Abstract
A vestigial, nonphotosynthetic plastid has been identified recently in protozoan parasites of the phylum Apicomplexa. The apicomplexan plastid, or “apicoplast,” is indispensable, but the complete sequence of both the Plasmodium falciparum and Toxoplasma gondii apicoplast genomes has offered no clue as to what essential metabolic function(s) this organelle might perform in parasites. To investigate possible functions of the apicoplast, we sought to identify nuclear-encoded genes whose products are targeted to the apicoplast in Plasmodium and Toxoplasma. We describe here nuclear genes encoding ribosomal proteins S9 and L28 and the fatty acid biosynthetic enzymes acyl carrier protein (ACP), β-ketoacyl-ACP synthase III (FabH), and β-hydroxyacyl-ACP dehydratase (FabZ). These genes show high similarity to plastid homologues, and immunolocalization of S9 and ACP verifies that the proteins accumulate in the plastid. All the putatively apicoplast-targeted proteins bear N-terminal presequences consistent with plastid targeting, and the ACP presequence is shown to be sufficient to target a recombinant green fluorescent protein reporter to the apicoplast in transgenic T. gondii. Localization of ACP, and very probably FabH and FabZ, in the apicoplast implicates fatty acid biosynthesis as a likely function of the apicoplast. Moreover, inhibition of P. falciparum growth by thiolactomycin, an inhibitor of FabH, indicates a vital role for apicoplast fatty acid biosynthesis. Because the fatty acid biosynthesis genes identified here are of a plastid/bacterial type, and distinct from those of the equivalent pathway in animals, fatty acid biosynthesis is potentially an excellent target for therapeutics directed against malaria, toxoplasmosis, and other apicomplexan-mediated diseases.
Keywords: apicomplexan parasites/organellar targeting/fatty acid biosynthesis/malaria
The phylum Apicomplexa is a group of obligate endoparasites that includes Plasmodium spp. (the causative agents of malaria), Toxoplasma gondii (an important opportunistic pathogen associated with AIDS and congenital birth defects), and several other parasites of medical and economic significance (Cryptosporidium, Eimeria, Babesia, and Theileria). It has been shown recently that these parasites contain a plastid (1–5), known as the apicoplast, and available evidence suggests that all of the 4,500 apicomplexan species, which parasitise diverse animal groups including mollusks, worms, insects, and vertebrates (6), probably harbor this organelle (4, 7). The apicoplast is indispensable in Toxoplasma (8), and Plasmodium (9), which probably explains its apparent ubiquity in Apicomplexa.
The origin of the apicoplast is uncertain (10). Thus far, all apicoplasts examined show common characters consistent with a shared origin (4, 11). The multiple surrounding membranes are consistent with a secondary endosymbiotic origin (3, 12, 13), and phylogenetic analysis of sequence data for the plastid protein TufA weakly allies apicoplasts with plastids of green algae and plants, suggesting a green algal endosymbiont (3). On the other hand, the organization of ribosomal protein genes is more congruent between apicoplasts and nongreen plastids (red algae, cryptomonads, and chromophytes), possibly suggesting a different endosymbiont (7). Dinoflagellates are thought to be the nearest relatives of the apicomplexa (6) but very little is known about their plastids, which may derive from either a primary (14) or secondary endosymbiosis (12).
A key question is why the apicoplast has been retained in a highly specialized group of intracellular parasites. The apicoplast genome, although clearly homologous to plastid genomes of plants and algae, is highly reduced and completely lacking in genes known to be involved in photosynthesis (1, 3, 7, 10, 15). Of the 68 genes in the malaria apicoplast, at least 60 can be ascribed to “house-keeping” functions such as transcription and translation (1, 15). One or more of the remaining eight unidentified ORFs may be involved in some function key to plastid retention. However, plastids also are dependent on many hundreds of nuclear-encoded proteins that are targeted to the organelle (16). To date, no nuclear-encoded plastid proteins have been reported in the apicomplexa despite the absence of numerous essential house-keeping genes from the apicoplast genome (1, 7). We have therefore taken advantage of sequence information from the Toxoplasma gondii expressed sequence tag (EST) databases, and the Plasmodium falciparum genomic sequence data, to identify candidate plastid proteins encoded by genes in the parasite nucleus. We show here that several of these proteins are indeed targeted to the apicoplast, and their identity provides the first clues to the possible function of this intriguing organelle.
MATERIALS AND METHODS
Cloning of cDNAs and gDNAs from T. gondii and P. falciparum.
