Abstract
The glial cells missing (gcm) gene in Drosophila encodes a transcription factor that determines the choice between glial and neuronal fates. We report here the isolation of two mammalian gcm homologs, Gcm1 and Gcm2, and the characterization of their expression patterns during embryonic development. Although Gcm2 is expressed in neural tissues at a low level, the major sites of expression for both of the mammalian genes are nonneural, suggesting that the functions of the mammalian homologs have diverged and diversified. However, when expressed ectopically, Gcm1 can substitute functionally for Drosophila gcm by transforming presumptive neurons into glia. Thus, certain biochemical properties, although not the specificity of the tissue in which the gene is expressed, have been conserved through the evolution of the Gcm gene family.
The Drosophila gene glial cells missing (gcm) controls the binary fate decision between neuronal and glial lineages (1–3). Loss-of-function mutations in gcm result in conversion of presumptive glial cells into neurons, whereas ectopic expression of gcm generates additional glial cells at the expense of neurons. Based on its nuclear localization and sequence-specific DNA-binding activity, GCM was proposed to be a transcriptional activator for glial-specific genes. Consistent with this, multiple copies of the proposed GCM-binding elements are found in the putative upstream regulatory region of a glial-specific gene, reverse polarity (repo) (4). GCM was also shown to be able to activate a reporter gene in a GCM-binding-site-dependent manner in transiently transfected cells (5).
Many vertebrate homologs of Drosophila transcription factors involved in neurogenesis have been isolated and shown to be expressed specifically in neural tissues (6, 7). This is particularly well established for proteins belonging to the basic helix–loop–helix family (8). However, no mammalian genes thus far discovered fulfill the role of a glial fate-determination gene. Thus, it was of significant interest to examine the possible role of mammalian homologs of the gcm gene in the neuron–glia fate decision. Recently, two mammalian gcm homologs have been identified (4, 9). Not surprisingly, the DNA-binding domain, now called the gcm motif, is conserved, whereas the rest, likely responsible for interaction with other proteins, is not. Although one report made a preliminary claim that one of the homologs is expressed in embryonic neural tissues (4), no detailed study of the expression pattern has been presented thus far.
We describe here the isolation of the two rodent Gcm genes and the characterization of their expression patterns by in situ hybridization and reverse transcription–PCR (RT-PCR). The results indicate that the major sites of expression in embryos are not neural for either of the rodent genes, although their expression is highly specific to certain nonneural tissues. Thus, their function is, for the most part, not conserved from that of Drosophila gcm. Interestingly, one of the homologs, Gcm1 but not Gcm2, was able to generate extra glial cells when expressed in Drosophila and could partially rescue the loss-of-function phenotype, indicating that Gcm1 shares conserved regulatory capabilities with gcm.
MATERIALS AND METHODS
Isolation of Rodent Homologs of gcm.
A pair of degenerate oligonucleotide primers (5′-CGGATCCAGACCT/CGCCATT/CTGT/CGACAAG-3′ corresponding to the sequence coding for amino acids RPAICDK and 5′-CGGAATTCTTCT/GGGTT/CTT/GGGA/GTGATCA/GTG-3′ corresponding to the complementary sequence coding for amino acids HDHPR/KPE) was used to screen by PCR amplification a rat placental cDNA library that was built in the plasmid vector pcDNA3 (Invitrogen) and divided into 50 groups. A positive pool was transformed into Escherichia coli, and the resulting transformants were screened with a 32P-labeled probe derived from the PCR product. Several positive clones were sequenced and identified as rat Gcm1 (rGcm1). The 3′ end of rGcm1 was determined by 3′ rapid amplification of cDNA ends by using a cDNA preparation from embryonic day 14.5 (E14.5) rat placental tissue primed with an anchored oligo(dT) primer (5′-CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTTTTTTTTTTTTT-3′) as the template. The primers used for the first 30 PCR cycles were 5′-CCTGTGGATTTCAGCAGC-3′ and 5′-CCAGTGAGCAGAGTGACG-3′. Primers for the second 30 cycles were 5′-TCGCTTACGGCTCTCATC-3′ and 5′-GAGGACTCGAGCTCAAGC-3′.
The same 32P-labeled probe was used to screen a 129SvEv mouse genomic DNA library built in the phage vector λ DASH II (Stratagene). The positive clones were divided into two groups based on the strength of hybridization. Partial sequence analyses confirmed that the strongly hybridizing group contained Gcm1 clones, whereas the other was composed of a distinct yet homologous gene called Gcm2. Several overlapping genomic clones were sequenced, and from over 5 kb of composite sequence, a partial cDNA sequence for mouse Gcm2 (mGcm2) was proposed (see Results).
