Abstract
Polytropic murine leukemia viruses (MLVs) are generated by recombination of ecotropic MLVs with members of a family of endogenous proviruses in mice. Previous studies have indicated that polytropic MLV isolates comprise two mutually exclusive antigenic subclasses, each of which is reactive with one of two monoclonal antibodies termed MAb 516 and Hy 7. A major determinant of the epitopes distinguishing the subclasses mapped to a single amino acid difference in the SU protein. Furthermore, distinctly different populations of the polytropic MLV subclasses are generated upon inoculation of different ecotropic MLVs. Here we have characterized the majority of endogenous polytropic MLV-related proviruses of NFS/N mice. Most of the proviruses contain intact sequences encoding the receptor-binding region of the SU protein and could be distinguished by sequence heterogeneity within that region. We found that the endogenous proviruses comprise two major groups that encode the major determinant for Hy 7 or MAb 516 reactivity. The Hy 7-reactive proviruses correspond to previously identified polytropic proviruses, while the 516-reactive proviruses comprise the modified polytropic proviruses as well as a third group of polytropic MLV-related proviruses that exhibit distinct structural features. Phylogenetic analyses indicate that the latter proviruses reflect features of phylogenetic intermediates linking xenotropic MLVs to the polytropic and modified polytropic proviruses. These studies elucidate the relationships of the antigenic subclasses of polytropic MLVs to their endogenous counterparts, identify a new group of endogenous proviruses, and identify distinguishing characteristics of the proviruses that should facilitate a more precise description of their expression in mice and their participation in recombination to generate recombinant viruses.
Ecotropic murine leukemia viruses (MLVs) exist as exogenous viruses and also as endogenous retroviruses expressed in certain mouse strains. They are infectious for cells of murine origin but not for cells of other species. Upon infection of susceptible mouse strains with exogenous ecotropic MLVs or expression of endogenous ecotropic MLVs, the viruses frequently undergo recombination with members of a large endogenous provirus family to generate polytropic MLVs (13, 14, 19, 21, 43, 44). Recombination invariably involves substitution of the 5′ end of the env gene, encoding the amino-terminal region of the SU protein (1, 4, 8, 9, 12, 14, 25, 41, 50). This region determines the binding of the SU protein to a specific receptor on the cell surface, and substitution of the ecotropic receptor-binding sequences with endogenous sequences results in the utilization of a receptor distinct from that utilized by the ecotropic MLV (38). In contrast to ecotropic MLVs, polytropic MLVs are capable of infecting cells derived from several different species as well as murine cells (11). These recombinant viruses are intimately involved in a number of pathological processes, including proliferative diseases of lymphoid and erythroid origin and the induction of neurological disease (7, 10, 19, 26, 39, 44, 50).
Previous studies from this laboratory have described two antigenic subclasses of polytropic virus isolates based on their reactivities with two different monoclonal antibodies termed Hy 7 and MAb 516 (28). Virtually all of the polytropic isolates were reactive with either Hy 7 or MAb 516; however, none of the viruses were reactive with both antibodies. We mapped a major determinant of the epitopes for both antibodies to a single amino acid residue in the receptor-binding region of the polytropic SU protein. Hy 7-reactive viruses contained a lysine at that position, and MAb 516-reactive viruses contained a glutamine. The reactivities to the antibodies could be changed by substitution of a single nucleotide in the coding sequence of the SU protein. Furthermore, it was found that different inoculated ecotropic MLVs gave rise to distinctly different populations of the antigenic subclasses (28). Mice inoculated with Moloney MLV (MMLV), on average, exhibited equal titers of MAb 516- and Hy 7-reactive recombinants, whereas mice inoculated with Friend MLV (FMLV) exhibited predominantly Hy 7-reactive recombinants, suggesting recombination of these MLVs with distinct groups of endogenous proviruses. Subsequent studies using chimeras between MMLV and FMLV indicated that this specificity was determined by a short sequence encoding the nucleocapsid protein and a small sequence of the protease (27). It was apparent that an understanding of the mechanism of this specificity would be facilitated by a more complete description of the endogenous proviruses involved in the generation of polytropic MLVs. A major objective of this study was to determine how the antigenic subclasses of polytropic MLVs are reflected in the population of endogenous proviruses from which they are derived.
Genomes of inbred mouse strains contain numerous proviral sequences which bear very close homology to the genes encoding the amino-terminal SU sequences found in recombinant polytropic MLVs (16, 17, 24, 46, 47). Stoye and Coffin (46) have described two major groups of endogenous polytropic MLV-related sequences, termed polytropic (PT) and modified polytropic (mPT) proviruses, that include all nonecotropic and nonxenotropic proviruses thus far identified in inbred mouse strains. A major feature distinguishing mPT from PT proviruses is a 27-bp deletion in the sequences encoding the SU proteins of mPT proviruses. It was clear from previous analyses of virus isolates that both MAb 516- and Hy 7-reactive viruses include viruses derived from PT proviruses (14, 28). Thus, the antigenic subclasses of polytropic virus isolates do not strictly correspond to mPT and PT types of recombinants. It was unclear, however, if the antigenic subclasses of recombinant virus isolates reflected major groups of endogenous proviruses or, alternatively, if one or both of the subclasses corresponded to recombination with minor populations that were the principal participants in recombination. Furthermore, if the antigenic subclasses of polytropic virus isolates reflect recombination with two major groups of endogenous viruses, it was of interest to determine how these groups relate to the PT and mPT proviruses.
We have approached these issues by the characterization of numerous clones homologous to the receptor-binding domain of polytropic MLVs from an NFS/N mouse genomic lambda library. Our findings indicate that all endogenous polytropic MLV-related proviruses identified encode either a lysine or a glutamine at the position in the SU protein corresponding to the major determinant defined for Hy 7 or MAb 516 reactivity. The Hy 7 group corresponds to the PT proviruses previously described, while the MAb 516 group comprises mPT proviruses as well as a third distinct group of polytropic proviruses that provide insight into the evolutionary relationships of polytropic MLV-related proviruses in mice.
MATERIALS AND METHODS
Mice.
The mice used in this study were NFS/N mice maintained as an inbred colony at Rocky Mountain Laboratories, Veterinary Branch. All animals were treated in accordance with National Institutes of Health regulations and the guidelines of the Animal Care and Use Committee of Rocky Mountain Laboratories.
Analysis of genomic DNA by blot hybridization.
The procedure for detection of endogenous polytropic proviruses by blot hybridization has been described in detail previously (26). Briefly, DNA from late-gestation NFS/N embryos was purified as described below and digested with BamHI for 4 h with 10 U of enzyme per μg of DNA. The digested DNA and a molecular weight marker labeled with digoxigenin (DIG) were electrophoresed on a 22-cm-long 0.8% agarose gel in Tris-borate-EDTA buffer at 85 V for 17 h. DNA was transferred from the gel to positively charged nylon membranes with a vacuum blotting apparatus (VacuGene XL; Pharmacia), cross-linked to the membrane by UV irradiation, and baked at 80°C for 1 h. The membrane was blocked and hybridized in blocking solution containing a DIG-labeled 600-bp BamHI-to-EcoRI fragment cloned from the spleen focus-forming virus. After hybridization, the membranes were washed and treated according to the manufacturer's instructions. The membranes were subsequently developed by addition of anti-DIG Fab fragments and incubated with 0.25 mM CDP-Star (Tropix). Exposure of the film for a 6-μg sample lasted approximately 12 h.
Generation of an NFS/N genomic DNA lambda library and isolation of clones containing polytropic MLV-related proviral DNA.
Late-gestation NFS/N embryos obtained from pregnant females were frozen and blended in liquid nitrogen to produce a frozen tissue powder. The frozen tissue was suspended in 8 volumes of lysing buffer from a DNA isolation kit (D5000A; Gentra Systems, Inc.), and the DNA was subsequently purified according to the manufacturer's instructions. Purified DNA was partially digested with MboI and sedimented on a 10 to 30% linear glycerol gradient in 100 mM NaCl-10 mM Tris HCl (pH 7.4)-1 mM EDTA. The gradient was fractionated, and a sample of each fraction was analyzed on a 0.4% agarose gel (Seakem GTG). Fractions containing fragments of approximately 15 to 25 kb were pooled, precipitated with ethanol, resuspended in 10 mM Tris-1 mM EDTA, and cloned into the Lambda Fix vector (Stratagene) according to the manufacturer's instructions. The library was screened by using a polytropic env probe excised from a plasmid containing a 620-bp BamHI-to-EcoRI fragment cloned from the Friend spleen focus-forming virus (32). The BamHI-to-EcoRI fragment is homologous to the receptor-binding domain of the SU-encoding sequences in polytropic genomes. Labeling of the probe for subsequent detection by chemiluminescence was performed by random-primed synthesis with DIG-labeled UTP by use of a DIG DNA labeling kit (Genius 2 kit; Roche). Development of the filters to identify plaques homologous to the probe was carried out as described for blot hybridization of DNAs (26).
Preparation of lambda DNA.