Publicly available Toxoplasma and Plasmodium databases were searched for likely plastid-targeted protein genes. T. gondii EST sequences (Toxoplasma Genome Web; http://www.ebi.ac.uk/parasites/toxo/toxpage.html) with similarity to plastid genes rps9, rpl28, acpP, and fabZ were used to recover full length cDNA clones from a T. gondii RH strain λZAP-II cDNA library. ORFs were identified with consideration of recognized start consensus sequences (17). Intron positions were determined for rps9, rpl28, and acpP by amplifying homologous genes from genomic DNA by using primers based on the cDNA sequence. Preliminary sequence data for P. falciparum acpP and fabH genes on chromosome 2 was obtained from the Institute for Genomic Research website (www.tigr.org). Sequencing of chromosome 2 was part of the International Malaria Genome Sequencing Project and was supported by awards from the National Institute of Allergy and Infectious Diseases and the U.S. Department of Defense. Gene sequences and intron positions were confirmed by sequencing cDNAs amplified from P. falciparum (strain 3D7) single stranded cDNA. Signal peptides in inferred protein sequences were identified by using prediction programs signalp and psort (18).
Phylogenetic Analysis.
Neighbor-joining trees and neighbor-joining bootstrap trees were constructed from distances corrected by the Dayhoff PAM substitution matrix by using the programs neighbor and protdist (19). Protein maximum likelihood was performed by using protml (20). Trees were searched exhaustively with the JTT-f substitution model, using constraints derived from nodes that found strong support from the neighbor-joining trees, or otherwise well-defined relationships. These constraints are indicated by asterisks in Fig. 1, with the exception that mycobacterial rps9 genes also were constrained. Bootstrap values for protein maximum likelihood were estimated by protml using the rell method, and collated by using mol2con. Trees also were constructed using the Fitch–Margoliash algorithm (fitch), unweighted parsimony (paup 3.1.1), and maximum likelihood quartet puzzling (puzzle 2.0). Multiple sequence alignments are available upon request.
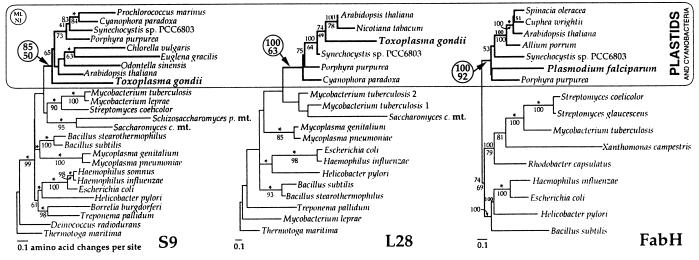
Figure 1.
Phylogenetic relationship of apicomplexan S9, L28, and FabH proteins to plastid and cyanobacterial homologs. Neighbor-joining (NJ) trees are shown with bootstrap confidence values >50% from both NJ (below the nodes) and protein maximum likelihood (ML) (above the nodes). Asterisks indicate constrained nodes in ML analyses. For all three data sets, the apicomplexan genes consistently group among genes from plastids and cyanobacteria (see circled bootstrap values) and distinct from mitochondria (mt data available only for S9 and L28) and other eubacterial groups.
Immunological Analysis of S9 and Acyl Carrier Protein (ACP).
T. gondii rps9 and acpP EST clones (GenBank accession nos. N82743 and N60936, respectively) were cloned in-frame into pGEX expression vectors (Pharmacia) and glutathione S-transferase (GST)-fusion proteins expressed in Escherichia coli. Affinity-purified fusion proteins were used to immunize rabbits, and antibodies were affinity purified by using immobilized fusion protein. Western blot analysis was performed on detergent extracts of RH strain T. gondii tachyzoites, using secondary antibodies conjugated to alkaline phosphatase. For immunofluorescence, T. gondii tachyzoites were rapidly frozen, thawed in 3% paraformaldehyde in PBS, and post-fixed 20 min in cold methanol; antibody incubations were carried out in PBS at room temperature, using fluorescein isothiocyanate-conjugated secondary antibodies. DNA counterstaining was carried out by using 5 μg/ml propidium iodide. Immunolabeling of ultra-thin sections for electron microscopy was performed similarly, using parasites embedded in LR-Gold (London Resin) and secondary antibodies conjugated to 20 nm gold particles (BioCell). Cells were examined by using a Leica TCS 4D confocal microscope or a Philips CM120 BioTWIN transmission electron microscope. Controls using preimmune sera, secondary antibodies only, or anti-glutathione S-transferase antibodies were negative.
Green Fluorescent Protein (GFP) Reporter Targeting.
GFP fusion protein constructs were made by using T. gondii acpP-coding sequences corresponding to either the entire acpP gene product or the first 104 aa only. These fragments were amplified from tachyzoite cDNA by using sense primer 5′-ggaagatctaaaATGGAGATGCATCCCCGCAACGC-3′ and antisense primers 5′-TGGACCTAGGCGCTGTAGCGCTCTTGGCTTTCTC-3′ or 5′-TGGACCTAGGCCGATCATCAGAACTCGCCTCGT-3′ (for TgACPfull-GFP or TgACPleader-GFP, respectively) and ligated as BglII–Avr II fragments in place of the P30 sequences in plasmid ptubP30-GFP/sag-CAT (21). T. gondii tachyzoites were transfected and selected for stable transformants as described (21). Fluorescence was observed by using a Zeiss Axiovert 35-inverted microscope equipped with a 100 W Hg-vapor lamp. Western blotting and immunolabeling of ACP–GFP fusion proteins was carried out as described (21) by using rabbit anti-GFP (CLONTECH) and anti-ACP, and colocalization with apicoplast DNA was examined by counterstaining with 4′,6-diamidino-2-phenylindole (3, 8).