To isolate a full-length mGcm2 cDNA, two successive 30-cycle rounds of PCR amplification were run with a cDNA preparation from E13.5 mouse pharyngeal tissue as the template. The oligonucleotide primers used were based on the genomic DNA sequence and the subsequently published mGcm2 cDNA sequence (GenBank accession no. D88611; ref. 4). For the first 30-cycle round, primers were 5′-CTCTTTCAGGGCCCTGACTAG-3′ and 5′-CATAAATGCACCCTTGGCGTG-3′; for the second 30 cycles, primers were 5′-GATCGAATTCAATGCCAGCAGACAGCACG-3′ and 5′-GATCGGATCCCTGGATTTCTCTTAAAAGTCC-3′. The resulting full-length product was ligated to pCR2.1 with a TA Cloning kit (Invitrogen; now called pG2–24). To generate a mGcm2 construct containing the 5′ UTR (untranslated region), a pair of primers (5′-GATCGAGCTCCTTTGTGTGTATATCTGCCTC-3′ and 5′-TGTCCCACGTGAGTTTCATCC-3′) was used to PCR amplify from genomic DNA a 300-bp product containing 220 bases of 5′ UTR and parts of the first exon. This product was digested with SacI and PmlI and inserted into pG2–24 (p5G2–24). To generate a construct with a MYC-epitope tag, a three-way ligation was performed with the following DNA fragments: (i) p5G2–24 digested with BsgI and XhoI, (ii) a PCR product amplified from p5G2–24 with primers 5′-TCGAGTCCATATTCCACCCTG-3′ and 5′-GATCGGATCCAAAGTCCTCATTGTCAAAGC-3′ digested with BsgI and BamHI, and (iii) a PCR product amplified from pCS2+MT (10) with primers 5′-TGCAGGATCCCATCGATTTAAAGC-3′ and 5′-GCATCTCGAGTTAGGTGAGGTCGCCCAAGCTCTC-3′ digested with BamHI and XhoI. The resulting construct has the 5′ UTR, mGcm2 ORF, and five MYC-epitope tags in tandem (p5G2-myc).
In Situ Hybridization.
In situ hybridization on frozen sections and whole-mount in situ hybridization were performed as described with minor modifications (11). Detailed protocols are available on request. For Gcm1, the antisense probe was derived from the longest rGcm1 cDNA clone (see above). For the mGcm2 probe template, a pair of primers (5′-ATGCGAATTCGCAAGAAGCACTCAGGAC-3′ and 5′-CTAGTCTAGAGTCCTCATTGTCAAAGCTAAAGGGC-3′) was used to PCR amplify a 927-base fragment corresponding to the 3′ end exon of mGcm2 from mouse genomic DNA. After EcoRI and XbaI digestion, the fragment was inserted into the pBluescript KS (+) plasmid. The template for the parathyroid hormone (PTH) gene probe (a kind gift from B. Lanske) has been described (12).
RT-PCR.
Total RNA preparations were extracted from ≈50 mg of B6D2F2 mouse embryonic tissues with RNAzol B (Leedo Medical Laboratories, Houston, TX) following the manufacturer’s protocol. Oligo(dT)-primed reverse transcription was performed on the total RNA preparations with the SuperScript Preamplification System (GIBCO/BRL) following the manufacturer’s protocol. First-strand cDNAs thus generated served as templates for PCR amplification. For the amplification of actin cDNA, a pair of primers (5′-CACACITTCTACAATGAGCTGCGTGT-3′ and 5′-GGTGAGGATCTTCATGAGGTAGTC-3′) was used for a single 30-cycle round of PCR. For mGcm1 and mGcm2, two successive 30-cycle rounds of PCR were run. For mGcm1, the first-round primers were 5′-GCACGAATTCAATGGAACTGGACGACTTTG-3′ and 5′-TAGCTGCTCAGATCCACAGA-3′, and the second-round primers were 5′-CTGCAATGGACCCCTGAAACTAATTCCC-3′ and 5′-CTGCTTCTAGCTTGGTCTCCGGCCTGGG-3′. For mGcm2, the first-round primers were 5′-ATGCGAATTCGCAGCCAGGAGAAGAAGG-3′ and 5′-CTAGTCTAGACAGGGCAGCTCTAGGTTG-3′, and the second-round primers were 5′-TGGGCCATGCGCAACACCAAC-3′ and 5′-GGGAAGCTGCTATCAGCAGTC-3′.
Drosophila Stocks.
The gcm null allele gcmΔP1 and UAS-gcm (upstream activating sequence-gcm) reporter lines have been described (2). The sca-Gal4 line was obtained from C. Klämbt (13).
Generation and Analysis of UAS-Gcm Transgenic Reporter Lines in Drosophila.
