Abstract
Heterogeneous nuclear ribonucleoprotein (hnRNP) A1 has previously been shown to bind mouse hepatitis virus (MHV) RNA at the 3′ end of both plus and minus strands and modulate MHV RNA synthesis. However, a mouse erythroleukemia cell line, CB3, does not express hnRNP A1 but still supports MHV replication, suggesting that alternative proteins can replace hnRNP A1 in cellular functions and viral infection. In this study, we set out to identify these proteins. UV cross-linking experiments revealed that several CB3 cellular proteins similar in size to hnRNP A1 interacted with the MHV RNA. These proteins were purified by RNA affinity column with biotinylated negative-strand MHV leader RNA and identified by mass spectrometry to be hnRNP A2/B1, hnRNP A/B, and hnRNP A3, all of which belong to the type A/B hnRNPs. All of these proteins contain amino acid sequences with strong similarity to the RNA-binding domains of hnRNP A1. Some of these hnRNPs have previously been shown to replace hnRNP A1 in regulating RNA splicing. These proteins displayed MHV RNA-binding affinity and specificity similar to those of hnRNP A1. hnRNP A2/B1, which is predominantly localized to the nucleus and shuttles between the nucleus and the cytoplasm, was shown to relocalize to the cytoplasm in MHV-infected CB3 cells. Furthermore, overexpression of hnRNP A/B in cells enhanced MHV RNA synthesis. Our findings demonstrate that the functions of hnRNP A1 in MHV RNA synthesis can be replaced by other closely related hnRNPs, further supporting the roles of cellular proteins in MHV RNA synthesis.
Mouse hepatitis virus (MHV) belongs to the Coronaviridae family of positive-sense, single-stranded RNA viruses. The replication of the viral RNA genome and transcription of viral mRNAs occur exclusively in the cytoplasm of infected cells. The viral RNA polymerase first replicates the viral genomic RNA into negative-strand RNA, which is then used as the template to replicate genomic RNA and transcribe subgenomic mRNAs, all of which contain an identical 5′ nontranslated leader sequence of 72 to 77 nucleotides (18). RNA replication and transcription are regulated by several viral RNA elements, including the 5′- and 3′-end untranslated regions, leader RNA (22, 41), and intergenic sequences (24). These regulatory RNA elements bind several cellular proteins, including heterogeneous nuclear ribonucleoprotein (hnRNP) A1 and polypyrimidine tract-binding protein (PTB), also known as hnRNP I (10, 12, 13, 20, 21, 38, 39).
There is considerable biochemical evidence suggesting possible interactions between the various RNA regulatory elements through these cellular and, possibly, viral proteins. Indeed, hnRNP A1 mediates the formation of a ribonucleoprotein complex containing the MHV negative-strand leader and intergenic sequences (40). Both hnRNP A1 and PTB have been shown to be involved in MHV RNA replication and transcription (20, 21, 42). hnRNP A1 is relocated from the nucleus to the cytoplasm in MHV-infected cells (21). Furthermore, the overexpression of a C-terminally truncated hnRNP A1, which was localized predominantly in the cytoplasm, caused strong dominant-negative inhibitory effects on viral genomic RNA replication and subgenomic mRNA transcription (34). In contrast, overexpression of wild-type hnRNP A1 accelerated the synthesis of all viral RNAs, suggesting that an intact hnRNP A1 is important for regulating viral RNA synthesis (34).
hnRNPs constitute a large group of RNA-binding proteins which are classified into several families and subfamilies based on shared structural and functional motifs (9, 16). The most abundant hnRNPs are those belonging to the A/B type, with a molecular mass around 40 kDa, such as A1/A1B, A2/B1, B2, and A3. This family consists of a large number of isoforms with extensive posttranslational modifications. These proteins have a common molecular structure, with a highly conserved N-terminal domain but a divergent glycine-rich auxiliary domain at the C terminus. The N-terminal part consists of two tandemly repeated RNA-binding domains (RBD), which are directly involved in RNA binding. The C-terminal glycine-rich domain contributes to the cooperative binding of the proteins to RNA (8, 29), protein-protein interactions (7), and modulation of RNA conformation (3). Due to their significant structural similarities, these proteins are typically referred to as 2×RBD-Gly proteins (4, 5). They have been shown to participate in a variety of cellular functions, including mRNA splicing, trafficking, and turnover (9, 16, 23). hnRNP A1 and A2/B1 are known to shuttle between the nucleus and the cytoplasm, mediated by the M9 sequence located near the C terminus of the proteins (26, 30, 35, 36). A mouse hnRNP A/B protein, also known as CA-rich G box-binding factor A (1, 15, 27), and its rat homologues (19) can also regulate gene transcription.
Our previous studies have shown that hnRNP A1 is involved in MHV RNA replication in the mouse astrocytoma cell line DBT (34). However, a more recent report showed that mouse erythroleukemia cell line CB3, which is devoid of hnRNP A1, could nevertheless support MHV RNA replication (33). It was thus claimed that hnRNP A1 was not involved in MHV RNA synthesis (33). However, the very fact that hnRNP A1 deletion is not lethal to CB3 cells does not invalidate the important role of hnRNP A1 in cellular functions. Rather, it is likely that alternative factors can replace hnRNP A1 in this cell line. Indeed, several hnRNP A1-related molecules can replace hnRNP A1 in regulating alternative splicing of cellular pre-mRNAs (25) and human immunodeficiency virus pre-mRNA splicing (6). It is very likely that this is also true for MHV RNA synthesis in CB3 cells.