Thiolactomycin Growth Inhibition of P. falciparum.
Thiolactomycin was synthesized as described (22). Asynchronous cultures of P. falciparum-infected erythrocytes (strains D10 and W2 mef; 0.5% parasitemia) were inoculated into microtiter wells containing serial twofold dilutions of thiolactomycin in culture medium (final concentrations: 250–0.5 μg/ml = 1189–2.3 μM). 3H-labeled hypoxanthine was added in fresh hypoxanthine-free medium at 28 hr, and incorporation was assessed 18 hr later as described (23). Inhibition was plotted as the percentage of growth in drug-free medium (average of six replicates), and IC50 values were estimated from this graph.
RESULTS AND DISCUSSION
Identification of Nuclear-Encoded Apicoplast Proteins.
Searching of the T. gondii and P. falciparum databases for entries exhibiting similarity to nuclear-encoded genes that are known to be targeted to plastids in plant and algal systems (16) identified several candidate sequences, including ribosomal protein genes rps9 and rpl28 (encoding S9 and L28, respectively) and fatty acid biosynthetic genes acpP, fabH, and fabZ (encoding ACP, β-ketoacyl-ACP synthase III and β-hydroxyacyl-ACP dehydratase, respectively). The rps9 and rpl28, acpP, fabH, and fabZ genes are undoubtedly nuclear because they (i) are absent from the apicoplast genomes (1, 3), (ii) were recovered from polyA+ cDNA, and (iii) harbor spliceosomal introns. Moreover, Southern analysis of P. falciparum pulsed field gel electrophoresis blots shows that acpP and fabH are located on chromosome 2 (data not shown). Molecular phylogenies for rps9, rpl28, and fabH revealed that these nuclear genes are unequivocally prokaryotic in nature and cluster with plastid and cyanobacterial homologs (Fig. 1). Phylogenetic analysis of ACP proteins also indicates a plastid origin for the Toxoplasma and Plasmodium acpP genes (data not shown), but these trees were less well resolved, due in part to the short protein length (≈85 aa). β-hydroxyacyl-ACP dehydratase (FabZ) is present in plant chloroplasts (24) but has not been cloned yet from plants or algae, preventing informative phylogenetic analysis.
These data suggest that all of the above genes now present in the nucleus of apicomplexan parasites derive from the endosymbiont, (either directly from the plastid genome or from the endosymbiont nucleus). It is noteworthy that (β-ketoacyl-ACP synthase III) FabH, ACP, L28, and S9 are plastid encoded in red algae but nuclear encoded in plants (16). A corollary of such intracellular gene relocation is that protein products of the nuclear genes acquired from the endosymbiont are likely to be targeted back to the organelle of origin. This is especially likely for rps9 and rpl28, which are not encoded in the apicoplast genome of Toxoplasma (3) or Plasmodium (1), but which are likely to be required for protein translation.
Antisera raised against recombinant Toxoplasma rps9 and acpP gene products were used to test for apicoplast targeting of these proteins in Toxoplasma tachyzoites. As shown in Fig. 2, immunolocalization of S9 and ACP in intact Toxoplasma cells (Fig. 2A–C) and ultra-thin sections (Fig. 2 D and E) confirms accumulation of these gene products within the apicoplast. Labeling appears evenly spread throughout the organelle, but little or no signal is detectable in the parasite cytoplasm, nucleus, mitochondrion, or endomembrane system (Fig. 2A–E).
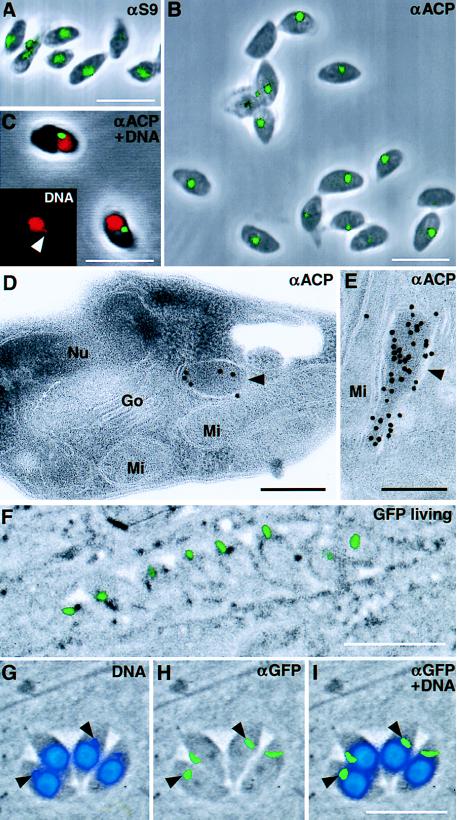
Figure 2.