A 1.4-kb rGcm1 cDNA was subcloned as an EcoRI fragment into the EcoRI site of pUAST, a UAS reporter P-element vector (14). The mGcm2 cDNA (from p5G2–24) and the mGcm2-myc cDNA (from p5G2-myc) were first subcloned as SacI–XbaI fragments into pUC18, then reisolated as EcoRI–XbaI fragments, and subcloned into the EcoRI and XbaI sites of pUAST. Transgenic lines were generated by P-element-mediated transformation by standard procedures. Panneural expression was achieved by crossing these lines with sca-Gal4. Expression of UAS-gcm and UAS-rGcm1 in a gcm null background was achieved by first recombining second-chromosome inserts with a gcmΔP1 second chromosome to create the following stocks: w; P[w+ UAS-gcm]2 gcmΔP1/CyO and w; P[w+ UAS-rGcm1]2B gcmΔP1/CyO. These lines were crossed against w; gcmΔP1 sca-Gal4/CyO.
Immunohistochemical Detection of REPO Protein in Drosophila Embryos.
Recombinant REPO-fusion protein was produced in E. coli with the QIAexpress system (Qiagen, Chatsworth, CA). A 1.4-kb BamHI-HindIII fragment from the repo cDNA pcrepo-2.6 (15) encoding for amino acids 219–612 was cloned into the pQE-30 expression vector. The resulting fusion protein has a 22-aa N-terminal addition that contains six histidine residues, allowing for a one-step purification by immobilized Ni2+ chelate-affinity chromatography. Mice were injected with 50 μg of protein emulsified in RIBI adjuvant (Immunochem Research, Hamilton, MO) and were boosted at 2-week intervals. Horseradish peroxidase immunohistochemistry and embryo dissections were carried out as described (16). Anti-REPO antiserum was used at 1:1000 dilution, followed by horseradish-peroxidase-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch) at 1:300 dilution. The diaminobenzidine reaction was enhanced by the addition of 0.064% NiCl to give a black color. mGCM2-MYC protein was detected with anti-c-MYC mAb 1-9E10.2 (17) at 1:5 dilution followed by horseradish-peroxidase-conjugated goat anti-mouse secondary antibody.
RESULTS
Cloning of Rat Gcm1.
A database search with the sequence of Drosophila gcm gene led to the identification of a human expressed sequence tag clone (GenBank accession no. R62635) exhibiting a region of significant homology from a placental cDNA library. A pair of degenerate primers derived from amino acids conserved between the human and Drosophila genes was used to screen a subdivided rat E14.5 placental cDNA library by PCR amplification of the corresponding portion of the rat homolog. The PCR product was used subsequently to screen bacterial transformants of a positive pool leading to the isolation of rGcm1 cDNA clones. The longest clone had a 1.4-kb insert comprising 160 bases of 5′ untranslated region and an ORF encoding a predicted peptide of 423 amino acids. The C-terminal sequence was obtained from a 3′ rapid amplification of cDNA ends by using rat placenta as the source of cDNA. The complete ORF of rGcm1 encodes a 436-aa protein (Fig. 1A). As expected, the region corresponding to the DNA-binding domain, the gcm motif, shows a high degree of conservation with Drosophila GCM (60% identity at the amino acid level). Additionally, rGcm1 is homologous to mGcm1 (mGcm1: GenBank accession no. U59876; mGCMa: GenBank accession no. D88612; refs. 4 and 9) throughout the ORF (87% identity at the amino acid level; Fig. 1A).
Figure 1.
(A) Comparison of rGCM1 and mGCM1. The deduced amino acid sequence of rGCM1 is aligned with that of mGCM1 in the database (GenBank accession no. U59876). Conserved residues are boxed. The extent of the gcm motif is indicated by arrowheads. (B) Comparison of the strain-specific mGCM2 sequences. The sequence for 129SvEv GCM2 is based on the partial cDNA sequence proposed from the genomic DNA sequence. For C57BL/6, the sequence is based on a full-length cDNA clone. D88611 (GenBank accession no. D88611) represents the published BALB/c GCMb sequence. Missense alterations found in C57BL/6 and BALB/c sequences with respect to residues in 129SvEv are boxed. (C) Comparison of mGCM2 and human GCM2 (hGCM2; GenBank accession no. AA782779). Conserved residues are boxed.
Strain-Specific Sequence Variations Among Murine Gcm2 Genes.