In this study, we identified cellular proteins that specifically interact with MHV RNA in CB3 cells. These proteins were identified as proteins of the hnRNP type A/B family, including hnRNP A/B, hnRNP A2/B1, and hnRNP A3, all of which are highly related to hnRNP A1. Biochemical and functional studies showed that they interact with MHV RNA and function in its replication much as hnRNP A1 does. Our results provide further evidence that the hnRNP type A/B proteins are important in MHV RNA synthesis.
MATERIALS AND METHODS
Cells and viruses.
CB3 cells, a mouse erythroleukemia cell line (2), were grown in suspension culture in Eagle's minimal essential alpha medium supplemented with 10% fetal bovine serum. DBT cells were cultured in Eagle's minimal essential medium supplemented with 10% newborn calf serum and 10% tryptose phosphate buffer. MHV strain A59 (31) was propagated in DBT cells and maintained in virus growth medium containing 1% newborn calf serum.
RNA affinity column purification.
CB3 cells were lysed in an equal volume (to the cell pellet) of buffer A (10 mM HEPES [pH 7.8], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 20% glycerol) without detergent on ice for 20 min and passed through a 25-gauge needle 20 times. The lysate was centrifuged at 1,000 × g for 20 min at 4°C, and the supernatant (cytosolic fraction) and pellet (crude nuclear fraction) were collected in separate tubes. The crude nuclear pellet was resuspended in an equal volume (to the nuclear pellet) of buffer C (20 mM HEPES [pH 7.8], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 20% glycerol) and incubated on ice for 30 min. Nuclear debris was removed by centrifugation at 4,000 × g for 20 min at 4°C. The supernatant (nuclear fraction) was mixed with the cytosolic fraction to constitute the CB3 whole-cell lysates.
The minus-strand MHV leader RNA was transcribed in vitro from plasmid pNX1-182 (10) by T3 polymerase in the presence of biotin-UTP with the T3 MegaScript system (Ambion, Austin, Tex.). The biotin-labeled RNA was purified by lithium chloride precipitation according to the manufacturer's instructions. The RNA affinity column was prepared by incubating an excess amount of the biotin-labeled RNA with streptavidin beads (Pierce, Rockford, Ill.) in binding buffer (5 mM HEPES [pH 7.5], 25 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 3.8% glycerol, 2 mM dithiothreitol) at 4°C overnight. The beads were blocked with 1% bovine serum albumin in binding buffer at 30°C for 30 min and washed five times with binding buffer before use.
CB3 total cell lysates were incubated with the RNA affinity beads in binding buffer at 30°C for 30 min. The beads were then washed five times with binding buffer and boiled in sodium dodecyl sulfate (SDS) sample buffer before loading onto a 10% polyacrylamide gel containing SDS. After electrophoresis, the gel was stained with Coomassie blue; individual bands were excised from the gel and submitted to the W. M. Keck Facility at Yale University, New Haven, Conn., for protein identification by mass spectrometry.
Plasmid construction.
The cDNA of the murine hnRNP A/B prepared by reverse transcription-PCR of RNA extracted from CB3 cells was cloned into pGEX-4T-1 (Amersham Biosciences, Piscataway, N.J.) for the bacterial expression of glutathione S-transferase (GST)-hnRNP A/B fusion proteins. For expression in mammalian cells, hnRNP A/B was Flag tagged at the N terminus by PCR and cloned into pcDNA3.1 (Invitrogen, Carlsbad, Calif.).
UV cross-linking assay.
The UV cross-linking assay was performed as previously described (13). In brief, CB3 or DBT cell extracts (30 μg of protein) or recombinant GST-hnRNP A/B fusion proteins (10 ng), 200 μg of tRNA per ml, and 106 cpm of an in vitro-transcribed, 32P-labeled MHV RNA were incubated for 10 min at 30°C. The reaction mixture was placed on ice and UV irradiated in a UV Stratalinker 2400 (Stratagene, La Jolla, Calif.) for 10 min, followed by digestion with 400 μg of RNase A per ml for 15 min at 37°C. The protein-RNA complexes were then separated on an SDS-10% polyacrylamide gel and visualized by autoradiography.
Antibodies.
The monoclonal antibodies against hnRNP A1 and A2/B1 were generous gifts of G. Dreyfuss, University of Pennsylvania. The chicken polyclonal antibody against hnRNP A1 was produced by Aves Labs, Inc. (Tigard, Oreg.) by immunizing chickens with the purified mouse hnRNP A1 protein expressed in Escherichia coli (21). The polyclonal anti-Flag antibody was purchased from Affinity Bioreagents (Golden, Colo.). The polyclonal antibody against green fluorescent protein (GFP) was obtained from Chemicon (Temecula, Calif.).
Immunofluorescence staining.
Cells were washed in phosphate-buffered saline and fixed in 4% formaldehyde for 20 min at room temperature, followed by 5 min in acetone at −20°C. Primary antibodies were diluted in 5% bovine serum albumin and incubated with cells for 1 h at room temperature. After three washes in phosphate-buffered saline, fluorescein-conjugated secondary antibodies were added to cells at 1:200 dilution for 1 h at room temperature. Fluorescein isothiocyanate- or tetramethyl rhodamine isothiocyanate-conjugated secondary antibodies were used to generate green and red fluorescence, respectively. Cells were then washed in phosphate-buffered saline and mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, Calif.).