Apicoplast localization of nuclear-encoded proteins. (A and B) Immunodetection of S9 and ACP (respectively) in intact T. gondii cells demonstrates that these proteins are restricted to a distinct region of the parasite similar to the location of the apicoplast. Some cells show two plastids, which are probably division stages. (C) Counterstaining with propidium iodide (red) confirms colocalization of ACP (green) with the extranuclear apicoplast DNA (Inset shows DNA staining only; arrowhead indicates apicoplast DNA). (D and E) Detection of ACP by immunogold labeling of ultra-thin sections shows strong labeling of the apicoplast (arrowhead). Nu, nucleus; Mi, mitochondrion; Go, Golgi apparatus. (F) The N-terminal domain of ACP (TgACPleader-GFP) is sufficient to target GFP to the apicoplast, and the recombinant protein can be visualized in living cells. (G–I) In fixed cells labeled with anti-GFP (green) and counterstained with DAPI (blue), TgACPleader-GFP can be seen to colocalize with the apicoplast DNA (arrowheads indicate apicoplasts in two cells). Color images were collected independently and overlaid on top of phase-contrast micrographs. (White scale bars = 10 μm, black scale bars = 200 nm).
Nuclear-Encoded Apicoplast Proteins Contain Cleavable Presequences that Mediate Targeting.
Plastid targeting can be classified into two broad categories depending on evolutionary origin and ultrastructure, particularly the number of membranes surrounding the plastid. In plants, green algae, red algae, and glaucophytes, whose plastids are enclosed by two membranes, targeting is mediated by an N-terminal presequence known as the transit peptide, which is removed after import (Fig. 3) (25). Plant and green algal transit peptides are the best characterized of these, and although no consensus sequence or secondary structure is evident, they are typically 25–125 aa in length, basic, and rich in serine and threonine (25, 26).
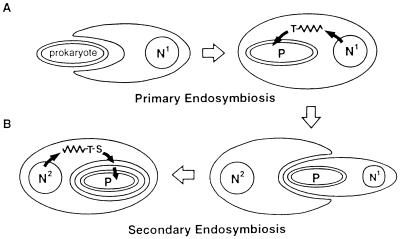
Figure 3.
Plastid origins and protein targeting. (A) Primary endosymbiosis describes the uptake of a prokaryote by a eukaryote. Plastids derived by primary endosymbiosis are generally surrounded by two membranes, and targeting of nuclear-encoded gene products to the endosymbiont is effected by an N-terminal transit peptide (T). (B) Secondary endosymbiotic plastid origin involves a heterotrophic eukaryote phagocytozing a photosynthetic eukaryote produced by primary endosymbiosis. The secondary endosymbiont’s cytoplasm and nucleus (N1) are typically lost or heavily reduced, and the resulting plastid is surrounded by four membranes, the outermost of which derives from the phagocytotic membrane. Sometimes one of the two outer membranes is lost at this point, resulting in a total of three. Targeting of nucleus-encoded (N2) gene products to secondary plastids requires a signal peptide (S) to mediate protein passage across the outer membrane(s) followed by a transit peptide (T) for import across the inner membrane pair.
Targeting presents a more complex problem in organisms with more than two membranes around the plastid. Euglenoids and dinoflagellates have three plastid membranes, and certain other algal groups (heterokont and haptophyte algae, chlorarachniophytes, and cryptomonads) have four plastid membranes. Plastids with four membranes were almost certainly acquired by secondary endosymbiosis, in which a phagotrophic eukaryote engulfed (or was invaded by) a plastid-containing eukaryote (Fig. 3) (12). Plastids with three membranes may have been acquired either by secondary (12) or primary (14) endosymbiosis. The details of protein trafficking to plastids containing multiple membranes is not well understood, but because the outermost membrane is part of the host’s endomembrane system, targeting apparently commences via the secretory pathway into the endoplasmic reticulum (ER) courtesy of a classic signal peptide (27–29). Subsequent targeting across the inner pair of plastid membranes involves a downstream transit peptide (Fig. 3) (27–29).
The nuclear-encoded apicoplast genes identified here are all predicted to encode substantial N-terminal extensions when compared with the equivalent plastid and bacterial proteins (Fig. 4). The extreme N-terminal regions (16–34 aa) of these extensions resemble classic signal peptides, containing a hydrophobic domain followed by a “von Heijne” cleavage site (30). Downstream of the predicted signal peptides, the Toxoplasma proteins exhibit the general features of transit peptides: net positive charge and enrichment for serine and threonine residues. Putative Toxoplasma transit peptides range from 57–107 aa in length. The putative Plasmodium transit peptides also have a net positive charge, are somewhat shorter (30–42 aa), and rich in lysine and asparagine, probably reflecting the strong A/T bias of the Plasmodium genome (>80%) as well as the apparently loose constraints of transit peptides (26). No apicoplast transit peptide cleavage motif has been identified to date. Several targeting peptide boundaries are located near introns (Fig. 4), possibly indicating acquisition of these motifs by exon shuffling (31). It is also interesting to note that the locations of two introns are shared in T. gondii and P. falciparum acpP genes (Fig. 4), suggesting these genes are orthologous and that the gene transfer from endosymbiont to parasite nucleus and acquisition of targeting sequences occurred prior to the divergence of these species.