Screening several cDNA libraries did not lead to the isolation of additional gcm homologs. However, the results from screening a mouse genomic DNA library indicated that at least one more member of the Gcm family existed. Partial sequencing of the genomic clones that hybridized to the aforementioned Gcm1 probe led to the identification of mGcm2. We obtained the sequence of over 5 kb of genomic DNA from overlapping phage clones, and from this composite sequence, generated a potential partial cDNA sequence for mGcm2 (Fig. 1B; 129SvEv). A comparison with the subsequently published murine cDNA sequence (mGCMb: GenBank accession no. D88611) confirmed that our proposed sequence accounts for the C-terminal 474 amino acids of mGCM2 (4).
We independently isolated a full-length mGcm2 cDNA from the C57BL/6 strain. Interestingly, the sequence of this cDNA clone showed variations from the sequence of the genomic DNA that originated from the 129SvEv strain (Fig. 1B). Specifically, 14 nucleotide differences, of which 7 lead to changes in amino acid residues, were discovered. All of these missense alterations are found outside the gcm motif. To determine whether these differences reflect strain-specific variations, RNA editing, or the existence of multiple Gcm2 alleles, we compared genomic mGcm2 sequences of the two mouse strains (129SvEv and C57BL/6) by using PCR amplification of genomic DNA with both intron- and exon-derived oligonucleotide primers. The data indicated that a single and distinct Gcm2 allele is present for each strain. We also noted that the sequence of the published Gcm2 cDNA isolated from BALB/c strain (GenBank accession no. D88611; ref. 4) is at variance with those of the two mGcm2 alleles we isolated (Fig. 1B). A genomic DNA sequence analysis of the BALB/c Gcm2 gene indicated that its sequence is in full agreement with that of 129SvEv. Furthermore, although all four differences (of which three lead to amino acid changes) between these genomic DNAs and the published BALB/c cDNA are A to G transitions, we found no evidence of RNA editing, as Gcm2 cDNAs isolated from both neural and pharyngeal tissues of BALB/c mice had sequences identical to the BALB/c genomic DNA sequence (data not shown). Although the simplest explanation is that these A to G transitions represent sequencing errors, we cannot exclude the possibility that the published BALB/c mGcm2 cDNA is derived from an mRNA that underwent RNA-editing in the specific tissue (male-adult brain) from which the mRNA was obtained.
Expression of Gcm1 and Gcm2.
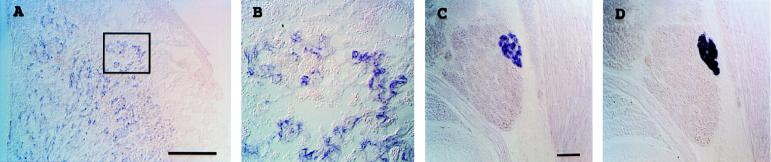
We used RNA in situ hybridization to examine the expression patterns of Gcm1 and Gcm2 in developing murine embryos. Gcm1 was detected in a subset of cells in the placenta (Fig. 2 A and B). The location of the positive cells within the placenta suggests that they are labyrinthine trophoblasts. No other tissue examined was positive for Gcm1 transcripts by in situ hybridization. The expression of Gcm2 also appeared to be highly restricted. Only parathyroid tissue was positive for Gcm2 (Fig. 2C). A comparison to PTH gene expression (Fig. 2D) on adjacent sections indicated that the cells expressing Gcm2 are PTH-secreting cells. The expression of Gcm2 in parathyroid tissue is consistent with the isolation of a human Gcm2 cDNA from an adult parathyroid adenoma (GenBank accession no. AA782779; Fig. 1C). A developmental time course analysis of Gcm2 expression showed that it is expressed as early as E10, preceding the expression of PTH (data not shown).
Figure 2.
(A and B) Expression of rGcm1 in rat E14.5 placenta. The section was hybridized with an antisense cDNA probe for rGcm1. A positive region in A is shown in enlarged form in B. (C) Sagittal section of the pharyngeal region of an E16.5 mouse showing mGcm2 expression in the parathyroid tissue. (D) The adjacent section was positive for parathyroid hormone gene transcript. (Scale bars: 500 μm for A and 100 μm for C.)
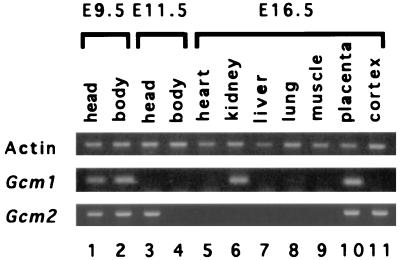
In addition to the placenta, Gcm1 has been reported to be expressed in the embryo proper during early stages of development (9). To examine the expression of Gcm1 in nonplacental embryonic tissues, we used RT-PCR to amplify Gcm1 transcripts. The Gcm1 mRNA was detected at E9.5 in both the head and body but was not detected at E11.5 (Fig. 3). Additionally, the Gcm1 message could be amplified from a E16.5 kidney cDNA preparation. With the exception of the placenta, the detection of the Gcm1 message by RT-PCR required extended cycles of amplification. Taken together with the fact that no transcripts were detected by in situ hybridization to embryonic tissue even at stages where signals were detectable by RT-PCR, the data suggest that the primary if not the exclusive site of embryonic Gcm1 expression is the placenta.