Lambda DNA was prepared by utilizing various features of reported procedures (29, 49) along with a lambda DNA preparation kit (catalog no. 12543; Qiagen). A phage stock containing 20 × 103 to 50 × 103 PFU was plated with the MRF′ bacterial strain (Stratagene) on a 100-mm-diameter NZY plate to obtain complete lysis after overnight incubation at 37°C. Phage were eluted from the plate in 5.0 ml of suspension medium (0.1 M NaCl, 0.008 M MgSO4, 0.05 M Tris-HCl, pH 7.5, and 2% gelatin) plus 10 mM MgSO4 by rocking for 2 h at room temperature. A 50-ml culture of bacteria in Luria-Bertani medium containing 10 mM MgSO4 was initiated by inoculation of 0.3 ml of an overnight culture of bacteria grown in the same medium. The culture was grown at 37°C to an A600 of 0.5 to 0.9, at which time the entire phage lysate was added to the culture and incubated at 39°C until lysis had occurred. If lysis was not apparent after 2 to 3 h, 1 ml of chloroform was added and the culture was incubated an additional 10 min. The lysate was cleared of bacterial debris by centrifugation at 5,000 × g for 10 min, and the DNA was prepared by using the lambda DNA preparation kit with the following modifications. After treatment of the lysate with DNase and RNase (solution L1), the lysate was diluted with an equal volume of distilled water and warmed at 37°C for 10 min. The lysate was then adjusted to 0.04 M ZnCl2 by addition of a freshly prepared filtered 2.0 M solution, incubated at 37°C for 5 min, and then centrifuged at 5,000 × g for 5 min at room temperature. The precipitate was resuspended in 1.5 ml of solution L3 from the kit and further processed according to the manufacturer's instructions. This procedure typically yielded 50 to 100 μg of lambda DNA.
PCR analyses and DNA sequencing.
Lambda DNA was used as the template for PCRs. PCR amplification of a region of the polytropic provirus encoding the receptor-binding region of the SU protein was accomplished with a primer set consisting of oligonucleotides termed FORPOLY (GCAGTACAACGAGAGGTCTGG) and REVPOLY (GGGTCAAAGAGAACCGGGTCAC). FORPOLY is a sequence in the polymerase gene of the recombinant polytropic MLV MCF1233 (GenBank accession no. U13766) (45) from nucleotide 5519 to 5539. REVPOLY is the reverse complement of sequences in the published envelope genes of the endogenous PT provirus MX27 (GenBank accession no. M17327) and the mPT provirus MX33 (GenBank accession no. M17326) from nucleotide 710 to 731 (46) and in MCF1233 from nucleotide 6443 to 6454. PCRs in preparation for sequencing were conducted by using the puReTaq Ready-To-Go PCR system (catalog no. 27-9558-01; Amersham Pharmacia Biotech Inc.) with Pfu Turbo DNA polymerase (catalog no. 600153; Stratagene) or Platinum Pfx DNA polymerase (catalog no. 11708-021; Invitrogen) according to the manufacturer's instructions.
PCRs to distinguish PT from mPT envelope sequences utilized oligonucleotides 510MX33 (CCCTTAAGCGAGGAAACACCCCTCGG) and 897MX33RC (GCTTGGTAGGCTCCATCTACCAGGT). 510MX33 corresponds to a sequence present in MX33 and MX27 from position 510 to 535. 897MX33RC corresponds to the reverse complements of a sequence in MX33 from position 897 to 921 and a sequence in MX27 from position 924 to 948. Additional oligonucleotides used for the generation of PCR products and as sequencing primers included MIDREVPOLY (ACATTGAAGACCTGATGAGGG), MIDFORPOLY (AGCTAATGCTACCTCCCTCCTG), 820MX33 (CATGCTCCCCAGGCCTCCTC), 1125MX33 (CCCATCAGGCCCTGTGTAATACC), 1203MX33RC (AGCCCGGTGTTGCAAGCCC), 1555MX33 (GGTGGTCCTACAGAACCGGA), 1645MX33RC (CTRACGCCAGTGTGGTCCGCG), 1882MX33 (GGTGGTGCAGGCCCTGGTTC), 2016MX33RC (CTGCAGCTAGCTTGCTAAGCC), 2024MX33 (AAGCTAGCTGCAGTAACGTCCATTTTGC), 2224MX33RC (CTTCTGTGTCTGTTGCTGGTTCCGC), 2328MX33RC (CCAGAGCACTGTGCACCTC), 2359MX33RC (CAGAACATTGGCAGACACACAGG), and 2580MX33RC (GCGCGCCGAGTGTGGGAGTT). All oligonucleotides utilized in this study were synthesized by Sigma Genosys, and sequence analyses of the PCR products were carried out on an ABI model 377 DNA sequencer at the DNA Sequencing and Synthesis Facility at Iowa State University.
Alignment and phylogeny of DNA sequences.
Alignment and phylogenetic analyses were performed by using the ClustalX sequence alignment program (51) in conjunction with the PAUP phylogenetic analysis program (version 4; Sinauer Associates, Sunderland, Mass.) for neighbor joining (NJ), maximum parsimony, and maximum likelihood analyses. Bayesian analysis of phylogeny was performed with the MrBayes program.
Nucleotide sequence accession numbers.
The sequences in this report have been deposited in GenBank under accession no. AY219536 through AY219567.
RESULTS
Identification of endogenous polytropic MLV-related proviruses that contain undeleted receptor-binding domains.
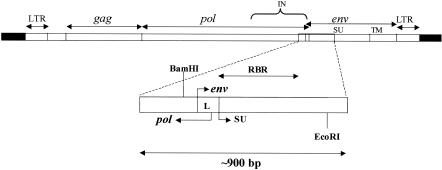
The generation of polytropic MLVs involves substitution of the receptor-binding domain in the SU protein of ecotropic MLVs with endogenous polytropic MLV-related proviruses of the mouse. Inbred mouse strains contain about 20 to 40 endogenous proviral sequences homologous to the receptor-binding domain acquired by polytropic MLVs (18, 46, 47), and NFS/N mice appear to harbor approximately 30 such sequences in their genome (Fig. 1). We derived a lambda library of NFS/N genomic DNA and isolated numerous clones containing sequences homologous to those encoding the polytropic receptor-binding domain. Previous reports have indicated that some of the polytropic MLV-related proviruses contained defects, including deletions in their env genes that would preclude their participation by a simple recombination process in the generation of replication-competent polytropic MLVs (6, 24, 46). We wanted to limit our analyses to proviruses that could potentially contribute intact sequences to recombinant polytropic MLVs. Accordingly, the clones were screened by PCR to identify sequences that had not undergone major deletions in the region encoding the receptor-binding domain of the SU protein. For this purpose we chose primers corresponding to highly conserved sequences that flanked a region encompassing a 3′ portion of the pol gene as well as a 5′ portion of the env gene. The sequence flanked by the primers comprises about 900 bp in intact polytropic proviruses and encompasses the minimal substitution that has been found in recombinant polytropic MLV isolates (14). This includes sequences of the pol gene that encode a portion of the integrase protein and those of the env gene that encode the receptor-binding domain of the SU protein (Fig. 2).
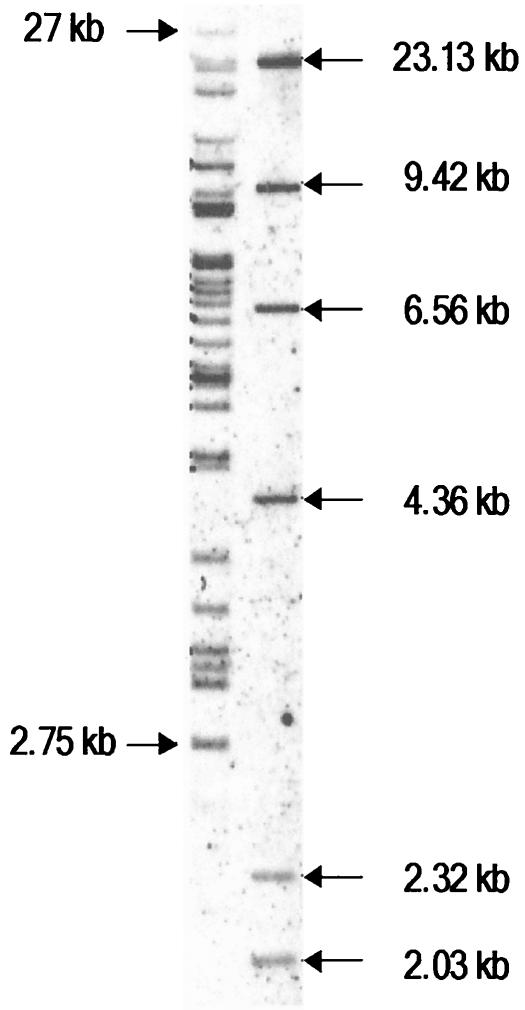
FIG. 1.
Endogenous PT and mPT sequences of NFS/N mice. DNA from a prenatal NFS/N mouse (6 μg) was digested with BamHI and analyzed by electrophoresis on a 0.8% agarose gel. The gel was then blotted and hybridized to the DIG-labeled polytropic MLV env probe to detect homologous sequences. Nearly all polytropic MLVs have a BamHI site derived from the endogenous sequence near the 3′ end of the pol gene and no additional BamHI sites in the env gene or LTR. Thus, the bands correspond to fragments between the BamHI site in the endogenous pol gene and the nearest BamHI site after the 3′ end of the provirus.