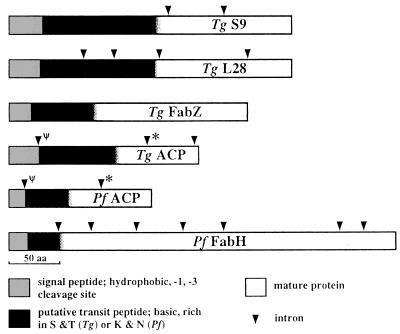
Figure 4.
Schematic of inferred proteins of T. gondii (Tg) genes rps9, rpl28, fabZ, and acpP and P. falciparum (Pf) genes acpP and fabH, showing N-terminal extensions resembling signal plus transit peptides (shaded and black boxes, respectively). Putative signal peptide cleavage sites were predicted by using signalp and psort (18); transit peptide/mature protein boundaries were imprecisely defined based on Western data (see Fig. 5) and sequence similarity with other mature proteins. Intron positions are indicated (triangle) (except for FabZ); “ψ” and “∗” indicate introns shared between T. gondii and P. falciparum homologs, suggesting shared ancestry.
Processing of nuclear-encoded apicoplast proteins was examined by Western blot analysis, as shown in Fig. 5. S9 and ACP antisera recognized protein bands of the expected size for the mature proteins (lower bands in Fig. 5B), suggesting that the predicted N-terminal extensions are removed. Interestingly, both antisera also recognize larger proteins corresponding to the predicted size of the relevant mature protein plus transit peptide in each case. These larger bands probably represent processing intermediates associated with apicoplast targeting, either in the process of traversing the multiple apicoplast membranes, or awaiting proteolytic cleavage in the apicoplast stroma. As signal peptides are usually removed during cotranslational import into the ER, it is not surprising that full length precursors including the signal peptide were not detected.
Figure 5.
Processing of apicoplast-targeted proteins in T. gondii. (A) Schematic of T. gondii S9 and ACP proteins, and GFP fusion proteins (ACPfull-GFP and ACPleader-GFP), showing protein band sizes before and after predicted processing (mature ACP carries a phosophopantetheine prosthetic group that adds 4 kDa to its expected gel mobility (45). (B) Western blot analysis of T. gondii S9 and ACP proteins (lanes i and ii, respectively). The observed antigens are in close agreement with the expected sizes for fully processed protein (lower bands) and full-length protein including the transit peptide but lacking the signal sequence (upper bands). (C) Western blot analysis of TgACPfull-GFP and TgACPleader-GFP proteins (lanes iii and iv). Observed molecular masses correspond to the predictions shown in (A) (the faint lower molecular mass band of ≈30 kDa in the ACPfull-GFP lane is not seen in untransfected parasites and probably represents a GFP degradation product). Note that similar amounts and relative levels of processed and unprocessed apicoplast proteins are observed for the native and recombinant proteins.
To test the hypothesis that the N-terminal extensions present on nuclear-encoded apicoplast proteins mediate targeting, GFP fusion proteins were constructed as shown in Fig. 5A by using either the entire T. gondii acpP gene or only the putative N-terminal targeting information (signal and transit peptides). These constructs, designated TgACPfull-GFP and TgACPleader-GFP, respectively, were transfected into T. gondii. As indicated in Fig. 2F–I, apicoplast-specific targeting was observed in both transient and stable transfectants, using both constructs. Successful targeting of the truncated ACP fusion protein (TgACPleader-GFP) demonstrates that the 104-aa N-terminal presequence of TgACP is sufficient to target a protein into the apicoplast. Interestingly, Western blot analyses of GFP fusion proteins by using anti-GFP antisera reveal both processed and unprocessed pools of apicoplast-targeted proteins of the expected size and in ratios comparable to native proteins (Fig. 5C).
A Model for Targeting Nuclear-Encoded Proteins to the Apicomplexan Plastid.
The identification of at least two nuclear genes (rps9 and acpP) whose products accumulate in the apicoplast, and the demonstration that the presequence of ACP is sufficient to effect apicoplast targeting, provides a preliminary model for apicoplast targeting in these parasites. Because rpl28, fabH, and fabZ also encode proteins with similar, bi-partite N-terminal leaders (Fig. 4), and molecular phylogenies support a plastid origin for rpl28 and fabH (Fig. 1), we predict that all of these proteins are targeted to the apicoplast.
The presence of an apparent signal peptide implies that routing to the apicoplast occurs via the endomembrane system, as proposed for other multi-membrane plastids (28, 29). Indeed, the bi-partite apicoplast targeting information provides support for a secondary endosymbiotic origin of the apicoplast, and is consistent with a previous observation of four membranes surrounding this organelle (3). Reports describing only two- or three-bounding membranes (2) were based on material with inferior ultrastructural preservation (7) and now appear to be incorrect. The precise molecular mechanisms targeting proteins into plastids with multiple-bounding membranes have been difficult to determine in other systems and remain poorly understood (28, 29). The GFP reporter system, coupled with the accessibility of T. gondii to molecular genetic manipulation (21), provides a convenient means to dissect plastid targeting in a secondary endosymbiont.