Kinetics of MHV RNA synthesis in hnRNP A/B-transfected cells.
Flag-hnRNP A/B or GFP was cotransfected with the plasmid encoding the MHV receptor into HEK-293 cells. At 24 h posttransfection, cells were infected with MHV A59 at a multiplicity of infection of 1 (at 37°C for 1 h), washed with serum-free medium twice, and incubated in medium containing 1% fetal calf serum. To label newly synthesized viral RNA, actinomycin D (5 μg/ml) was added 1 h in advance and 50 μCi of [3H]uridine per ml (NEN, Boston, Mass.) was added to the medium at specific time points for 1 h. The labeled cells were then collected, washed twice with cold phosphate-buffered saline, and lysed in NTE buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.5], and 1 mM EDTA) containing 0.5% NP-40, 0.5 mM dithiothreitol, and 40 U of RNasin per ml on ice for 20 min. After centrifugation, cytoplasmic extracts were spotted on 3MM paper. The paper was washed with 10% trichloroacetic acid three times and 100% ethanol once; the remaining radioactivity was counted in a scintillation counter.
RESULTS
Presence of hnRNP A1-related proteins in CB3 cells.
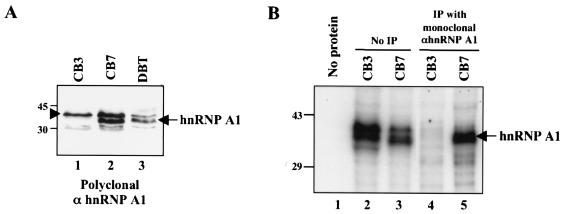
To identify the cellular proteins that can potentially participate in MHV RNA synthesis in CB3 cells, we first examined the expression of hnRNP A1 in CB3 cells and compared it with that in its parental cell line, CB7, and DBT cells. The polyclonal antibody against hnRNP A1 detected a major band of 35 kDa (arrow in Fig. 1A) and several proteins of slightly higher and lower molecular masses in both CB7 and DBT cells. This major band has previously been identified as hnRNP A1 by using a monoclonal antibody against hnRNP A1 (21). The remaining proteins are likely proteins related to hnRNP A1. The hnRNP A1 band was absent from CB3 cells. Northern blot analysis with an hnRNP A1-specific probe also failed to detect any hnRNP A1 transcript in CB3 cells (data not shown). Therefore, we conclude that CB3 cells are indeed devoid of hnRNP A1. However, the remaining protein bands that were detected in CB7 and DBT cells were also present in CB3 cells, suggesting that hnRNP A1-like proteins are likely present in CB3 cells.
FIG. 1.
Presence of hnRNP A1-related proteins in CB3 cells. (A) Immunoblotting of CB3, CB7, and DBT cell lysates with chicken polyclonal antibody against hnRNP A1 (21). The arrow indicates hnRNP A1 detected in CB7 and DBT cells but not in CB3 cells; the arrowhead indicates the protein band present in all three cell lines. (B) UV cross-linking studies of CB3 and CB7 cell lysates with MHV minus-strand leader RNA (lanes 2 and 3). The cross-linked proteins were immunoprecipitated (IP) with a monoclonal antibody against hnRNP A1 (lanes 4 and 5). Lane 1 contained RNA only. Sizes are shown in kilodaltons.
We then performed UV cross-linking assays to determine whether any cellular proteins may interact with MHV RNA in CB3 or CB7 cells. At least two cellular proteins in CB7 cells were found to bind to the MHV minus-strand leader RNA (Fig. 1B, lane 3); one of these proteins was precipitated with the monoclonal antibody against hnRNP A1 (Fig. 1B, lane 5). Interestingly, in CB3 cells, at least three proteins comparable to hnRNP A1 in size also cross-linked to MHV minus-strand leader RNA (Fig. 1B, lane 2); however, none of them could be precipitated with the monoclonal antibody against hnRNP A1 (Fig. 1B, lane 4). Since the polyclonal antibody against hnRNP A1 did not work in immunoprecipitation (data not shown), we could not determine unequivocally whether these proteins were related to hnRNP A1. Nevertheless, this result indicates that multiple cellular proteins with molecular masses similar to that of hnRNP A1 can interact with MHV minus-strand leader RNA in CB3 cells.
Purification and identification of MHV RNA-binding proteins in CB3 cells.
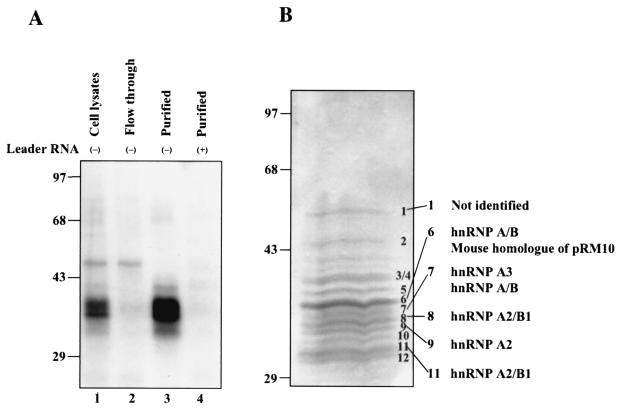
To identify the cellular proteins that interact with MHV RNA in CB3 cells, we performed RNA affinity column purification with MHV minus-strand leader RNA as a probe. By the one-step purification procedure (see Materials and Methods), almost all of the MHV RNA-binding proteins were removed from the CB3 cell lysates (flowthrough fraction, Fig. 2A, lane 2). The proteins recovered from the affinity-purified fractions interacted with minus-strand leader RNA, but not plus-strand MHV leader RNA (Fig. 2A, lanes 3 and 4). It has previously been shown that some cellular proteins such as PTB bind to MHV plus-strand leader RNA (13, 20); thus, the lack of any cellular protein binding to MHV plus-strand RNA in the purified fractions indicates that our purification procedure specifically purified the binding proteins of the MHV minus-strand leader.