Figure 3.
RT-PCR analyses of the expression of Gcm genes. Agarose-gel electrophoresis of the PCR products is shown. Oligo(dT) primed cDNA derived from mouse tissues were amplified with gene-specific primers. Actin cDNA was amplified as the control product. Oligonucleotide primers that span exon–intron junctions were used so as to be able to distinguish spliced messages from genomic DNA contamination. In addition, for each of the PCRs, a control reaction that used a cDNA preparation without reverse transcription was performed and shown to generate no product (data not shown).
A similar RT-PCR analysis was performed for Gcm2. The message was detected in both head and body at E9.5 but was restricted to the head region by E11.5 (Fig. 3). At E16.5, we detected the message from the placenta and the cortex of the embryo. The detection of the Gcm2 message in embryonic neural tissues is consistent with the reported isolation of Gcm2 cDNA from an adult brain cDNA library (4). Nevertheless, the detection of Gcm2 in neural tissue also required extended cycles of amplification, suggesting that the levels of expression are extremely low in all tissues except the parathyroid gland.
Expression of rGcm1 in Drosophila.
It has been shown that the gcm gene in Drosophila controls the choice between glial and neuronal fates in most glial lineages (1–3). In Drosophila, the GCM protein is expressed transiently in all embryonic glia except the mesectodermally derived midline glia. GCM-positive glia are also characterized by the expression of the homeodomain protein, REPO (15, 18, 19). Transient expression of GCM in developing glial cells is followed by maintained expression of REPO (Fig. 4A). In gcm-homozygous null mutant embryos, nearly all presumptive glia fail to differentiate into glia, and virtually all REPO expression is eliminated (Fig. 4B). This phenotype is associated with an increase in markers normally expressed in differentiated neuronal cells, suggesting that glial-cell precursors have been transformed into neurons (1, 2). Conversely, when GCM is expressed ectopically in neural precursors, there is a dramatic increase in the number of REPO-expressing cells (1, 2) (Fig. 4C). These REPO-positive cells exhibit glial morphologies at the expense of neuronal markers and morphologies, suggesting that presumptive neurons have been transformed into glia (1).
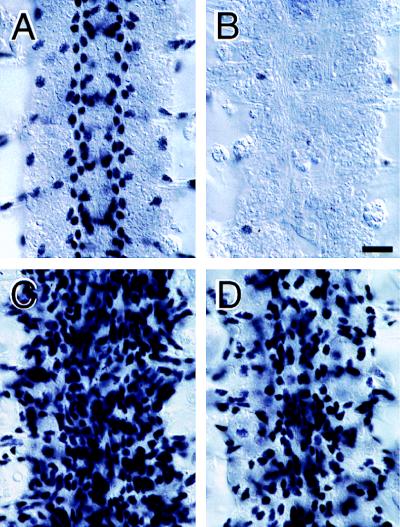
Figure 4.
Panneural expression of rGCM1 promotes glial-cell differentiation in Drosophila. Photomicrographs of the central nervous system (CNS) in stage 16 embryos showing four adjacent segmental neuromeres as stained with anti-REPO antisera. (A) Wild-type embryo. Anti-REPO stains the nuclei of all glial cells except midline glia. (B) gcmΔP1 loss-of-function mutant embryo. Virtually no cells express REPO. (C) Panneural expression of Drosophila GCM. In sca-Gal4/UAS-gcm; UAS-gcm/+ embryos (expressing two copies of UAS-gcm), panneural expression of GCM causes nearly all CNS cells to express REPO. (D) Panneural expression of rGCM1. In UAS-rGcm1/+; sca-Gal4/UAS-rGcm1 embryos (expressing two copies of UAS-rGcm1), panneural expression of rGCM1 also causes an increase in REPO expression in the CNS. Anterior is up. (Scale bar: 10 μm.)
Drosophila embryos that express rGcm1 and mGcm2 in neural precursors were generated with the GAL4-UAS system to test whether vertebrate homologs of gcm are conserved functionally (14). We constructed fusion genes that place rGcm1 and mGcm2 cDNAs under the control of a UAS, which allows them to be activated by the GAL4 transcriptional activator in specific tissues where Gal4 is expressed. Ectopic expression in the CNS was achieved by crossing these UAS reporter lines with a GAL4 effector line (sca-Gal4) that drives Gal4 expression panneurally (13).