By using this targeting model, we have identified three additional Plasmodium genes whose products are likely to be targeted to the apicoplast: clpP (a plastid protease GenBank accession no. AL009009), EF-TS (elongation factor TS, Sanger ID MP03010), and cpn60 (a plastid chaperonin GenBank accession no. X75420). Conversely, we can now predict proteins not likely to be directed to the apicoplast. For example, despite previous speculation that the apicoplast might harbor a shikimate pathway for essential amino acid synthesis (7, 10, 32), the lack of N-terminal extensions on both P. falciparum and T. gondii genes encoding chorismate synthase (32) suggests a cytoplasmic localization, as found in fungi (33). Characterization of presequences thus provides a powerful predictive tool for compiling a picture of apicoplast proteins and metabolic pathways from the ever-expanding genomics databases.
Does the Apicoplast Synthesize Fatty Acids?
Identification of nuclear-encoded apicoplast proteins with unequivocal similarity to the fatty acid biosynthetic genes acpP, fabH, and fabZ offers a new insight into the metabolic activity of this enigmatic organelle. By analogy with plant and algal plastids, apicoplasts have been suggested to perform three possible anabolic pathways: essential amino acid biosynthesis, heme biosynthesis, and fatty acid biosynthesis (1, 3, 7, 10). Although essential amino acid biosynthesis and heme biosynthesis in apicoplasts cannot yet be discounted, available evidence suggests these functions are fulfilled by cytosolic pathways (refs. 1, 3, 7, 15 and see above). In contrast, the presence of ACP and (very probably) FabH and FabZ in the apicoplast indicates that at least some fatty acid biosynthesis occurs in the apicoplast. All three proteins are members of the fatty acid synthase multi-enzyme complex (24, 34). ACP plays a central role in fatty acid biosynthesis by holding the forming acyl chain, whereas FabH and FabZ are involved in the condensation and dehydration steps, respectively, of acetyl addition during acyl chain elongation (24, 34). Partial sequence also has been obtained for another fatty acid synthase component from P. falciparum, fabF, which encodes β-ketoacyl-ACP synthase II (34), further implicating a fatty acid biosynthesis pathway in these parasites.
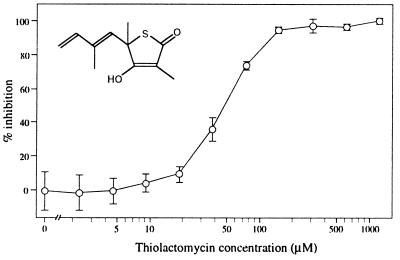
There are two types of fatty acid biosynthesis. Type I is found in the cytosol of animals and fungi (35). Type II is widespread among bacteria but in eukaryotes is restricted to the plastids of plants and algae (24, 34) and perhaps the mitochondria of yeast (36), which reflects the origin of these organelles from endosymbiotic bacteria (16). The Toxoplasma and Plasmodium fatty acid biosynthesis genes characterized here are of type II, which is consistent with localization of the protein products to the apicomplexan plastid. Importantly, the different types of fatty acid biosynthesis allow for selective perturbation. The antibiotic thiolactomycin is a selective inhibitor of type II fatty acid biosynthesis (37). In E. coli, thiolactomycin inhibits the condensing enzymes FabB, FabF, and FabH (34, 38), and it inhibits the equivalent plastid enzymes in plants (24, 39). In contrast, thiolactomycin has no effect on fatty acid biosynthesis (type I) of Saccharomyces cerevisiae, Candida albicans, and rat liver (37). We examined the effect of thiolactomycin on the malaria parasite. In vitro growth inhibition assays using a racemic mixture of (R and S) thiolactomycin with Plasmodium falciparum show an IC50 of ≈50 μM (Fig. 6). This level of inhibition is comparable with that seen for fatty acid biosynthesis in isolated pea and spinach plastids (IC50 of 10–100 μM; refs. 40 and 41), as well as for the individual condensing enzymes (5- >25 μM) (24, 39). Thiolactomycin inhibition of malaria thus provides additional supporting evidence for a type II fatty acid biosynthetic pathway in apicoplasts. Proof of this hypothesis awaits biochemical demonstration of fatty acid production in the apicoplast, perhaps by demonstrating presence of type II signature lipids (24, 42) in apicomplexa.
Figure 6.
Log concentration-growth inhibition curves for the type II fatty acid biosynthesis inhibitor, thiolactomycin, tested against P. falciparum. The IC50 for both D10-strain and the multi-drug-resistant strain W2 mef (data not shown) P. falciparum was calculated as ≈50 μM. Vertical bars indicate standard deviations from six samples. (Inset) the structure of thiolactomycin.