FIG. 2.
Amplified region of endogenous proviruses encompassing the receptor-binding region of the SU protein. The diagram at the top represents an intact endogenous polytropic provirus (open rectangles) and the flanking sequences (solid rectangles). Approximate locations of the LTR, the gag, pol, and env genes, and sequences encoding the integrase (IN), the surface glycoprotein (SU), and the transmembrane protein (TM) are indicated. The expanded region at the bottom represents the PCR product of approximately 900 bp that was used for initial sequence comparisons among the endogenous proviruses. The product contains pol gene sequences encoding a portion of the IN protein, the env gene leader sequence (L), and the receptor-binding region (RBR) of the SU protein. The positions of the BamHI and EcoRI sites that define the polytropic MLV probe are also indicated.
Of 57 lambda clones containing polytropic env sequences, 32 yielded PCR products of approximately 900 bp. Thirty of the 32 clones that yielded the ∼900-bp PCR product contained the remaining proviral env gene sequences and the 3′ long terminal repeat (LTR). Our initial sequence analyses focused on the ∼900-bp PCR products as well as the U3 region of the LTRs. The results of these analyses prompted the determination of the remaining intervening env sequences between the receptor-binding region and the LTR to assess the possibility that some clones represented endogenous recombinant proviruses.
The NFS/N mouse genome contains numerous sequences encoding intact polytropic MLV receptor-binding regions.
The PCR analyses identified clones that may contain intact receptor-binding sequences, based on the absence of large deletions in this region. However, the analyses would not distinguish sequences that were defective as a result of changes such as point mutations or small deletions that introduced a frameshift or a premature termination. We found that 27 of the 32 clones contained intact coding sequences devoid of frameshift or termination mutations within the receptor-binding region. Twenty-four of these were colinear with published sequences of recombinant virus isolates (Fig. 3), while three others contained additional small sequences of 9 bp (NJ1 and NJ2) or 12 bp (NO1) compared to the consensus sequence (Fig. 3). Five clones containing mutations that would preclude the synthesis of an intact SU protein were identified. The nature and location of these mutations are indicated in Fig. 3. It is perhaps noteworthy that one clone (ND1) had a tRNAIle rather than a tRNAMet codon as the initiator of the env gene. Endogenous proviruses in HRS/J and RFM/Un mice containing an identical mutation in their initiation codon have been described previously (35, 46), suggesting that they may represent allelic forms of the same proviral locus in different mouse strains. We also identified two distinct proviruses that had identical mutations resulting in a termination codon in the env gene, each of which was identified twice in different lambda clones (Fig. 3, NF1/NF2 and NH1/NH2).
FIG. 3.
Sequence properties of the ∼900-bp PCR products of lambda clones containing nondeleted receptor-binding regions. Diagrams depict the amplified region shown in Fig. 2. Vertical arrows indicate the positions at which additional sequences (given below) were located within the SU-encoding sequences. The approximate locations and natures of mutations rendering five of the proviruses defective are indicated. These include a mutation at nucleotide 251 in ND1 resulting in a tRNAIle as the initiator codon of env and a termination codon in pol, a frameshift mutation at nucleotide 785 in ND1, a premature termination codon in pol at nucleotide 252 in NF1 and NF2, a premature termination codon at nucleotide 385 in env in NF1 and NF2, and an identical mutation at nucleotide 385 in NH1 and NH2.
As noted earlier, the intervening env gene sequences were obtained for almost all of the clones included in these analyses. These analyses revealed additional mutations in two of the proviruses which would preclude the synthesis of an intact env polyprotein. NB1 and NB2 exhibit a frameshift mutation resulting in a termination codon at amino acid 438 of the SU protein. NL2, one of the mPT proviruses, exhibited a large deletion of 116 bp beginning at nucleotide 1191 of the env gene. It is unclear if mutations that do not involve the receptor-binding region influence the participation of the endogenous proviruses in recombination to generate polytropic MLVs.
Sequence heterogeneity among endogenous polytropic MLV-related proviruses.
Endogenous polytropic MLV-related proviruses are very closely homologous. Although the PT and mPT subclasses of polytropic MLV-related proviruses differ by several bases within the receptor-binding region, proviruses within each of these groups are nearly identical in sequence, even when they are obtained from different mouse strains. We wished to determine if the sequence heterogeneity in this region was sufficient to uniquely identify a substantial number of the polytropic MLV-related proviruses. If not, our eventual goal of unambiguously identifying proviruses that participate in recombination would not be feasible. Among the 32 clones we have characterized, we identified 17 different proviruses distinguishable by base differences within the receptor-binding region. Most of the sequences were found only once (nine clones) or twice (six clones), while other sequences had as many as six identical counterparts in different clones. We have designated the clones in an alphanumeric fashion indicating the mouse strain (N for NFS/N), sequence identity within the ∼900-bp region (A to Q), and a number to identify the individual clone (clones 1 to 6) (Table 1). Sequence analyses of the remainder of the env gene and the LTR (see below) indicated that some λ clones that exhibited identical ∼900-bp sequences were, in fact, derived from different endogenous proviruses. In this regard, the NA group is composed of two proviruses (NA2 differs from the remaining members of the NA group) and the NC group is composed of three different proviruses (NC1 and NC5 are identical, NC3 and NC4 are identical, and NC2 is unique). Thus, our sequence analyses have identified 20 distinct proviruses. Seventeen of these are nondefective within the minimal region found in polytropic MLVs and could potentially generate replication-competent recombinant viruses.
TABLE 1.
Properties of ∼900-bp PCR products from lambda clones
| Lambda clone(s)a | PCR product length (bp)b | Intact coding sequence | Group | PT or mPT |
|---|---|---|---|---|
| NA1-NA6c,d | 893 | Yes | 516 | mPT |
| NB1-NB2 | 893 | Yes | 516 | PT |
| NC1-NC5e | 893 | Yes | Hy 7 | PT |
| ND1 | 894 | No | Hy 7 | PT |
| NE1-NE2 | 893 | Yes | Hy 7 | PT |
| NF1-NF2 | 893 | No | Hy 7 | PT |
| NG1 | 893 | Yes | Hy 7 | PT |
| NH1-NH2 | 893 | No | Hy 7 | PT |
| N11-N12 | 893 | Yes | Hy 7 | PT |
| NJ1-NJ2 | 902 | Yes | 516 | PT |
| NK1 | 893 | Yes | 516 | mPT |
| NL1 | 893 | Yes | 516 | mPT |
| NM1 | 893 | Yes | 516 | mPT |
| NN1 | 893 | Yes | Hy 7 | PT |
| NO1 | 905 | Yes | 516 | PT |
| NP1 | 893 | Yes | Hy 7 | PT |
| NQ1 | 893 | Yes | Hy 7 | PT |
Each set (NA to NP) represents a unique sequence. Sets with more than one clone represent different clones with identical sequences.
The length represents the PCR product minus the sequences corresponding to the primers.
Lambda clone NA6 did not yield a PCR product with primers designed to identify PT and mPT proviruses.
The NA set of clones comprises at least two distinct proviruses.
The NC set of clones comprises at least three distinct proviruses.
Distribution of sequences corresponding to Hy 7- and MAb 516-reactive polytropic viral isolates.
Virtually all recombinant polytropic virus isolates can be grouped into one of two distinct subgroups based on their reactivity to antibody Hy 7 or MAb 516 (28). Reactivity to these antibodies is dependent on the presence of a lysine (Hy 7) or a glutamine (MAb 516) at position 173 in the SU protein. We found that all of the endogenous polytropic MLV-related sequences examined encode either a lysine or a glutamine at the analogous position in their SU proteins (Table 1). For the purposes of this report, we have designated the two groups of proviruses the Hy 7 proviruses and the 516 proviruses, respectively. Further, the difference between the two groups in all instances is the result of a single-nucleotide difference in the first base of the codon for this amino acid residue (A for the Hy 7 group and C for the MAb 516 group). Among the 17 distinct proviruses identified above that could potentially generate replication-competent recombinant viruses, 9 correspond to the Hy 7 group and 8 correspond to the 516 group, in very close agreement with the distribution of the antigenic groups found in polytropic MLV isolates. Thus, the antigenic subclasses of polytropic MLV isolates reflect two groups of proviruses that, together, are inclusive of all polytropic MLV-related proviruses examined.
Identification of PT and mPT envelope sequences.
Two groups of endogenous polytropic MLV-related proviral sequences have been described for inbred mouse strains; these groups are termed PT and mPT proviruses (46). A major distinction between PT and mPT env gene sequences is the presence of a 27-bp deletion in a region of the env gene encoding the SU protein of mPT proviruses. This deletion is located approximately 190 bases in the 3′ direction from the nucleotide distinguishing Hy 7 from MAb 516 proviruses. We found that 23 of the clones corresponded to PT proviruses, while 8 corresponded to mPT proviruses (Table 1). One clone (NA6) that yielded a ∼900-bp sequence appears to have been truncated during the generation of the lambda library. The other clones in this group (NA1 to NA5) yielded PCR products indicative of mPT proviruses. Interestingly, the Hy 7 group was entirely composed of PT proviruses whereas the 516 group included all of the mPT proviruses as well as some PT proviruses.