FIG. 2.
Affinity purification of MHV minus-strand leader-RNA-binding proteins from CB3 cells. (A) UV cross-linking studies of the purified proteins. CB3 whole-cell lysates (lane 1), affinity-column flowthrough (lane 2) and affinity-purified proteins (lane 3) were cross-linked to MHV minus-strand leader RNA. The purified proteins were also cross-linked to MHV plus-strand leader RNA (lane 4). (B) Detection of MHV minus-strand leader RNA-binding proteins. The affinity-purified proteins were separated on a 12.5% polyacrylamide gel containing SDS and stained with Coomassie blue. Individual bands were excised from the gel and analyzed by mass spectrometry for protein identification. The identities of these proteins are indicated. Sizes are shown in kilodaltons.
The affinity-purified fractions were separated by polyacrylamide gel electrophoresis and stained with Coomassie blue. Approximately 13 proteins, ranging in molecular mass from 30 to 50 kDa, were detected. Six of the major bands were selected for further identification by mass spectrometry. Remarkably, except for band 1, which could not be identified, all of the proteins were hnRNPs specifically belonging to the hnRNP type A/B family (Fig. 2B). Band 6 also contained a possible mouse homologue of the rat nucleic acid-binding factor pRM10, whose function is unknown (GenBank accession no. AF108653). Figure 3 shows an amino acid sequence alignment of these proteins. It is evident that these proteins show extensive sequence similarity, particularly in the N-terminal half, where the RNA-binding domains reside (9). The C-terminal half, which contains the protein-interacting domain, is more divergent but retains the Gly-rich motif (9). hnRNPs A1 and A2 have the highest degree of sequence identity. Most of these proteins are known to be involved in cellular RNA metabolism (Table 1). These proteins likely have very similar RNA-binding properties and similar functions. We conclude that multiple hnRNP A1-related proteins bind to MHV minus-strand leader RNA in CB3 cells. These proteins may perform functions in MHV RNA synthesis in CB3 cells similar to those of hnRNP A1 in DBT cells.
FIG. 3.
Amino acid sequence alignment of the hnRNP type A/B proteins. Conserved amino acids are highlighted in shaded boxes. The M9 sequence present at the C terminus of hnRNP A1 is underlined.
TABLE 1.
hnRNP A/B proteins that interact with MHV minus-strand leader RNA
| MHV RNA-binding protein | Size (kDa) | Putative function | Reference(s) |
|---|---|---|---|
| hnRNP A1 | 34/36 | RNA processing and transport | 9, 11 |
| Mouse homologue of rat pRM10 | ∼34 | Unknown | GenBank accession no. AF108653 |
| hnRNP A/B | 38/40 | Transcription regulation | 1, 15, 19, 27 |
| hnRNP A2/B1 | 38 | RNA processing and transport | 14, 25, 28, 32 |
| hnRNP A3 | 40 | RNA transport | 23 |
In vitro interaction of hnRNP A/B proteins with MHV RNA.
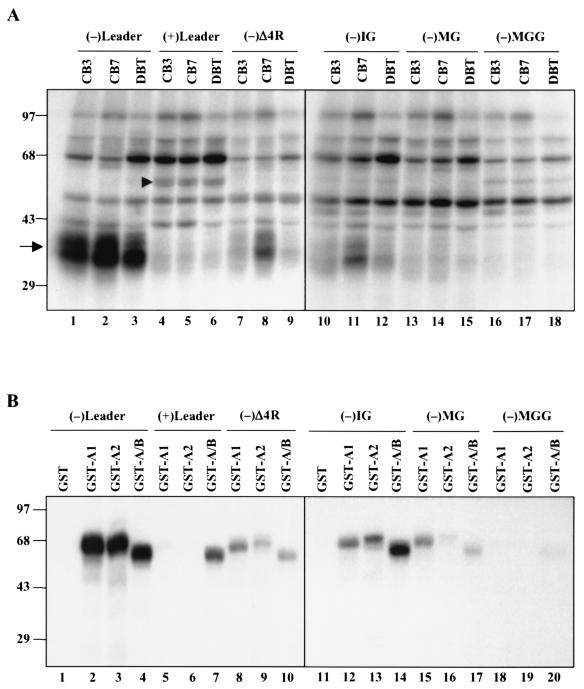
To establish that the hnRNP A1-related proteins in CB3 cells have properties similar to those of hnRNP A1 in DBT cells, we first performed UV cross-linking studies of MHV RNA with lysates of these cell lines. The whole-cell lysates of CB3, CB7, and DBT cells had very similar binding properties; a group of proteins of approximately 35 to 40 kDa bound to the minus-strand leader RNA but not to the plus-strand leader RNA (Fig. 4A). These proteins bound very poorly to minus-strand leader RNA that lacked the UCUAA repeats, (−)Δ4R (38). Furthermore, these proteins bound weakly to the negative-strand intergenic sequence (containing consensus sequence UCUAAAC) but not to the same sequences containing one or two nucleotide mutations (negative-strand MG containing UCGAAAC and negative-strand MGG containing GCUAAAG) (38). These binding properties are very similar to those reported previously for DBT cell extracts and for purified hnRNP A1 (21, 38). The CB7 lysates bound slightly more strongly to some RNA species than either the CB3 or DBT lysates did; the reason is not clear. It is noteworthy that plus-strand leader RNA bound an additional protein of approximately 55 kDa, which is likely PTB (lanes 4 to 6, Fig. 4A) (20), indicating the specificity of the UV cross-linking assay.