Why would the apicoplast synthesize lipids? Plant plastids require unusual fatty acids (43), and it is possible that apicoplast fatty acid biosynthesis merely provides an extended house-keeping function for the organelle. However, in plants and certain algae, the plastid exports lipids to the rest of the cell and is the sole site of fatty acid biosynthesis (24, 42). A fatty acid, or lipid, could thus be the mysterious “factor X” proposed to be produced by Toxoplasma apicoplasts (44), and perhaps such lipids are an essential component for parasitophorous vacuole formation because poisoning the apicoplast inhibits parasite replication at an early stage after infection (8).
Concluding Remarks.
We have characterized the first nuclear-encoded plastid proteins in apicomplexan parasites. We have demonstrated that an N-terminal extension on these proteins mediates targeting, probably via the endomembrane system. Several of the proteins are members of the type II fatty acid biosynthesis complex; a pathway not recognized previously in these parasites. Preliminary trials with an agent known to perturb the type II pathway suggests that fatty acid biosynthesis may be an excellent new target for combating apicomplexan-mediated diseases.
Acknowledgments
We are grateful to J. W. Ajioka for a T. gondii RH cDNA library, the WashU-Merck Toxoplasma Expressed Sequence Tag Project for T. gondii Expressed Sequence Tag clones, M. E. Wickham for P. falciparum cDNA, M. Guy for thiolactomycin synthesis, V. Su and M. Reed for technical assistance, J. Thompson for P. falciparum Southern analysis, M. Reith for informatics assistance, and A. Stoltzfus for mol2con. This research was supported by grants from the Australian Research Council and the U.S. National Institutes of Health. R.F.W. was supported by an Australian Postgraduate Research Award, P.J.K. by a Fellowship from the Medical Research Council of Canada, and D.S.R. is a Burroughs Wellcome Scholar in Molecular Parasitology. Preliminary sequence data for P. falciparum chromosome 2 was obtained from The Institute for Genomic Research website (www.tigr.org). Sequencing of chromosome 2 was part of the International Malaria Genome Sequencing Project and was supported by awards from the National Institute of Allergy and Infectious Diseases and the U.S. Department of Defense.
ABBREVIATIONS
- ACP
acyl carrier protein
- FabH
β-ketoacyl-ACP synthase III
- FabZ
and β-hydroxyacyl-ACP dehydratase
- GFP
green fluorescent protein
Footnotes
References
- 1. Wilson R J M, Denny P W, Preiser P R, Rangachari K, Roberts K, Roy A, Whyte A, Strath M, Moore D J, Moore P W, et al. J Mol Biol. 1996;261:155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- 2.McFadden G I, Reith M, Munholland J, Lang-Unnasch N. Nature (London) 1996;381:482. doi: 10.1038/381482a0. [DOI] [PubMed] [Google Scholar]
- 3.Köhler S, Delwiche C F, Denny P W, Tilney L G, Webster P, Wilson R J M, Palmer J D, Roos D S. Science. 1997;275:1485–1489. doi: 10.1126/science.275.5305.1485. [DOI] [PubMed] [Google Scholar]
- 4.Denny P, Preisser P, Williamson D, Wilson I. Protist. 1998;149:51–59. doi: 10.1016/S1434-4610(98)70009-4. [DOI] [PubMed] [Google Scholar]
- 5.Blunt D, Kjramtsov N, Upton S, Montelone B. Clin Diagn Lab Immunol. 1997;4:11–13. doi: 10.1128/cdli.4.1.11-13.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine N D. The Protozoan Phylum Apicomplexa. Boca Raton, FL: CRC Press; 1988. [Google Scholar]
- 7.McFadden G I, Waller R F, Reith M, Munholland J, Lang-Unnasch N. Plant Syst Evol Suppl. 1997;11:261–287. [Google Scholar]
- 8.Fichera M E, Roos D S. Nature (London) 1997;390:407–409. doi: 10.1038/37132. [DOI] [PubMed] [Google Scholar]
- 9.McConkey G A, Rogers M J, McCutchan T F. J Biol Chem. 1997;272:2046–2049. doi: 10.1074/jbc.272.4.2046. [DOI] [PubMed] [Google Scholar]
- 10.McFadden G I, Waller R F. BioEssays. 1997;19:1033–1040. doi: 10.1002/bies.950191114. [DOI] [PubMed] [Google Scholar]
- 11.Lang-Unnasch, N., Reith, M., Munholland, J. & Barta, J. (1998) J. Int. Parasitol., in press. [DOI] [PubMed]
- 12.Delwiche C F, Palmer J D. Plant Syst Evol Suppl. 1997;11:51–86. [Google Scholar]