The proportion of mPT proviruses among the clones was somewhat smaller than that indicated by previous reports of the relative frequencies of these two types of sequences in different mouse strains (46, 47). As noted above, sequence analyses of the remaining env gene sequences and the LTR revealed two distinguishable proviruses within the NA set of clones as well as three distinguishable proviruses within the NC set. Thus, of the 20 distinct proviruses identified, 15 correspond to PT proviruses while 5 correspond to mPT proviruses.
Phylogenetic relationships of endogenous proviral 3′ pol and 5′ env sequences.
The Hy 7 proviruses described above were composed entirely of PT proviruses, whereas the 516 proviruses comprised both PT and mPT proviruses. Furthermore, PT proviruses of the Hy 7 group were all nearly identical in sequence to the prototypic PT provirus MX27, while the mPT proviruses, all of which belonged to the 516 group, were nearly identical to the prototypic mPT provirus MX33 (46). In contrast, the remaining proviruses of the 516 group, which were PT in that they did not exhibit the deletion characteristic of mPT proviruses, exhibited numerous sequence differences distinguishing them from previously described PT or mPT proviruses. Sequence comparisons of the proviruses with the xenotropic MLV NZB-9-1 (37) indicated that several of the differences that distinguished the 516 PT proviruses from the Hy 7 PT proviruses and the mPT proviruses were shared with the xenotropic MLV. These included several specific nucleotides as well as the presence of a 12-bp xenotropic sequence beginning at nucleotide 262 of the env gene of one of the proviruses (NO1) that was uniformly deleted in the remaining proviruses. These observations were further assessed by phylogenetic analyses of the proviral sequences.
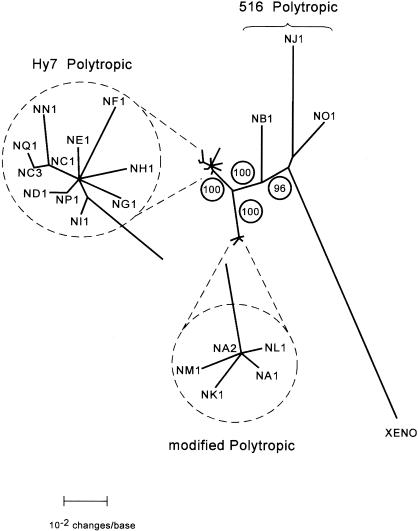
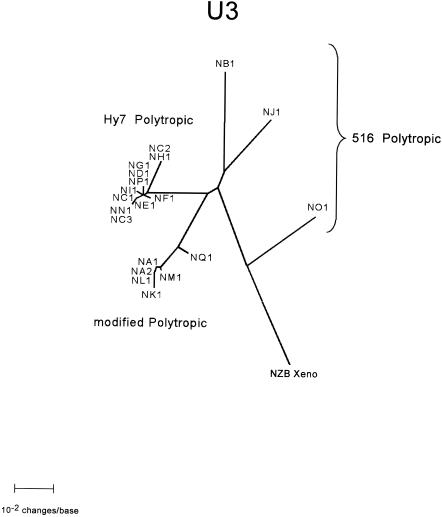
Phylogenetic analyses were performed on alignments of the xenotropic MLV (20, 37) and 19 polytropic MLV-related proviruses that could be distinguished by sequence heterogeneity within the region we have analyzed. The region compared in the analyses included the ∼900-bp PCR product (Fig. 2 and 3) extended to include the region of the env gene immediately past the 27-bp deletion present in mPT proviruses for a total of ∼1,100 bp. We found that the proviruses segregated into three distinct phylogenetic sets (clades) (Fig. 4). All members of the Hy 7 group segregated into a single closely related clade of PT proviruses, while proviruses of the 516 group segregated into two distinct clades. Several of the 516 proviruses segregated into a clade whose members were very closely related and comprised all mPT proviruses identified. The remaining 516 proviruses, corresponding to the PT members of this group, segregated into a clade that exhibited a much higher degree of divergence among its members than the Hy 7 PT or 516 mPT clades. Also included as the most divergent member of this clade was the xenotropic MLV. It is noteworthy that the xenotropic MLV, like the PT members of the clade, encodes the major determinant for MAb 516. The confidence limits of the phylogenetic segregation into the three clades were maximal by bootstrapping analysis. Moreover, essentially identical phylogenetic trees were obtained by using NJ (Fig. 4), maximum parsimony, maximum likelihood, and Bayesian analyses (with the MrBayes and PAUP programs). It is apparent from these analyses that the 516 PT proviruses reflect sequence characteristics of phylogenetic intermediates between the xenotropic MLV and the Hy 7 PT and mPT proviruses. The provirus present in clone NB1 appears to be somewhat more closely related to the Hy 7 PT and mPT proviruses than those in NJ1 and NO1.
FIG. 4.
Phylogenetic relationships of NFS/N endogenous proviruses. An unrooted phylogram generated by NJ phylogenetic analysis of a sequence encompassing a 3′ pol gene sequence and approximately 850 bases of the env gene is presented. This sequence corresponds to the ∼900-bp PCR product initially sequenced (Fig. 3) plus an additional ∼200 bp obtained by sequence analyses of the PCR products flanking the 27-bp deletion (Fig. 4). The analyses included the 19 proviruses distinguishable by sequence in this region as well as the analogous sequence from the xenotropic MLV NZB-9-1 (XENO). Branch lengths are drawn to scale. The identity of each clone is indicated at the branch termini. The very closely related clades containing the Hy 7 PT proviruses and the mPT proviruses are expanded (within dashed circles) in order to present the branch termini of these clades clearly. Group 516 PT viruses (NB1, NO1, and NJ1) are indicated by a brace. The robustness of the map was assessed by bootstrap analysis repeated 1,000 times. Bootstrap values are shown as circled numbers on the major branches.
Analyses of endogenous proviral LTRs.
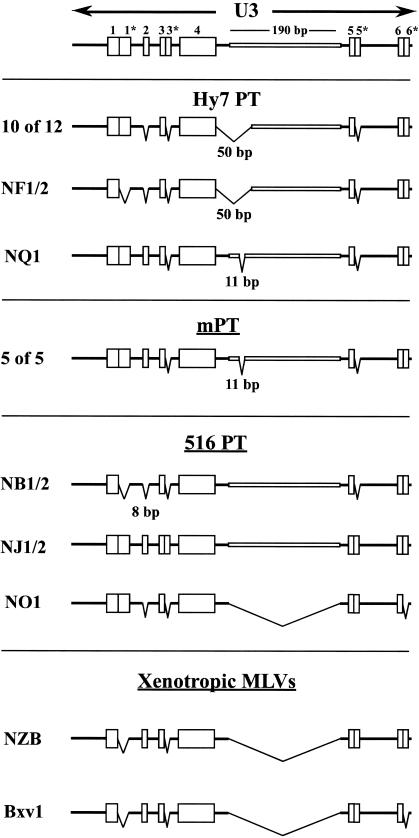
The LTRs of endogenous polytropic MLV-related proviruses exhibit a large insertion within the U3 region of the LTR relative to xenotropic or ecotropic MLVs. The insertion has a range of sizes due to deletions and is commonly referred to as the 190-bp insertion (23). The largest insertion described is 212 bp and may represent the complete undeleted insert (53). Additional polymorphisms consisting of directly repeated or unique sequences within the U3 regions distinguish several structural classes of polytropic MLV-related proviral LTRs that have been designated P-I through P-V by Tomonaga and Coffin (53) (Fig. 5). P-I and P-II correspond to U3 regions characteristic of PT and mPT MLV-related proviruses, respectively, originally identified in inbred mouse strains. The U3 regions of both the PT and mPT proviruses exhibit deletions when compared to the structures of additional polytropic MLV-related U3 regions identified in wild mouse subspecies as well as some laboratory strains of mice (P-III, P-IV, and P-V). It has been suggested that the latter structures may reflect properties of LTRs ancestral to mPT and PT LTRs (53). The deletion exhibited in mPT proviruses is 11 bp, whereas the PT proviruses have undergone a 50-bp deletion. PT and mPT U3 regions are further distinguished by a 5-bp sequence found in mPT but deleted in PT proviruses (Fig. 5, region 2).
FIG. 5.
U3 regions of endogenous polytropic MLV-related proviruses. Schematic diagrams are shown depicting distinguishing structural features of the U3 regions of polytropic MLV-related proviruses described by Tomanaga and Coffin (52, 53). The diagrams are adapted from similar diagrams presented in their reports. U3 types P-I and P-II correspond to the U3 regions found in PT and mPT proviruses, respectively, identified in inbred mouse strains (46). Types P-III, P-IV, and P-V correspond to U3 regions of proviruses identified in wild mouse subspecies and may reflect features that are ancestral to the P-I and P-II U3 regions. The regions exhibiting the distinguishing features, numbered 1 through 6, are indicated by boxes and include direct repeats (indicated by a repeated number with an asterisk), unique sequences, and deletions exhibited by the proviruses. In addition, the 190-bp insertion is indicated. V's extending down from the horizontal axes of the diagrams indicate regions that are absent from the various U3 types and include deleted sequences identified in P-I and P-II. The diagrams do not indicate the precise locations or sizes of the various structural features.