FIG. 4.
UV cross-linking of cell lysates (A) and recombinant GST-hnRNP type A/B proteins (B) with various MHV RNAs. Six different RNA species, i.e., MHV minus-strand leader [(−)Leader] and its mutant lacking the four UCUAA repeats [(−)Δ4R], plus-strand leader [(+)Leader], and the minus-strand intergenic region [(−)IG] and two of its mutants [(−)MG and (−)MGG], were UV cross-linked with 30 μg of whole-cell extract (A) or 10 ng of E. coli-expressed GST-hnRNP A/B proteins (B). The arrow in panel A identifies hnRNP A1-like proteins. The arrowhead identifies PTB. Sizes are shown in kilodaltons.
We then chose two of the hnRNP A/B molecules, hnRNP A2 and hnRNP A/B, for further characterization. They were fused to GST, and the purified GST fusion proteins were used for UV cross-linking studies with various MHV RNA species (Fig. 4B). GST-hnRNP A2 and GST-hnRNP A/B showed the same patterns of binding to MHV RNA as those of GST-hnRNP A1 and whole-cell lysates. They bound strongly to the wild-type negative-strand leader RNA but not the plus-strand leader RNA; deletion of the UCUAA repeats [(−)Δ4R] reduced but did not completely abolish the binding. Similarly, they bound strongly to negative-strand IG sequences, but only weakly to the mutant negative-strand IGs with one or two nucleotide mutations [(−)MG and (−)MGG]. hnRNP A/B also bound to plus-strand leader RNA; thus, it had a slightly lower specificity. Overall, our results showed that these hnRNP A1-related proteins have RNA-binding properties similar to those of hnRNP A1.
Relocalization of hnRNP A/B proteins during MHV infection.
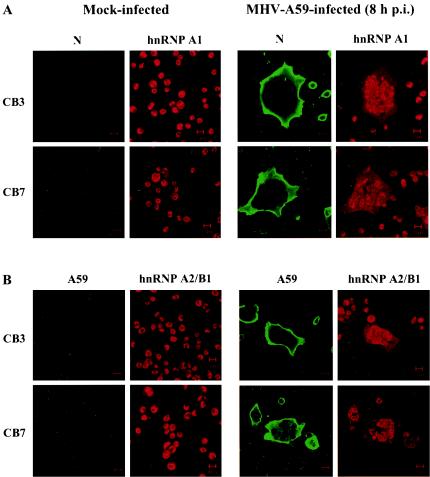
MHV RNA synthesis occurs exclusively in the cytoplasm of virus-infected cells. In order for hnRNP proteins to participate directly in viral RNA synthesis, they have to be recruited to the site of RNA synthesis. We have previously demonstrated that hnRNP A1 relocates from the nucleus to the cytoplasm of MHV-infected cells (21, 34). To determine whether the other type A/B hnRNPs participate in MHV RNA synthesis, we performed immunostaining of CB3 cells after MHV infection, with either a polyclonal anti-hnRNP A1 antibody or a monoclonal anti-hnRNP A2/B1 antibody to localize hnRNP type A/B proteins. Since CB3 cells do not express hnRNP A1 and the polyclonal antibody detected hnRNP A1-related proteins (Fig. 1A), the signal detected by the polyclonal hnRNP A1 antibody in CB3 cells most likely represents other members of the hnRNP A/B family.
In both CB3 and CB7 cells, a significant increase in the cytoplasmic levels of hnRNP type A/B proteins and a corresponding decrease in their nuclear levels were observed in virus-infected cell syncytia (Fig. 5). There was virtually no difference in staining between the CB3 and CB7 cells. Double staining with the monoclonal antibody against the MHV N protein and the polyclonal antibody against the whole MHV A59 virion showed that these cells also expressed the viral structural proteins in the cytoplasm. By comparison, in the uninfected cells, which did not have viral protein staining, the hnRNP type A/B proteins were predominantly localized to the nucleus. We have previously shown that, under similar conditions, other nuclear proteins, including Sam68 and RCC1, stayed in the nucleus of infected cells (21).
FIG. 5.
Relocalization of the hnRNP type A/B proteins in MHV-infected CB3 and CB7 cells. CB3 and CB7 cells were infected with MHV A59 at a multiplicity of infection of 2. The cells were fixed at 8 h postinfection (p.i.) and double stained with a monoclonal antibody against the viral N protein and a chicken polyclonal anti-hnRNP A1 antibody (A) or with a monoclonal antibody against hnRNP A2/B1 and a rabbit polyclonal anti-MHV A59 antibody (B).
Role of hnRNP A1-related proteins in MHV RNA synthesis.
