme L, Pays A, Tebabi P, Alexandre S, Pays E. Mol Cell Biol. 1995;15:5598–5606. doi: 10.1128/mcb.15.10.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cross G A. Parasitology. 1975;71:393–417. doi: 10.1017/s003118200004717x. [DOI] [PubMed] [Google Scholar]
- 34.Brun R, Schonenberger M. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 35.Hirumi H, Hirumi K. J Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 36.McCulloch R, Rudenko G, Borst P. Mol Cell Biol. 1997;17:833–843. doi: 10.1128/mcb.17.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanhamme L, Berberof M, Le Ray D, Pays E. Nucleic Acids Res. 1995;23:1862–1869. doi: 10.1093/nar/23.11.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zomerdijk J C. Ph.D. thesis. The Netherlands: Univ. of Amsterdam; 1992. pp. 129–140. [Google Scholar]
- 39.Shotton D. Histochem Cell Biol. 1995;104:97–138. doi: 10.1007/BF01451571. [DOI] [PubMed] [Google Scholar]
- 40.Chung H M, Shea C, Fields S, Taub R N, Van der Ploeg L H, Tse D B. EMBO J. 1990;9:2611–2619. doi: 10.1002/j.1460-2075.1990.tb07443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zomerdijk J C, Ouellette M, ten Asbroek A L, Kieft R, Bommer A M, Clayton C E, Borst P. EMBO J. 1990;9:2791–2801. doi: 10.1002/j.1460-2075.1990.tb07467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zomerdijk J C, Kieft R, Duyndam M, Shiels P G, Borst P. Nucleic Acids Res. 1991;19:1359–1368. doi: 10.1093/nar/19.7.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Netten H, van Vliet L J, Vrolijk H, Sloos W, Tanke H J, Young I T. Bioimaging. 1996;4:93–106. [Google Scholar]
- 44.Laird P W, Zomerdijk J C, de Korte D, Borst P. EMBO J. 1987;6:1055–1062. doi: 10.1002/j.1460-2075.1987.tb04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seebeck T, Whittaker P A, Imboden M A, Hardman N, Braun R. Proc Natl Acad Sci USA. 1983;80:4634–4638. doi: 10.1073/pnas.80.15.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomashow L S, Milhausen M, Rutter W J, Agabian N. Cell. 1983;32:35–43. doi: 10.1016/0092-8674(83)90494-4. [DOI] [PubMed] [Google Scholar]
- 47.Rudenko G, Bishop D, Gottesdiener K, Van der Ploeg L H. EMBO J. 1989;8:4259–4263. doi: 10.1002/j.1460-2075.1989.tb08611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Konig E, Delius H, Carrington M, Williams R O, Roditi I. Nucleic Acids Res. 1989;17:8727–8739. doi: 10.1093/nar/17.21.8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clayton C E, Fueri J P, Itzhaki J E, Bellofatto V, Sherman D R, Wisdom G S, Vijayasarathy S, Mowatt M R. Mol Cell Biol. 1990;10:3036–3047. doi: 10.1128/mcb.10.6.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung H M. Ph.D. thesis. New York: Columbia University; 1994. pp. 103–118. [Google Scholar]
- 51.Hadjiolov A A. Nucleolus Ribosome Biogenesis in Cell Biology Monographs. Vol. 12. New York: Springer; 1985. pp. 1–268. [Google Scholar]
- 52.Roeder R G, Rutter W J. Proc Natl Acad Sci USA. 1970;65:675–682. doi: 10.1073/pnas.65.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheer U, Rose K M. Proc Natl Acad Sci USA. 1984;81:1431–1435. doi: 10.1073/pnas.81.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jess W, Hammer A, Cornelissen A W. FEBS Lett. 1989;249:123–128. doi: 10.1016/0014-5793(89)80029-8. [DOI] [PubMed] [Google Scholar]
- 55.Sollner-Webb B, Mougey E B. Trends Biochem Sci. 1991;16:58–62. doi: 10.1016/0968-0004(91)90025-q. [DOI] [PubMed] [Google Scholar]
- 56.Nogi Y, Yano R, Nomura M. Proc Natl Acad Sci USA. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]