Panneural expression of rGCM1 caused a dramatic increase in the number of REPO-positive cells (Fig. 4D) similar to the phenotype obtained with GCM (Fig. 4C). In embryos carrying two copies of UAS-rGcm1 and one copy of sca-Gal4, ≈70% of CNS cells per abdominal segment expressed REPO, compared with a wild-type level of 23% (compare Fig. 4D with Fig. 4A). Most of these ectopic REPO-positive cells display the elongated morphology typical of glial cells. This phenotype is not as penetrant as that obtained by ectopically expressing two copies of Drosophila GCM panneurally, which causes nearly all CNS cells to express REPO (Fig. 4C).
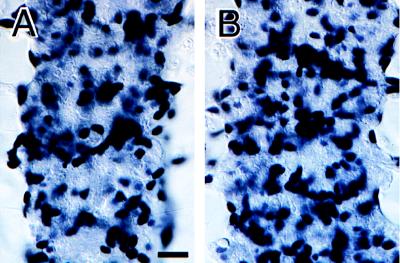
We were curious to find out whether the endogenous gcm locus is required for these transformations. In particular, rGCM1 might exert its effect on repo expression indirectly via activation of endogenous gcm. We therefore generated flies that carry one copy of UAS-rGcm1, together with a loss-of-function mutation in gcm on the second chromosome. These were crossed against flies that carry sca-Gal4 and a gcm loss-of-function mutation also on the second chromosome. Of the resulting F1 embryos, one-fourth should express rGCM1 panneurally in a gcm-homozygous null background. In the absence of endogenous GCM expression, rGCM1 was capable of inducing REPO expression (Fig. 5B; compare with gcm loss-of-function in Fig. 4B). Many of these REPO-positive cells display glial phenotypes, though they lack the patterning imposed by endogenous GCM expression. These embryos are almost indistinguishable from embryos in which one copy of native Drosophila gcm gene is expressed under the control of sca-Gal4 in a gcm-homozygous null background (Fig. 5A). Thus, the function of rGCM1 in flies is independent of the endogenous gcm gene.
Figure 5.
rGCM1 rescues glial differentiation in gcm loss-of-function mutant embryos. Photomicrographs of the CNS in stage 16 embryos showing four adjacent segmental neuromeres as stained with anti-REPO antisera. (A) Panneural expression of Drosophila GCM in a gcm loss-of-function mutant embryo (UAS-gcm gcmΔP1/gcmΔP1 sca-Gal4) promotes REPO expression and glial-cell development. (B) Panneural expression of rGCM1 in a gcm loss-of-function mutant embryo (UAS-rGcm1 gcmΔP1/gcmΔP1 sca-Gal4) also promotes REPO expression and glial-cell development. Anterior is up. (Scale bar: 10 μm.)
We were unable to detect any phenotypes associated with panneural expression of UAS-mGcm2. The embryos showed no ectopic REPO expression and developed into normal viable adults. To confirm that mGcm2 was properly translated, we engineered an MYC-epitope-tagged version of UAS-mGcm2 and assayed for its expression in embryos with anti-c-MYC mAb 1-9E10.2 (17). An 81-aa MYC-tag-encoding DNA fragment was cloned in frame with the 3′ end of the ORF of the mGcm2 cDNA, such that the entire mGcm2 ORF must be translated for the MYC epitope to be detected. Transgenic flies carrying UAS-mGcm2-myc were generated and were crossed to the sca-Gal4 activator line. Strong nuclear expression of MYC-tagged mGCM2 protein was detected in the nervous system of embryos generated from this cross. This expression confirms the translation of mGcm2 cDNA, but these embryos exhibit no neural phenotype (data not shown).
DISCUSSION
Molecular analyses of transcription factors involved in embryonic development have shown that homologs not only exist across the metazoa but are often expressed in analogous tissues and play comparable roles (7). In fact, searching for mammalian homologs of Drosophila genes has been a fruitful approach in discovering genes involved in specific developmental programs (8, 20, 21). Thus far, no mammalian gene has been shown to fulfill the role of a glial determination factor. Thus, it was of significant interest to isolate and characterize the mammalian homologs of Drosophila gcm, a transcription factor whose activity promotes glial over neuronal fates in multipotential neural precursors (1–3).