To establish that the hnRNP A1-related proteins identified in this study play a role in regulating MHV RNA synthesis, we attempted to overexpress these proteins in mammalian cells and then examine their effects on MHV RNA synthesis. We first attempted to establish stable CB3 cell lines overexpressing Flag-tagged hnRNP A/B. However, the amounts of the protein expressed in these stable cell lines were not large enough for our purpose. Therefore, we performed transient transfection of Flag-tagged hnRNP A/B together with a plasmid expressing the MHV receptor (37) into HEK-293 cells. As a control, parallel cell cultures were transfected with a comparable vector expressing GFP and the MHV receptor.
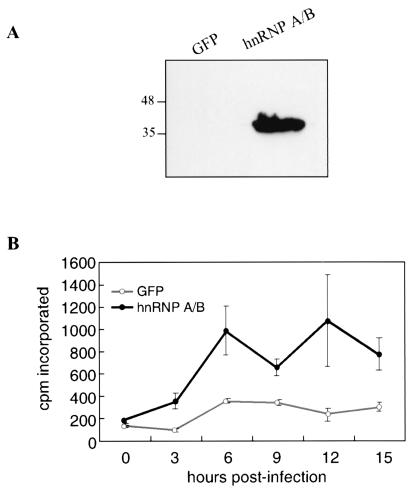
Immunoblotting studies with an anti-Flag antibody showed that hnRNP A/B was well expressed in the transfected HEK-293 cells (Fig. 6A). At 24 h posttransfection, cells were infected with MHV A59 and, at various time points after infection, labeled with [3H]uridine for 1 h in the presence of actinomycin D to determine the rate of viral RNA synthesis. The results showed that at every time point examined, [3H]uridine incorporation in cells expressing hnRNP A/B was much higher than that in the control cells expressing GFP (Fig. 6B). Previously, we have shown that GFP does not affect the rate of MHV RNA synthesis (data not shown). These results suggest that the overexpression of hnRNP A/B has a stimulatory effect on MHV RNA synthesis. Therefore, hnRNP A/B likely participates directly in MHV RNA synthesis, similar to hnRNP A1, as reported previously (34).
FIG. 6.
(A) Expression of hnRNP A/B in HEK-293 cells. Cells were transfected with plasmids expressing the MHV receptor and either GFP or hnRNP A/B. At 24 h posttransfection, cells were harvested, and immunoblotting with the anti-Flag antibody was performed. Sizes are shown in kilodaltons. (B) Kinetics of viral RNA synthesis. Cells transfected as for panel A were infected with MHV A59 at 24 h posttransfection at a multiplicity of infection of 1. Cells were then labeled with [3H]uridine for 1 h at various time points in the presence of actinomycin D. The trichloroacetic acid-precipitable counts were then determined.
DISCUSSION
Our findings presented here showed that in the absence of hnRNP A1, MHV minus-strand leader RNA interacts with several alternative cellular proteins. Remarkably, almost all of these proteins are related to hnRNP A1 and belong to the hnRNP A/B family. These results further suggest that the previously reported interaction of hnRNP A1 with MHV RNA (21) is very specific. All of the proteins have RNA-binding properties similar to those of hnRNP A1 and may perform similar functions in MHV RNA synthesis. These proteins are translocated to the cytoplasm during MHV replication, and the overexpression of hnRNP A/B in the cells stimulates MHV RNA synthesis. Thus, it appears that the hnRNP A/B family of cellular proteins collectively play an important role in MHV RNA replication and transcription.
It is possible that different hnRNP A/B proteins can replace each other in MHV RNA replication. The participation of multiple hnRNP A/B proteins in viral replication is similar to what has been observed for the roles of these proteins in other viral and cellular RNA splicing events. These hnRNP A/B family members have been shown to replace each other in these functions (6, 25). It is interesting that even in DBT and CB7 cells, both of which express hnRNP A1, there appear to be multiple UV-cross-linked proteins with MHV minus-strand leader RNA (Fig. 1B). These proteins were not characterized in the previous studies (21) because it was thought that they represented hnRNP A1 isoforms. In light of the results presented here, it is likely that these proteins belong to the hnRNP A/B family.
All of the hnRNP A1-related proteins are structurally similar and are particularly homologous in their RNA-binding domains at the N terminus. Therefore, it is not surprising that these proteins can participate in similar functions in MHV RNA synthesis. Our studies showed that hnRNP A/B has a stimulatory effect on MHV RNA replication in HEK-293 cells. Thus, even in cell lines that are rich in hnRNP A1, hnRNP A/B can exert its activity. Because of the poor transfection efficiency, we were not able to do similar studies in CB3 cells, which do not express hnRNP A1. Nevertheless, in a stable CB3 cell line expressing a small amount of the exogenous full-length hnRNP A/B, we did see a slight increase in MHV RNA synthesis over that in the control cells (data not shown).
We have also established several stable CB3 cell lines expressing a C-terminally truncated hnRNP A1 protein. In these cell lines, MHV RNA replication was inhibited compared to the control cells (unpublished observation). Thus, this truncated hnRNP A1 could serve as a dominant-negative mutant. Therefore, even in cell lines that do not express hnRNP A1, the overexpression of hnRNP A1 can have a modulatory effect on MHV RNA replication. The most likely explanation is that hnRNP A1 competes with other hnRNP A/B family members because of sequence homology. This result provides an additional piece of evidence in support of the roles of the hnRNP A/B family in MHV RNA replication.