Ten of 12 Hy 7 PT proviruses exhibited U3 structures identical to that of the P-I U3 region characteristic of previously described PT proviruses (Fig. 6). One exceptional provirus found in the NF1 and NF2 clones did not exhibit a 14-bp direct repeat (designated region 1*) observed in P-I and P-II U3 structures. This repeat is also missing in P-IV and P-V structures (Fig. 5); however, this provirus has a U3 structure otherwise identical to the P-I U3 structure typical of PT proviruses. A second striking exception was observed with the NQ1 provirus. NQ1 contained an unambiguous PT receptor-binding region yet exhibited a P-II U3 region characteristic of an mPT provirus, suggesting that this provirus is a recombinant between PT env and mPT LTR sequences. Although recombinant proviruses of this particular type have not been described previously, other endogenous recombinant proviruses in which the env and LTR sequences are derived from different groups have been reported to be common in wild mouse subspecies (52). Further analyses of the recombinant nature of this provirus are described below.
FIG. 6.
Structural features of U3 regions of endogenous polytropic MLV-related proviruses identified in NFS/N mice. Structural features of the U3 regions of Hy 7 PT, mPT, and 516 PT proviruses identified in this report are presented. The features are indicated as described in the legend to Fig. 5. The structures of the U3 regions of two distinguishable xenotropic MLVs are also included in order to facilitate comparison to the structure of the 516 PT provirus NO1, which lacks the 190-bp insertion characteristic of all other polytropic MLV-related proviruses.
Analyses of the U3 regions of the 516 PT proviruses provided further evidence supporting the intermediate nature of these proviruses (Fig. 6). NB1 exhibited a unique U3 structure that is similar, but not identical, to the P-IV U3 region (Fig. 5). As noted above, the P-IV U3 region has been suggested to be ancestral to the P-I, P-II, and P-III U3 regions (53). The U3 region of NJ1 also exhibited a unique structure when compared to other polytropic MLV-related LTRs. The structure of NJ1 is identical to that of the schematic depicting the features that distinguish the various U3 structures of polytropic MLV-related proviruses (Fig. 5 and 6, top). This includes the 5-bp insert designated region 2, found in mPT proviruses but not PT proviruses. In addition, NJ1 exhibits a 4-bp direct repeat that has been identified in the Core enhancer sequence of a single endogenous xenotropic-mP recombinant provirus (52). Importantly, neither NB1 nor NJ1 exhibits the 50- or 11-bp deletion that is characteristic of the U3 regions of PT and mPT proviruses, respectively; instead, they retain the apparently complete 212-bp insert.
NO1, the remaining 516 PT provirus identified in this study, exhibited an unexpected U3 structure. The U3 of NO1 lacked the 212-bp insertion and, as such, would be considered to possess a xenotropic U3 region (Fig. 6). In addition, the U3 of NO1 did not exhibit a small duplication near its 3′ end (region 6*). This duplication is common to PT, mPT and most xenotropic MLVs but is not present in the U3 region of the xenotropic MLV Bxv1, which is the origin of the LTR acquired by MCF recombinant MLVs in AKR/J and HRS/J mice (33, 48, 52). It is possible that NO1 represents an endogenous recombinant provirus containing a polytropic env gene and a xenotropic LTR. However, other features of the NO1 U3 region distinguish it from all previously described xenotropic LTRs. In contrast to replication-competent xenotropic MLV isolates from laboratory strains, NO1 exhibits the 14-bp repeated sequence designated 1*, as do the PT, mPT, and 516 PT proviruses (Fig. 6). Furthermore, the NO1 U3 exhibits the 5-bp deletion of region 2 that is characteristic of PT proviruses but not of the U3 regions of mPT or xenotropic MLVs. Thus, the LTR of this provirus, like its associated env gene, exhibits structural features that are intermediate between those of xenotropic MLVs and those of PT and mPT proviruses.
Phylogenetic analyses of the proviral LTRs and assessment of possible endogenous recombinant proviruses.
Phylogenetic analyses of alignments of the U3 regions of the proviruses (Fig. 7) yielded results similar to those of the analyses of the receptor-binding regions (Fig. 4), with some notable differences. With the exception of clone NQ1, the Hy 7 proviruses segregated into a single closely related clade of PT proviruses, while the mPT proviruses segregated into a separate distinct clade. As noted above (Fig. 6), NQ1, which possesses a PT env sequence, exhibits a U3 structure characteristic of an mPT provirus and, as expected, segregated with the mPT clade. The 516 PT proviral U3 regions of NB1 and NJ1 occupy intermediate phylogenetic positions, in agreement with the analyses of the receptor-binding regions of the proviruses (Fig. 4) as well as their structural organization, presented above (Fig. 6). The U3 region of NO1 also occupied an intermediate phylogenetic position; however, in contrast to the analysis of the env gene region (Fig. 4), the U3 region of NO1 segregated into a separate clade with the xenotropic U3 region (Fig. 7).
FIG. 7.
Phylogenetic relationships of the U3 regions of NFS/N endogenous proviruses. An unrooted phylogram generated by NJ phylogenetic analysis of alignments of the U3 regions of the proviruses is presented. U3 sequences extending from the 5′ end of the U3 up to, but not including, the TATA box were analyzed. The analyses included all 20 distinct proviruses identified in our analyses as well as the analogous sequence from the xenotropic MLV NZB-9-1. Branch lengths are drawn to scale.
Several endogenous proviruses have been described as recombinant proviruses in wild mouse subspecies based on the linkage of env and LTR sequences from different groups (52). NQ1 appears to be a clear example of such a recombinant, with a PT env gene and an mPT LTR. NO1 may also be a recombinant provirus with a 516 PT env gene and a xenotropic LTR. Recombination involving the joining of sequences that have separate evolutionary histories and segregate into distinct clades should be evident by a shift from one clade to the other at the points of recombination.
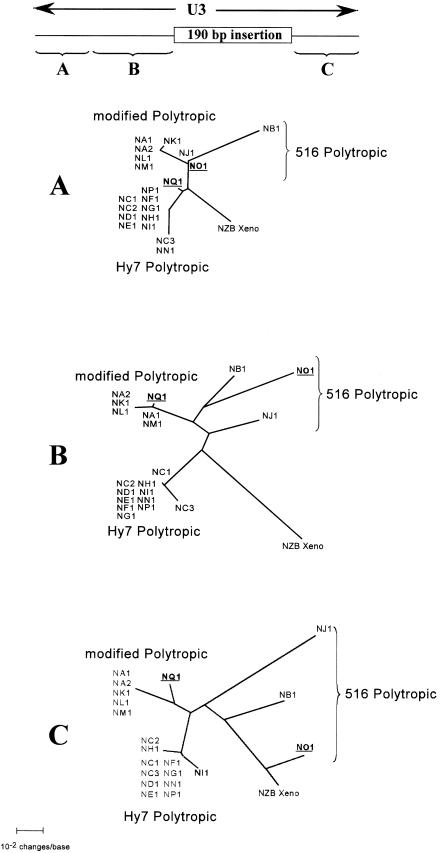
To assess the possibility of recombinant endogenous proviruses, the intervening env sequences between the ∼900 bp sequence and the U3 region were determined for 30 of the 32 clones included in this study. Determination of the NF2 and NA6 sequence was precluded by a truncation of the env gene during the cloning procedure. The sequences were aligned, and short (100- to 200-bp) segments of the alignment were subjected to phylogenetic analysis. This analysis yielded a likely recombination area for NQ1 within the 5′ portion of the U3 region. Throughout analysis of the entire envelope gene, the clones segregated into the three major clades consisting of Hy7 PT, mPT, or 516 PT proviruses quite similarly to their segregation in the analysis of the ∼900-bp region (Fig. 4 and data not shown). Notably, NQ1 segregated with the remaining Hy 7 PT proviruses, and NO1 segregated closely with NB1 and NJ1, the remaining 516 PT proviruses. These phylogenetic positions were also maintained in the first 100 bases of the U3 region (Fig. 8A); however, analysis of the adjoining 3′ sequences up to the point of the large insertion revealed a shift of NQ1 from the PT clade to the mPT clade (Fig. 8B).
FIG. 8.
Phylogenetic analysis of 5′ and 3′ segments of the U3 regions of NFS/N endogenous proviruses. Phylogenetic analyses were performed on alignments of two 5′ segments and a 3′ segment of the U3 region of the proviruses to assess shifts of putative recombinant provirus from one clade to another. The sequences included in the alignment are indicated by the braces in the diagram at the top of the figure, which is drawn to scale. Unrooted phylograms generated by NJ phylogenetic analysis of alignment of the segments of the U3 regions of the proviruses are presented. The analyses included all 20 distinct proviruses identified in our analyses as well as the analogous sequence from the xenotropic MLV NZB-9-1. The clones that are putative recombinant proviruses, NQ1 and NO1, are underlined and boldfaced for ease of identification. Branch lengths are drawn to scale.