We isolated two mammalian genes with extended homology to gcm, Gcm1 and Gcm2. As in other conserved families of transcription factors, the homology is limited to the DNA-binding domain, the gcm motif. While these studies were in progress, similar sequences were reported by two other laboratories (4, 9). The sequences designated by Hotta et al. as GCMa and GCMb (4) correspond to our GCM1 and GCM2 sequences, respectively. However, our C57BL/6 Gcm2 cDNA-derived amino acid sequence differs from that of the BALB/c-derived Gcmb (GenBank accession no. D88611) sequence at 10 different positions. Our comparison of C57BL/6 and BALB/c genomic Gcm2 sequences showed that 7 of 10 of these differences represent strain-specific variations. The remaining three nucleotide differences that result in amino acid changes and one silent nucleotide difference likely represent sequencing errors, although the possibility of tissue-specific RNA editing cannot be excluded (see Results). The two published mouse Gcm1/a cDNA sequences (GenBank accession nos. U59876 and D88612) also differ from each other at four amino acids, though we have not investigated whether these represent strain-specific differences. All of the differences among Gcm1/a as well as the strain-specific differences among Gcm2/b are outside the gcm motif, suggesting that selection pressure is lower outside the DNA-binding domain.
Our studies on the expression pattern of the two genes by RNA in situ hybridization and RT-PCR indicate that the roles of these genes in mammals have diverged and diversified compared with their Drosophila homolog. Although Gcm2 is expressed weakly in embryonic neural tissues, both Gcm1 and Gcm2 are most highly expressed in specific nonneural tissues. For example, the expression of Gcm1 is highest in a subset of placental labyrinthine trophoblasts. Similarly, Gcm2 appears to be the first described transcription factor that is expressed specifically in the PTH-secreting cells of the developing parathyroid gland. Such specific expression of Gcm2 suggests that it plays an important role in the development of the parathyroid gland and possibly in the transcription of the PTH gene.
Why should the neural expression of Drosophila transcription factors such as achaete-scute and atonal be conserved in vertebrates, whereas the neural expression of gcm is not? Many terminal differentiation genes specifically expressed in neurons are conserved between Drosophila and mammals, including ion channels, synaptic vesicle proteins, adhesion molecules, and neurotransmitter-synthesizing enzymes. Strong evolutionary-selection pressure to conserve the mechanisms that regulate their expression in neurons would explain why the tissue specificity of neuronal transcription-factor gene expression is conserved in parallel with sequences of their DNA-binding domains. By contrast, none of the terminal differentiation genes expressed in vertebrate glia, such as myelin proteins or glial fibrillary acidic protein, appear to have counterparts in Drosophila. This lack of molecular conservation of terminal differentiation genes may indicate that Drosophila glia are functionally much more diverged from mammalian glia than fly neurons are from mammalian neurons. In that case, there would be little selection pressure to maintain tissue specificity of expression for a fly glial determination gene. Rather, the gcm motif would be conserved as a mechanism to control tissue-specific gene expression, but would be evolutionarily coopted by nonglial tissues.
These arguments not withstanding, it should be noted that the data presented here do not exclude the possibility that Gcm1 and Gcm2 do have some function in neurogenesis, because both appear to be expressed in embryonic neural tissues during early embryonic stages, albeit at levels undetectable by in situ hybridization. Loss of function analyses of Gcm1 and Gcm2 may be required to establish the significance of their low-level expression in neural tissues. It is also possible that there are additional members of the mammalian Gcm family, some of which may be more closely related in function to Drosophila gcm than Gcm1 or Gcm2.
Ectopic expression of homologous vertebrate transcription factors in flies can lead to phenotypes similar to those observed in gain-of-function mutations or can result in rescue of loss-of-function phenotypes (22–25). Such results suggest a conservation of functional properties at the biochemical level among the homologs in question. The fact that rGcm1 can substitute partially for gcm when expressed in Drosophila suggests that key properties of rGCM1 as a transcription factor are conserved with respect to those of GCM. Thus, we can safely conclude that rGCM1 is a transcriptional activator with a DNA-binding specificity similar to that of GCM. In fact, recently it has been shown that mGCM1 and Drosophila GCM bind to the same DNA elements (26). Surprisingly, mGcm2, whose degree of homology to gcm in the DNA-binding domain is similar to that of rGcm1, failed to substitute for gcm. Several possibilities may explain this result: (i) GCM2 may have a binding site distinct from Drosophila GCM and GCM1; (ii) GCM2 may not be a transcriptional activator but a transcriptional repressor; or (iii) Drosophila GCM and GCM1 may use a common cofactor that GCM2 does not. Further analyses, including the identification of GCM2 binding sites and domain swapping between GCM1 and GCM2, should help to distinguish amongst these possibilities.
Acknowledgments
We thank Beate Lanske for the PTH probe, Craig Montell for the repo cDNA (pcrepo-2.6), Christian Klämbt for sca-Gal4 line, Sherry Perez and Joel Pomerantz for their comments on the manuscript, and Janet Rossant and Jay Cross for helpful discussions. This work was supported by fellowships from the National Institutes of Health (to J.K.), from the American Cancer Society (to B.W.J.), from the Howard Hughes Medical Institute (to B.W.J. and Z.C.), and from the Human Frontiers Science Program (to C.Z.). D.J.A. and C.S.G. are Investigators of the Howard Hughes Medical Institute.