So far, several cellular hnRNP proteins have been shown to interact with MHV RNA directly and modulate MHV RNA synthesis. These include hnRNPs A1, A/B, and I (PTB). Thus, cellular and viral proteins involved in various RNA-processing activities may be crucial for MHV RNA replication. These findings further strengthen the potential roles of cellular factors in viral RNA replication and transcription, as postulated earlier (17). We believe that these studies have also settled a controversy regarding the role of cellular factors, particularly hnRNP A1, in MHV RNA synthesis (33, 34). Clearly, multiple hnRNP A/B proteins are involved in MHV RNA synthesis.
Acknowledgments
We thank G. Dreyfuss at the University of Pennsylvania for generously providing monoclonal antibodies against hnRNP A1 and A2/B1.
This work was partially supported by National Institutes of Health research grant AI19244. S.T.S. was supported by a postdoctoral fellowship from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. M.M.C.L. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Bemark, M., H. Olsson, D. Heinegard, and T. Leanderson. 1998. Purification and characterization of a protein binding to the SP6 kappa promoter. A potential role for CArG-box binding factor-A in kappa transcription. J. Biol. Chem. 273:18881-18890. [DOI] [PubMed] [Google Scholar]
- 2.Ben-David, Y., M. R. Bani, B. Chabot, A. De Koven, and A. Bernstein. 1992. Retroviral insertions downstream of the heterogeneous nuclear ribonucleoprotein A1 gene in erythroleukemia cells: evidence that A1 is not essential for cell growth. Mol. Cell. Biol. 12:4449-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biamonti, G., and S. Riva. 1994. New insights into the auxiliary domains of eukaryotic RNA binding proteins. FEBS Lett. 340:1-8. [DOI] [PubMed] [Google Scholar]
- 4.Burd, C. G., M. S. Swanson, M. Gorlach, and G. Dreyfuss. 1989. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: a diversity of RNA binding proteins is generated by small peptide inserts. Proc. Natl. Acad. Sci. USA 86:9788-9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buvoli, M., F. Cobianchi, M. G. Bestagno, A. Mangiarotti, M. T. Bassi, G. Biamonti, and S. Riva. 1990. Alternative splicing in the human gene for the core protein A1 generates another hnRNP protein. EMBO J. 9:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartegni, L., M. Maconi, E. Morandi, F. Cobianchi, S. Riva, and G. Biamonti. 1996. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J. Mol. Biol. 259:337-348. [DOI] [PubMed] [Google Scholar]
- 8.Casas-Finet, J. R., J. D. Smith, Jr., A. Kumar, J. G. Kim, S. H. Wilson, and R. L. Karpel. 1993. Mammalian heterogeneous ribonucleoprotein A1 and its constituent domains. Nucleic acid interaction, structural stability and self-association. J. Mol. Biol. 229:873-889. [DOI] [PubMed] [Google Scholar]
- 9.Dreyfuss, G., M. J. Matunis, S. Pinol-Roma, and C. G. Burd. 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62:289-321. [DOI] [PubMed] [Google Scholar]
- 10.Furuya, T., and M. M. C. Lai. 1993. Three different cellular proteins bind to complementary sites on the 5′-end-positive and 3′-end-negative strands of mouse hepatitis virus RNA. J. Virol. 67:7215-7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton, B. J., C. M. Burns, R. C. Nichols, and W. F. C. Rigby. 1997. Modulation of AUUUA response element binding by heterogeneous nuclear ribonucleoprotein A1 in human T lymphocytes. The roles of cytoplasmic location, transcription, and phosphorylation. J. Biol. Chem. 272:28732-28741. [DOI] [PubMed] [Google Scholar]
- 12.Huang, P., and M. M. Lai. 2001. Heterogeneous nuclear ribonucleoprotein a1 binds to the 3′ untranslated region and mediates potential 5′-3′-end cross talks of mouse hepatitis virus RNA. J. Virol. 75:5009-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, P., and M. M. C. Lai. 1999. Polypyrimidine tract-binding protein binds to the complementary strand of the mouse hepatitis virus 3′ untranslated region, thereby altering RNA conformation. J. Virol. 73:9110-9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchison, S., C. LeBel, M. Blanchette, and B. Chabot. 2002. Distinct sets of adjacent heterogeneous nuclear ribonucleoprotein (hnRNP) A1/A2 binding sites control 5′ splice site selection in the hnRNP A1 mRNA precursor. J. Biol. Chem. 277:29745-29752. [DOI] [PubMed] [Google Scholar]
- 15.Kamada, S., and T. Miwa. 1992. A protein binding to CArG box motifs and to single-stranded DNA functions as a transcriptional repressor. Gene 119:229-236. [DOI] [PubMed] [Google Scholar]
- 16.Krecic, A. M., and M. S. Swanson. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11:363-371. [DOI] [PubMed] [Google Scholar]
- 17.Lai, M. M. C. 1998. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology 244:1-12. [DOI] [PubMed] [Google Scholar]
- 18.Lai, M. M. C., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leverrier, S., E. Cinato, C. Paul, J. Derancourt, M. Bemark, T. Leanderson, and C. Legraverend. 2000. Purification and cloning of type A/B hnRNP proteins involved in transcriptional activation from the rat spi2 gene GAGA box. Biol. Chem. 381:1031-1040. [DOI] [PubMed] [Google Scholar]