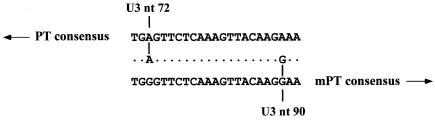
Examination of the alignment revealed a short sequence beginning with nucleotide 72 of the U3 region and extending to nucleotide 90 (Fig. 9). The consensus sequences of the PT and mPT proviruses differ at those positions, and NQ1 exhibits the PT consensus sequence from nucleotide 72 extending in the 5′ direction and the mPT consensus sequence from nucleotide 90 extending in the 3′ direction. Thus, it is apparent that recombination occurred within this short sequence. Similarly, NO1 segregated closely with NB1 and NJ1 in the 5′ U3 region (Fig. 8A and B) but exhibited a shift to a clade with the xenotropic virus in the analysis of the 3′ U3 region (Fig. 8C), suggesting that NO1 may also be a recombinant endogenous provirus. Examination of the alignment, however, did not reveal an obvious region of recombination. If NO1 is indeed a recombinant provirus between a 516 PT provirus and a xenotropic MLV, it is likely that recombination occurred in a region very near the 5′ border of the large insertion found in polytropic MLV-related proviruses. This could account for the lack of an insertion in the U3 of NO1 as well as its segregation with the remaining 516 PT proviruses in phylogenetic analyses of the 5′ portion of the U3 region. Examination of the alignment, however, did not reveal an obvious region of recombination near the 5′ border. The phylogenetic analysis was also performed using the xenotropic MLV sequence of Bvx-1, with essentially identical results (data not shown).
FIG. 9.
Recombination region in a PT env-mPT U3 endogenous recombinant provirus. An alignment of the consensus sequence of PT proviruses (top), the corresponding sequence of the putative recombinant provirus NQ1 (center), and the consensus sequence of mPT proviruses (bottom) is presented. A 17-bp sequence in the alignment is flanked by a PT provirus-specific base on the 5′ side and an mPT provirus-specific base on the 3′ side. Positions in the NQ1 sequence that are identical in both the PT and mPT consensus sequences are indicated by dots, and positions that are specific to the PT or mPT consensus sequence and shared by NQ1 are indicated by a vertical line connecting the base of NQ1 to the base of the matching consensus sequence. NQ1 sequences 5′ of the 17 bp exhibit nearly complete agreement with the PT consensus sequence, whereas sequences 3′ of the 17 bp exhibit nearly complete agreement with the mPT consensus sequence. Arrows directed toward the 5′ (PT) or 3′ (mPT) sequences are intended to convey this observation.
DISCUSSION
Polytropic MLV-related proviruses and the antigenic subclasses of polytropic MLVs.
Previous studies describing the antigenic subclasses of polytropic MLV isolates indicated that all polytropic MLVs from NFS/N or other mouse strains are reactive with MAb 516 or Hy 7 and that the reactivities to these antibodies correspond to alternate forms of a determinant that maps to a single amino acid in the SU protein (27, 28). One of the major goals of this study was to determine how the antigenic subclasses of polytropic MLVs are reflected in the endogenous polytropic MLV-related proviruses, particularly those that possess intact coding sequences for the receptor-binding region of their SU proteins. Previous reports have indicated that some of the proviruses contain defects, including defects in their env genes, that would preclude their participation by a simple recombination process in the generation of replication-competent polytropic MLVs (6, 24, 35, 46). It was possible that only a few of the endogenous proviruses would be candidates for the parents of recombinant viruses and that fewer still would possess the determinants for MAb 516 or Hy 7. Our results indicated that most (17 of 20) of the distinct proviruses identified in NFS/N mice contain intact sequences encoding the receptor-binding region of the SU protein. Furthermore, all of these encode the major determinant for either MAb 516 (eight proviruses) or Hy 7 (nine proviruses). Thus, there are a large number of endogenous proviruses that are potentially capable of donating sequences found in recombinant viruses corresponding to both antigenic subclasses. Our initial screening for proviruses that were not deleted in regions contributing to recombinant polytropic MLVs may have excluded a number of defective proviruses from our analyses. Nevertheless, it would appear that the majority of existing proviruses in NFS/N mice are potential parents of recombinant viruses.
Possible implications of identical mutations in different proviruses.
Among the proviruses that were apparently defective, it was noted that the same, presumably lethal mutations were identified in different proviruses. In one instance (ND1,) the provirus exhibited a tRNAIle rather than a tRNAMet codon as the initiator of the env gene, a mutation identical to those found in two other mouse strains (35, 46). In another instance, an identical termination codon in the env gene was identified in different lambda clones (Fig. 3, NF1/NF2 and NH1/NH2). These observations suggest the possibility that some viruses were introduced into the germ line as replication-defective viruses, probably through viral pseudotyping. This would, in turn, suggest that there were periods during their evolution in which many of the proviruses were replicating simultaneously in the host. In this regard, analyses of endogenous proviruses of wild mice suggest that polytropic MLV-related proviruses may have entered the germ line of mice prior to the speciation of Mus musculus and undergone extensive periods of replication (52, 53). Alternatively, identical mutations may have been introduced into different proviruses by gene conversion. Convincing evidence has recently been reported indicating gene conversion between 5′ and 3′ LTRs of human endogenous retroviruses (22). In view of the large number of retroviral elements in mammalian genomes, the possibility of recombination at the DNA level must be considered.
Phylogenetic relationships of polytropic MLV-related proviruses in NFS/N mice.
An unanticipated result of these analyses was the finding that the 516 PT proviruses differed extensively from the Hy 7 PT proviruses, particularly considering that the basis for grouping the proviruses relied on the identity of only one nucleotide (28). Analysis of the ∼900-bp sequence alignments that encompassed the receptor-binding region of the proviral env gene indicated that the 516 PT proviruses occupied a phylogenetic position intermediate between xenotropic proviruses, on the one hand, and the Hy7 PT and 516 mPT proviruses, on the other. This conclusion was strongly supported by analyses of the U3 regions of the proviruses. Two of the proviruses, NB1 and NJ1, exhibited large inserts of 212 bp and additional structural features similar to those of proviral U3 regions identified in wild mice that have been suggested as possible ancestors of the PT and mPT proviruses (53). The third 516 PT provirus exhibited env sequences that segregated as a phylogenetic intermediate but did not exhibit a large insertion in its U3 region and, as such, corresponded to a xenotropic U3 region. NFS mice reportedly have a single endogenous provirus containing a xenotropic LTR termed Nfxv-1 (20), and a single xenotropic MLV has been isolated from these mice (NFS-Thy-1). Restriction enzyme analyses of the envelope gene linked to the endogenous xenotropic LTR were not in concordance with analyses of the envelope contained in NFS-Thy-1, leading to the suggestion that NFS-Thy-1 was the result of recombination between the LTR of the endogenous virus and other sequences in the mouse. A portion of the env gene sequence and the LTR sequence of NFS-Thy-1 have been reported (23, 40) and are nearly identical to the sequence of the xenotropic MLV NZB. It is clear that the NFS-Thy-1 LTR does not correspond to the LTR identified in NO1, although it is quite likely that NO1 corresponds to Nfxv-1. Barring the existence of a second xenotropic LTR in NFS mice, the origin of NFS-Thy-1 is unclear. In this regard, the probe used to identify lambda clones containing polytropic MLV-related proviruses in this study would also hybridize to xenotropic env gene sequences.
Our analyses suggest that NO1 may be a recombinant between a 516 PT provirus and a xenotropic MLV, resulting in a loss of the large insertion. This is based on a shift toward the xenotropic MLV clade in phylogenetic comparisons of 5′ and 3′ segments of the U3 region. However, the 3′ U3 region of NB1 also appears to be more closely related to the xenotropic MLV, although to a lesser extent than NO1 (Fig. 8), suggesting that this may be a feature of the 516 PT proviruses. Rather than being a simple recombinant, NO1 could reflect features of an intermediate in a number of evolutionary schemes. It has been suggested that endogenous proviruses similar to NB1 or NJ1 are likely ancestral to the PT and mPT proviruses in a process that involved, among other changes, deletions in the U3 region of the LTR (53). Similarly, NO1 could represent an intermediate in an evolutionary process leading from a 516 PT provirus to xenotropic MLVs via a large (212-bp) deletion within the U3 region. Alternatively, NO1 could represent an earlier 516 PT provirus, possibly derived from a xenotropic MLV, that has not acquired the large inserted sequence characteristic of other polytropic MLV-like proviruses. In this regard it should be emphasized that the phylogenetic analyses do not establish an evolutionary direction progressing from xenotropic to polytropic MLV-related proviruses, as might be inferred from the phylograms (Fig. 4, 7, and 8). The common ancestor of the polytropic MLV-related proviruses and the xenotropic MLVs is not necessarily more closely related to xenotropic MLVs. The observation that the 516 PT proviruses reflect properties of phylogenetic intermediates between the xenotropic MLVs and the remaining polytropic MLV-related proviruses implies that the intermediates reflect properties of proviruses that were ancestral either to the Hy 7 PT and mPT proviruses or to the xenotropic MLVs, or conceivably to both groups (53).