ABBREVIATIONS
- CNS
central nervous system
- En
embryonic day n
- GCM
glial cells missing
- mGCM
mouse GCM
- PTH
parathyroid hormone
- REPO
reverse polarity
- rGCM
rat GCM
- RT-PCR
reverse transcription–PCR
- UAS
upstream activating sequence
Footnotes
References
- 1. Hosoya T, Takizawa K, Nitta K, Hotta Y. Cell. 1995;82:1025–1036. doi: 10.1016/0092-8674(95)90281-3. [DOI] [PubMed] [Google Scholar]
- 2.Jones B W, Fetter R D, Tear G, Goodman C S. Cell. 1995;82:1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- 3.Vincent S, Vonesch J, Giangrande A. Development (Cambridge, UK) 1996;122:131–139. doi: 10.1242/dev.122.1.131. [DOI] [PubMed] [Google Scholar]
- 4.Akiyama Y, Hosoya T, Poole A M. Proc Natl Acad Sci USA. 1996;93:14912–14916. doi: 10.1073/pnas.93.25.14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schreiber J, Sock E, Wegner M. Proc Natl Acad Sci USA. 1997;94:4739–4744. doi: 10.1073/pnas.94.9.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arendt D, Nubler-Jung K. BioEssays. 1996;18:255–259. doi: 10.1002/bies.950180314. [DOI] [PubMed] [Google Scholar]
- 7.Duboule D, Wilkins A D. Trends Genet. 1998;14:54–59. doi: 10.1016/s0168-9525(97)01358-9. [DOI] [PubMed] [Google Scholar]
- 8.Lee J E. Curr Opin Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- 9.Altshuller Y, Copeland N G, Gilbert D J, Jenkins N A, Frohman M A. FEBS Lett. 1996;393:201–204. doi: 10.1016/0014-5793(96)00890-3. [DOI] [PubMed] [Google Scholar]
- 10.Turner D L, Weintraub H. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 11.Birren S J, Lo L, Anderson D J. Development (Cambridge, UK) 1993;119:597–610. doi: 10.1242/dev.119.3.597. [DOI] [PubMed] [Google Scholar]
- 12.Li Y C, Pirro A E, Amling M, Delling G, Baron R, Bronson R, Demay M B. Proc Natl Acad Sci USA. 1997;94:9831–9835. doi: 10.1073/pnas.94.18.9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klaes A, Menne T, Stollewerk A, Scholz H, Klambt C. Cell. 1994;78:149–160. doi: 10.1016/0092-8674(94)90581-9. [DOI] [PubMed] [Google Scholar]
- 14.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 15.Xiong W, Okano H, Patel N, Blendy J, Montell C. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- 16.Lin D M, Goodman C S. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 17.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell G, Goring H, Lin T, Spana E, Andersson S, Doe C, Tomlinson A. Development (Cambridge, UK) 1994;120:2957–2966. doi: 10.1242/dev.120.10.2957. [DOI] [PubMed] [Google Scholar]
- 19.Halter D A, Urban J, Rickert C, Ner S, Ito K, Travers A, Technau G. Development (Cambridge, UK) 1995;121:317–332. doi: 10.1242/dev.121.2.317. [DOI] [PubMed] [Google Scholar]
- 20.Johnson J E, Birren S J, Anderson D J. Nature (London) 1990;346:858–861. doi: 10.1038/346858a0. [DOI] [PubMed] [Google Scholar]
- 21.Ma Q, Kintner C, Anderson D J. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 22.Nagao T, Leuzinger S, Acampora D, Simeone A, Finkelstein R, Reichert H, Furukubo-Tokunaga K. Proc Natl Acad Sci USA. 1998;95:3737–3742. doi: 10.1073/pnas.95.7.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malicki J, Schughart K, McGinnis W. Cell. 1990;63:961–967. doi: 10.1016/0092-8674(90)90499-5. [DOI] [PubMed] [Google Scholar]
- 24.McGinnis N, Kuziora M A, McGinnis W. Cell. 1990;63:969–976. doi: 10.1016/0092-8674(90)90500-e. [DOI] [PubMed] [Google Scholar]
- 25.Lutz B, Lu H C, Eichele G, Miller D, Kaufman T C. Genes Dev. 1996;10:176–184. doi: 10.1101/gad.10.2.176. [DOI] [PubMed] [Google Scholar]
- 26.Schreiber J, Enderich J, Wegner M. Nucleic Acids Res. 1998;26:2337–2343. doi: 10.1093/nar/26.10.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]