- 13.Wilson R J, Williamson D H, Preiser P. Infect Agents Dis. 1994;3:29–37. [PubMed] [Google Scholar]
- 14.Cavalier-Smith T. Biol J Linn Soc. 1982;17:289–306. [Google Scholar]
- 15.Wilson R J M, Williamson D H. Microbiol Mol Biol Rev. 1997;61:1–16. doi: 10.1128/mmbr.61.1.1-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin W, Stoebe B, Goremykin V, Hansmann S, Hasegawa M, Kowallik K. Nature (London) 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- 17.Seeber F. Parasitol Res. 1997;83:309–311. doi: 10.1007/s004360050254. [DOI] [PubMed] [Google Scholar]
- 18.Claros M, Brunak S, von Heijne G. Curr Opinion Struct Biol. 1997;7:394–398. doi: 10.1016/s0959-440x(97)80057-7. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. Syst Zool. 1978;27:401–410. [Google Scholar]
- 20.Adachi J, Hasegawa M. Computer Science Monographs. Tokyo: Institute of Statistical Mathematics; 1996. , No. 28. [Google Scholar]
- 21.Striepen B, He C Y, Matrajt M, Soldati D, Roos D S. Mol Biochem Parasitol. 1998;92:325–338. doi: 10.1016/s0166-6851(98)00011-5. [DOI] [PubMed] [Google Scholar]
- 22.Wang C-L, Salvino J. Tetrahedron Lett. 1984;25:5243–5246. [Google Scholar]
- 23.Desjardin R E, Canfield C J, Haynes J D, Chulay J D. Antimicrob Agents Chemother. 1979;16:710–718. doi: 10.1128/aac.16.6.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harwood J. Biochim Biophys Acta. 1996;1301:7–56. doi: 10.1016/0005-2760(95)00242-1. [DOI] [PubMed] [Google Scholar]
- 25.von Heijne G, Hirai T, Klösgen R-B, Steppuhn J, Bruce B, Keegstra K, Herrmann R. Plant Mol Biol Rep. 1991;9:104–126. [Google Scholar]
- 26.von Heijne G. FEBS Lett. 1991;278:1–3. doi: 10.1016/0014-5793(91)80069-f. [DOI] [PubMed] [Google Scholar]
- 27.Sulli C, Schwartzbach S D. J Biol Chem. 1995;270:13084–13090. doi: 10.1074/jbc.270.22.13084. [DOI] [PubMed] [Google Scholar]
- 28.Reith M. In: Oxygenic Photosynthesis: The Light Reactions. Ort D, Yocum C, editors. The Netherlands: Kluwer; 1996. pp. 643–657. [Google Scholar]
- 29.Bodyl A. Bot Acta. 1997;110:395–400. [Google Scholar]
- 30.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 31.Sahrawy M, Hecht V, Lopez-Jaramillo J, Chueca A, Chartier Y, Meyer Y. J Mol Evol. 1996;42:422–431. doi: 10.1007/BF02498636. [DOI] [PubMed] [Google Scholar]
- 32.Roberts F, Roberts C, Johnson J, Kyle D, Krell T, Coggins J, Coombs G, Milhous W, Tzipori S, Ferguson D, et al. Nature (London) 1998;393:801–806. doi: 10.1038/31723. [DOI] [PubMed] [Google Scholar]
- 33.Bode R, Birnbaum D. Z Allg Mikrobiol. 1981;21:417–422. doi: 10.1002/jobm.3630210602. [DOI] [PubMed] [Google Scholar]
- 34.Cronan J E, Rock C O. In: Escherischia coli and Salmonella. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 612–636. [Google Scholar]
- 35.Smith S. FASEB J. 1994;8:1248–1259. [PubMed] [Google Scholar]
- 36.Schneider R, Brors B, Burger F, Camrath S, Weiss H. Curr Genet. 1997;32:384–388. doi: 10.1007/s002940050292. [DOI] [PubMed] [Google Scholar]
- 37.Hayashi T, Yamamoto O, Sasaki H, Kawaguchi A, Okazaki H. Biochem Biophys Res Comm. 1983;115:1108–1113. doi: 10.1016/s0006-291x(83)80050-3. [DOI] [PubMed] [Google Scholar]
- 38.Nishida I, Kawaguchi A, Yamada M. J Biochem. 1986;99:1447–1454. doi: 10.1093/oxfordjournals.jbchem.a135614. [DOI] [PubMed] [Google Scholar]
- 39.Jones A, Dancer J, Harwood J. Phytochemistry. 1995;39:511–514. [Google Scholar]
- 40.Jones A, Dancer J, Harwood J. Biochem Soc Trans. 1994;22:202. doi: 10.1042/bst022258s. [DOI] [PubMed] [Google Scholar]
- 41.Jaworski J, Clough R, Barnum S, Post-Beitenmiller D, Ohlrogge J. In: Plant Lipid Biochemistry, Structure and Utilization. Quinn P, Harwood J, editors. London: Portland Press; 1990. pp. 97–104. [Google Scholar]
- 42.Somerville C, Browse J. Science. 1991;252:80–87. doi: 10.1126/science.252.5002.80. [DOI] [PubMed] [Google Scholar]
- 43.Slabas T. Trends Plant Sci. 1997;2:161–162. doi: 10.1016/s1360-1385(99)01544-7. [DOI] [PubMed] [Google Scholar]
- 44.Tomavo S, Boothroyd J C. Int J Parasitol. 1995;25:1293–1299. doi: 10.1016/0020-7519(95)00066-b. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez M D, Lamppa G K. Plant Cell. 1990;2:195–206. doi: 10.1105/tpc.2.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]