The provirus isolated in clone NQ1 is a clear example of an endogenous recombinant provirus. We were able to unambiguously identify a 17-base sequence in the 5′ portion of the U3 as the region of recombination. The existence of endogenous recombinant proviruses likely reflects the simultaneous infection and replication of viruses of different groups during their evolution. However, one must again consider the possibility of gene conversion as an alternative to active replication (52).
Phylograms nearly identical to that shown in Fig. 6 were obtained from analyses of short segments of the sequences, including the analyses of sequences limited to pol gene or U3 sequences. Thus, it is apparent that the term “516 PT” has little descriptive value. Accordingly, we propose that the 516 PT proviruses be termed intermediate polytropic (iPT) proviruses. This nomenclature indicates their relationship to phylogenetic intermediates and distinguishes them from the PT and mPT proviruses. Furthermore, it serves to eliminate reference to the epitope, which is simply one of multiple structural features of this group.
It seems likely that at least some iPT proviruses participate in recombination to generate polytropic MLVs. The distribution of MAb 516- and Hy 7-reactive polytropic MLV isolates is about equal (28), yet few mPT isolates have been identified. Considering that the mPT proviruses uniformly encode the major determinant for the MAb 516 epitope, many of the MAb 516-reactive polytropic MLVs may be generated by recombination with iPT proviruses. In this regard, the polytropic FMLV FrNx (1) differs from the iPT provirus NB1 at only two positions in the ∼1,100-bp region we have analyzed. In contrast, FrNx differs from each of the PT and mPT proviruses at more than 30 positions in this sequence. As noted above, the provirus cloned in NB1 and NB2 carries an inserted A within the SU-encoding region, resulting in a frameshift mutation which introduces a termination codon. The PT sequences of FrNx are limited to sequences 5′ of this mutation, suggesting that the endogenous parents of polytropic MLVs do not require intact coding sequences outside of the region of recombination.
Identification of distinct proviruses by sequence heterogeneity.
One of the original goals of this study was to determine the extent of heterogeneity of the endogenous polytropic MLV-related provirus sequences in the minimal region of recombination and to identify single-nucleotide polymorphisms within this region that differentiate the proviruses. It is possible that the specificity of recombination observed with FMLV and MMLV (27, 28) simply reflects preferential replication of the ecotropic viruses in different cell types in which different endogenous proviruses are expressed (15, 42). However, we found that the specificity was dependent on sequences encoding the ecotropic nucleocapsid protein rather than on sequences one might expect to influence tissue tropism (27). This suggested that the specificity of recombination might reflect structural features of the endogenous proviruses rather than the tissue tropism of the ecotropic MLVs. A necessary prelude to the identification of features of the proviruses that might influence this specificity is the precise identification of the proviruses that actually participate in recombination. This determination remains elusive, due largely to the close homology of the endogenous sequences themselves and to the rate of evolution of retroviral sequences during viral replication. Furthermore, the region of comparison to assess the precise origin of the MLV sequences is limited to those sequences acquired by recombination and in many instances is restricted to 3′ pol and 5′ env gene sequences. A completely unambiguous identification of endogenous sequences participating in recombination requires the characterization of all of the endogenous proviruses in a mouse strain. Our analyses are not yet exhaustive, and there likely remain several endogenous proviruses not represented in our clones. Based on an estimate of 30 endogenous proviruses reactive with our probe and the 57 positive clones we have thus far isolated, we expect that approximately 85% of the endogenous viruses of NFS/N mice are represented in our clones. To achieve a 99% certainty that any individual provirus is represented will require the isolation of ∼135 clones. We are continuing to characterize the clones that did not yield 900-bp PCR products and are isolating additional clones to attain this level of confidence.
We found that almost all of the endogenous proviruses we have identified thus far can be distinguished by sequence heterogeneity within the minimal region of recombination found in polytropic MLVs. This includes 17 proviruses distinguishable within this region and 3 additional proviruses (NA2, NC2, and NC3) distinguishable by sequences immediately downstream of this region. Alignments of the proviruses reveal numerous single-nucleotide polymorphisms that are unique to individual proviruses as well as several that are diagnostic of each group of proviruses (Fig. 10). These markers can be detected in samples by a variety of techniques (2, 3, 34) and should greatly facilitate the identification of proviruses that participate in recombination. Furthermore, endogenous proviral genes are expressed in a tightly controlled fashion at various times and in various tissues of mice (5, 30, 31, 36, 54). The expression of these sequences may have a physiological role and certainly has physiological consequences in some mouse strains, such as those that exhibit a high incidence of leukemia. Little is known about the control of their expression or, in the case of polytropic proviruses, the precise identity of the proviruses involved. The polymorphisms defined in these analyses provide valuable markers for the detailed examination of provirus expression.
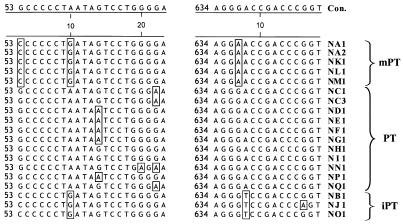
FIG. 10.
Single-nucleotide polymorphisms characteristic of PT, mPT, and iPT proviruses and of specific proviruses. Two windows in the alignment of 19 distinct polytropic MLV-related proviruses with their consensus sequence (Con.) are shown and illustrate examples of nucleotide polymorphisms. Numbers on the left of each window indicate the nucleotide position starting from the beginning of the initiation codon for env. Nucleotides differing from the consensus sequence are boxed. A single-nucleotide polymorphism specific for PT proviruses is located at position 60. Single-nucleotide polymorphisms characteristic of mPT proviruses are located at positions 53 and 637, and a single-nucleotide polymorphism characteristic of iPT proviruses is located at position 638 in NB1. Single-nucleotide polymorphisms specific for individual proviruses are found at positions 70 (NN1) and 646 (NJ1).
Acknowledgments
We thank Derrick Dimcheff, Karen Peterson, and Kim Hasenkrug for helpful discussions and Frank Malik for technical assistance.
REFERENCES
- 1.Adachi, A., K. Sakai, N. Kitamura, S. Nakanishi, O. Niwa, M. Matsuyama, and A. Ishimoto. 1984. Characterization of the env gene and long terminal repeat of molecularly cloned Friend mink cell focus-inducing virus DNA. J. Virol. 50:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi, W., and P. J. Stambrook. 1997. CCR: a rapid and simple approach for mutation detection. Nucleic Acids Res. 25:2949-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi, W., and P. J. Stambrook. 1998. Detection of known mutation by proof-reading PCR. Nucleic Acids Res. 26:3073-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosselman, R. A., F. van Straaten, C. Van Beveren, I. M. Verma, and M. Vogt. 1982. Analysis of the env gene of a molecularly cloned and biologically active Moloney mink cell focus-forming proviral DNA. J. Virol. 44:19-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britt, W. J., B. Chesebro, and J. L. Portis. 1984. Identification of a unique erythroleukemia-associated retroviral gp70 expressed during early stages of normal erythroid differentiation. J. Exp. Med. 159:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ch'ang, L. Y., W. K. Yang, F. E. Myer, C. K. Koh, and L. R. Boone. 1989. Specific sequence deletions in two classes of murine leukemia virus-related proviruses in the mouse genome. Virology 168:245-255. [DOI] [PubMed] [Google Scholar]
- 7.Chattopadhyay, S. K., M. W. Cloyd, D. L. Linemeyer, M. R. Lander, E. Rands, and D. R. Lowy. 1982. Cellular origin and role of mink cell focus-forming viruses in murine thymic lymphomas. Nature 295:25-31. [DOI] [PubMed] [Google Scholar]
- 8.Chattopadhyay, S. K., M. R. Lander, S. Gupta, E. Rands, and D. R. Lowy. 1981. Origin of mink cytopathic focus-forming (MCF) viruses: comparison with ecotropic and xenotropic murine leukemia virus genomes. Virology 113:465-483. [DOI] [PubMed] [Google Scholar]
- 9.Chien, Y. H., I. M. Verma, T. Y. Shih, E. M. Scolnick, and N. Davidson. 1978. Heteroduplex analysis of the sequence relations between the RNAs of mink cell focus-inducing and murine leukemia viruses. J. Virol. 28:352-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cloyd, M. W., J. W. Hartley, and W. P. Rowe. 1980. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J. Exp. Med. 151:542-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cloyd, M. W., M. M. Thompson, and J. W. Hartley. 1985. Host range of mink cell focus-inducing viruses. Virology 140:239-248. [DOI] [PubMed] [Google Scholar]
- 12.Donoghue, D. J., E. Rothenberg, N. Hopkins, D. Baltimore, and P. A. Sharp. 1978. Heteroduplex analysis of the nonhomology region between Moloney MuLV and the dual host range derivative HIX virus. Cell 14:959-970. [DOI] [PubMed] [Google Scholar]