- 20.Li, H. P., P. Huang, S. Park, and M. M. C. Lai. 1999. Polypyrimidine tract-binding protein binds to the leader RNA of mouse hepatitis virus and serves as a regulator of viral transcription. J. Virol. 73:772-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, H. P., X. Zhang, R. Duncan, L. Comai, and M. M. C. Lai. 1997. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc. Natl. Acad. Sci. USA 94:9544-9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao, C. L., and M. M. C. Lai. 1994. Requirement of the 5′-end genomic sequence as an upstream cis-acting element for coronavirus subgenomic mRNA transcription. J. Virol. 68:4727-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma, A. S., K. Moran-Jones, J. Shan, T. P. Munro, M. J. Snee, K. S. Hoek, and R. Smith. 2002. Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein. J. Biol. Chem. 277:18010-18020. [DOI] [PubMed] [Google Scholar]
- 24.Makino, S., M. Joo, and J. K. Makino. 1991. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J. Virol. 65:6031-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayeda, A., S. H. Munroe, J. F. Caceres, and A. R. Krainer. 1994. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 13:5483-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michael, W. M., H. Siomi, M. Choi, S. Pinol-Roma, S. Nakielny, Q. Liu, and G. Dreyfuss. 1995. Signal sequences that target nuclear import and nuclear export of pre-mRNA-binding proteins. Cold Spring Harb. Symp. Quant. Biol. 60:663-668. [DOI] [PubMed] [Google Scholar]
- 27.Mikheev, A. M., S. A. Mikheev, Y. Zhang, R. Aebersold, and H. Zarbl. 2000. CArG binding factor A (CBF-A) is involved in transcriptional regulation of the rat Ha-ras promoter. Nucleic Acids Res. 28:3762-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munro, T. P., R. J. Magee, G. J. Kidd, J. H. Carson, E. Barbarese, L. M. Smith, and R. Smith. 1999. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J. Biol. Chem. 274:34389-34395. [DOI] [PubMed] [Google Scholar]
- 29.Nadler, S. G., B. M. Merrill, W. J. Roberts, K. M. Keating, M. J. Lisbin, S. F. Barnett, S. H. Wilson, and K. R. Williams. 1991. Interactions of the A1 heterogeneous nuclear ribonucleoprotein and its proteolytic derivative, UP1, with RNA and DNA: evidence for multiple RNA binding domains and salt-dependent binding mode transitions. Biochemistry 30:2968-2976. [DOI] [PubMed] [Google Scholar]
- 30.Pollard, V. W., W. M. Michael, S. Nakielny, M. C. Siomi, F. Wang, and G. Dreyfuss. 1996. A novel receptor-mediated nuclear protein import pathway. Cell 86:985-994. [DOI] [PubMed] [Google Scholar]
- 31.Robb, J. A., and C. W. Bond. 1979. Pathogenic murine coronaviruses. I. Characterization of biological behavior in vitro and virus-specific intracellular RNA of strongly neurotropic JHMV and weakly neurotropic A59V viruses. Virology 94:352-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shan, J., K. Moran-Jones, T. P. Munro, G. J. Kidd, D. J. Winzor, K. S. Hoek, and R. Smith. 2000. Binding of an RNA trafficking response element to heterogeneous nuclear ribonucleoproteins A1 and A2. J. Biol. Chem. 275:38286-38295. [DOI] [PubMed] [Google Scholar]
- 33.Shen, X., and P. S. Masters. 2001. Evaluation of the role of heterogeneous nuclear ribonucleoprotein A1 as a host factor in murine coronavirus discontinuous transcription and genome replication. Proc. Natl. Acad. Sci. USA 98:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi, S. T., P. Huang, H. P. Li, and M. M. C. Lai. 2000. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J. 19:4701-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siomi, H., and G. Dreyfuss. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weighardt, F., G. Biamonti, and S. Riva. 1995. Nucleo-cytoplasmic distribution of human hnRNP proteins: a search for the targeting domains in hnRNP A1. J. Cell Sci. 108:545-555. [DOI] [PubMed] [Google Scholar]
- 37.Yokomori, K., and M. M. C. Lai. 1992. Mouse hepatitis virus utilizes two carcinoembryonic antigens as alternative receptors. J. Virol. 66:6194-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, X., and M. M. C. Lai. 1995. Interactions between the cytoplasmic proteins and the intergenic (promoter) sequence of mouse hepatitis virus RNA: correlation with the amounts of subgenomic mRNA transcribed. J. Virol. 69:1637-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, X., H. P. Li, W. Xue, and M. M. C. Lai. 1998. Cellular protein hnRNP-A1 interacts with the 3′-end and the intergenic sequence of mouse hepatitis virus negative-strand RNA to form a ribonucleoprotein complex. Adv. Exp. Med. Biol. 440:227-234. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, X., H. P. Li, W. Xue, and M. M. C. Lai. 1999. Formation of a ribonucleoprotein complex of mouse hepatitis virus involving heterogeneous nuclear ribonucleoprotein A1 and transcription-regulatory elements of viral RNA. Virology 264:115-124. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, X., C. L. Liao, and M. M. C. Lai. 1994. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription both in trans and in cis. J. Virol. 68:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, X. M., and M. M. C. Lai. 1995. Regulation of coronavirus RNA transcription is likely mediated by protein-RNA interactions. Adv. Exp. Med. Biol. 380:515-521. [DOI] [PubMed] [Google Scholar]