- 13.Evans, L. H., and M. W. Cloyd. 1984. Generation of mink cell focus-forming viruses by Friend murine leukemia virus: recombination with specific endogenous proviral sequences. J. Virol. 49:772-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, L. H., and M. W. Cloyd. 1985. Friend and Moloney murine leukemia viruses specifically recombine with different endogenous retroviral sequences to generate mink cell focus-forming viruses. Proc. Natl. Acad. Sci. USA 82:459-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, L. H., and J. D. Morrey. 1987. Tissue-specific replication of Friend and Moloney murine leukemia viruses in infected mice. J. Virol. 61:1350-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankel, W. N., and J. M. Coffin. 1994. Endogenous nonecotropic proviruses mapped with oligonucleotide probes from the long terminal repeat region. Mamm. Genome 5:275-281. [DOI] [PubMed] [Google Scholar]
- 17.Frankel, W. N., B. K. Lee, J. P. Stoye, J. M. Coffin, and E. M. Eicher. 1992. Characterization of the endogenous nonecotropic murine leukemia viruses of NZB/B1NJ and SM/J. inbred strains. Mamm. Genome 2:110-122. [DOI] [PubMed] [Google Scholar]
- 18.Frankel, W. N., J. P. Stoye, B. A. Taylor, and J. M. Coffin. 1989. Genetic identification of endogenous polytropic proviruses by using recombinant inbred mice. J. Virol. 63:3810-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartley, J. W., N. K. Wolford, L. J. Old, and W. P. Rowe. 1977. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc. Natl. Acad. Sci. USA 74:789-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoggan, M. D., R. R. O'Neill, and C. A. Kozak. 1986. Nonecotropic murine leukemia viruses in BALB/c and NFS/N mice: characterization of the BALB/c Bxv-1 provirus and the single NFS endogenous xenotrope. J. Virol. 60:980-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishimoto, A., A. Adachi, K. Sakai, T. Yorifuji, and S. Tsuruta. 1981. Rapid emergence of mink cell focus-forming (MCF) virus in various mice infected with NB-tropic Friend virus. Virology 113:644-655. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, W. E., and J. M. Coffin. 1999. Constructing primate phylogenies from ancient retrovirus sequences. Proc. Natl. Acad. Sci. USA 96:10254-10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan, A. S., and M. A. Martin. 1983. Endogenous murine leukemia proviral long terminal repeats contain a unique 190-base-pair insert. Proc. Natl. Acad. Sci. USA 80:2699-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan, A. S., W. P. Rowe, and M. A. Martin. 1982. Cloning of endogenous murine leukemia virus-related sequences from chromosomal DNA of BALB/c and AKR/J mice: identification of an env progenitor of AKR-247 mink cell focus-forming proviral DNA. J. Virol. 44:625-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koch, W., W. Zimmermann, A. Oliff, and R. Friedrich. 1984. Molecular analysis of the envelope gene and long terminal repeat of Friend mink cell focus-inducing virus: implications for the functions of these sequences. J. Virol. 49:828-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavignon, M., and L. Evans. 1996. A multistep process of leukemogenesis in Moloney murine leukemia virus-infected mice that is modulated by retroviral pseudotyping and interference. J. Virol. 70:3852-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavignon, M., J. Richardson, and L. H. Evans. 1997. A small region of the ecotropic murine leukemia virus (MuLV) gag gene profoundly influences the types of polytropic MuLVs generated in mice. J. Virol. 71:8923-8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavignon, M., J. L. Walker, S. M. Perryman, F. G. Malik, A. S. Khan, T. S. Theodore, and L. H. Evans. 1994. Characterization of epitopes defining two major subclasses of polytropic murine leukemia viruses (MuLVs) which are differentially expressed in mice infected with different ecotropic MuLVs. J. Virol. 68:5194-5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, S. H., and J. B. Clark. 1997. High-yield method for isolation of lambda DNA. BioTechniques 23:598-600. [DOI] [PubMed] [Google Scholar]
- 30.Levy, D. E., R. A. Lerner, and M. C. Wilson. 1982. A genetic locus regulates the expression of tissue-specific mRNAs from multiple transcription units. Proc. Natl. Acad. Sci. USA 79:5823-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy, D. E., R. A. Lerner, and M. C. Wilson. 1985. Normal expression of polymorphic endogenous retroviral RNA containing segments identical to mink cell focus-forming virus. J. Virol. 56:691-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linemeyer, D. L., S. K. Ruscetti, E. M. Scolnick, L. H. Evans, and P. H. Duesberg. 1981. Biological activity of the spleen focus-forming virus is encoded by a molecularly cloned subgenomic fragment of spleen focus-forming virus DNA. Proc. Natl. Acad. Sci. USA 78:1401-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massey, A. C., S. C. Lawrenz-Smith, D. J. Innes, and C. Y. Thomas. 1994. Origins of enhancer sequences of recombinant murine leukemia viruses from spontaneous B- and T-cell lymphomas of CWD mice. J. Virol. 68:3773-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newton, C. R., A. Graham, L. E. Heptinstall, S. J. Powell, C. Summers, N. Kalsheker, J. C. Smith, and A. F. Markham. 1989. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17:2503-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikbakht, K. N., L. R. Boone, P. L. Glover, F. E. Myer, and W. K. Yang. 1987. Characterization of a molecular clone of RFM/Un mouse chromosomal DNA that contains a full-length endogenous murine leukaemia virus-related proviral genome. J. Gen. Virol. 68:683-693. [DOI] [PubMed] [Google Scholar]
- 36.Oliver, P. L., and J. P. Stoye. 1999. Genetic analysis of Gv1, a gene controlling transcription of endogenous murine polytropic proviruses. J. Virol. 73:8227-8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Neill, R. R., C. E. Buckler, T. S. Theodore, M. A. Martin, and R. Repaske. 1985. Envelope and long terminal repeat sequences of a cloned infectious NZB xenotropic murine leukemia virus. J. Virol. 53:100-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ott, D., and A. Rein. 1992. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70SU. J. Virol. 66:4632-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Portis, J. L., S. Czub, S. Robertson, F. McAtee, and B. Chesebro. 1995. Characterization of a neurologic disease induced by a polytropic murine retrovirus: evidence for differential targeting of ecotropic and polytropic viruses in the brain. J. Virol. 69:8070-8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Repaske, R., R. R. O'Neill, A. S. Khan, and M. A. Martin. 1983. Nucleotide sequence of the env-specific segment of NFS-Th-1 xenotropic murine leukemia virus. J. Virol. 46:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rommelaere, J., D. V. Faller, and N. Hopkins. 1978. Characterization and mapping of RNase T1-resistant oligonucleotides derived from the genomes of Akv and MCF murine leukemia viruses. Proc. Natl. Acad. Sci. USA 75:495-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosen, C. A., W. A. Haseltine, J. Lenz, R. Ruprecht, and M. W. Cloyd. 1985. Tissue selectivity of murine leukemia virus infection is determined by long terminal repeat sequences. J. Virol. 55:862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rulli, K., P. A. Lobelle-Rich, A. Trubetskoy, J. Lenz, and L. S. Levy. 2001. Tissue distribution and timing of appearance of polytropic envelope recombinants during infection with SL3-3 murine leukemia virus or its weakly pathogenic SL3ΔMyb5 mutant. J. Virol. 75:522-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruscetti, S., L. Davis, J. Feild, and A. Oliff. 1981. Friend murine leukemia virus-induced leukemia is associated with the formation of mink cell focus-inducing viruses and is blocked in mice expressing endogenous mink cell focus-inducing xenotropic viral envelope genes. J. Exp. Med. 154:907-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sijts, E. J., C. J. Leupers, E. A. Mengede, W. A. Loenen, P. J. van den Elsen, and C. J. Melief. 1994. Cloning of the MCF1233 murine leukemia virus and identification of sequences involved in viral tropism, oncogenicity and T cell epitope formation. Virus Res. 34:339-349. [DOI] [PubMed] [Google Scholar]
- 46.Stoye, J. P., and J. M. Coffin. 1987. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J. Virol. 61:2659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoye, J. P., and J. M. Coffin. 1988. Polymorphism of murine endogenous proviruses revealed by using virus class-specific oligonucleotide probes. J. Virol. 62:168-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoye, J. P., C. Moroni, and J. M. Coffin. 1991. Virological events leading to spontaneous AKR thymomas. J. Virol. 65:1273-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su, M. T., T. V. Venkatesh, and R. Bodmer. 1998. Large- and small-scale preparation of bacteriophage lambda lysate and DNA. BioTechniques 25:44-46. [DOI] [PubMed] [Google Scholar]
- 50.Thomas, C. Y., R. Khiroya, R. S. Schwartz, and J. M. Coffin. 1984. Role of recombinant ecotropic and polytropic viruses in the development of spontaneous thymic lymphomas in HRS/J mice. J. Virol. 50:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomonaga, K., and J. M. Coffin. 1998. Structure and distribution of endogenous nonecotropic murine leukemia viruses in wild mice. J. Virol. 72:8289-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomonaga, K., and J. M. Coffin. 1999. Structures of endogenous nonecotropic murine leukemia virus (MLV) long terminal repeats in wild mice: implication for evolution of MLVs. J. Virol. 73:4327-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, W. K., L. Y. Ch'ang, C. K. Koh, F. E. Myer, and M. D. Yang. 1989. Mouse endogenous retroviral long-terminal-repeat (LTR) elements and environmental carcinogenesis. Prog. Nucleic Acid Res. Mol. Biol. 36:247-266. [DOI] [PubMed] [Google Scholar]