Abstract
Social behaviors are often targets of natural selection among higher organisms, but quantifying the effects of such selection is difficult. We have used the bacterium Myxococcus xanthus as a model system for studying the evolution of social interactions. Changes in the social behaviors of 12 M. xanthus populations were quantified after 1,000 generations of evolution in a liquid habitat, in which interactions among individuals were continually hindered by shaking and low cell densities. Derived lineages were compared with their ancestors with respect to maximum growth rate, motility rates on hard and soft agar, fruiting body formation ability, and sporulation frequency during starvation. Improved performance in the liquid selective regime among evolved lines was usually associated with significant reductions in all of the major social behaviors of M. xanthus. Maintenance of functional social behaviors is apparently detrimental to fitness under asocial growth conditions.
Social behaviors play a central role in the life histories of many higher organisms, and consequently are often important components of evolutionary fitness (1). In social insects, the fitness of fertile females depends on the activities of nonreproductive workers (2). In lions, wild dogs, and hawks, cooperative hunting is a major means of resource acquisition (3–5). However, studying the action of natural selection on social cooperation is generally difficult because of the complexity of eukaryotic organisms, their social structure, and selective conditions in the wild (6–8).
Of central importance to the evolution of any set of traits (social or otherwise) are the rate and extent of heritable changes in those traits under various selective conditions. For example, imagine a parameter, C, that quantifies the level of cooperation within groups of wolves during their predation on moose, as occurs on Isle Royale in Lake Superior, MI (9). Further imagine that several groups of wolves are established on several different islands that vary only in the degree to which they favor cooperation during predation. At one extreme, the selective environment would contain only the healthiest moose at low density, making resource acquisition difficult and thus favoring extensive cooperation among wolves. At the other extreme, the wolves might be provided with an excess of freshly killed moose, thereby eliminating selection for cooperative predation. How much would the value of C change as the result of prolonged evolution in these different selective environments, and how quickly would this change occur?
The hypothesis that sociality will decrease during evolution under asocial conditions is intuitively appealing, but it is not necessarily true. In particular, the genes that encode social functions may have become so tightly integrated into the organism’s overall physiology and genetic regulation that the optimal expression of other genes, which encode more basic functions, requires the expression of social functions. If so, then loss of social functions may be detrimental even under asocial conditions. That such integration of seemingly dispensable genes can occur has been demonstrated, in another context, by several studies that measured the “cost” to bacteria of possessing genes that encode unnecessary resistance to antibiotics (10–15). In these studies, resistance genes reduced fitness, in the absence of antibiotic, when they were first introduced into the genome. But after the bacteria evolved with the resistance genes, they rapidly became integrated into the overall physiology of the cell, so that the cost of resistance was reduced or eliminated; in some cases, the resistance genes became beneficial to the evolved bacteria even in the absence of antibiotic. Thus, one cannot automatically assume that social functions will be costly to social organisms, because that assumption ignores the possibility that these functions have become integrated into the overall physiology and genetic regulation of the organism.
We have used a relatively simple social organism, the bacterium Myxococcus xanthus, to conduct an evolution experiment analogous to (the negative extreme of) the thought experiment described above. Myxobacteria are an unusual group of soil microbes that congregate as swarms, feed collectively, and sporulate via a process of multicellular development that involves differentiation into distinct cell types, only some of which survive to reproduce again (16). These social behaviors, along with rapid growth and ease of laboratory culture, make myxobacteria excellent model systems for studying the evolution of social interaction and development (17). In our study, replicate populations of M. xanthus were propagated in a physically unstructured, nutrient-rich liquid habitat in which their social behaviors were presumably not under positive selection (see below). After 1,000 generations of evolution, the extent of their adaptation to the selective regime was quantified, as were changes in the social traits of the derived populations. Before describing this study further, we first describe the social behaviors of M. xanthus in greater detail.
M. xanthus motility is regulated by two genetically independent systems (18). The first, “S” (social) motility, requires cell-to-cell proximity (roughly 5 μm) as well as extracellular pili and fibrils to operate (19, 20). The second system, “A” (adventurous) motility, allows movement of isolated individuals but is also stimulated by cell-to-cell proximity (18, 21, 22). The precise mechanisms of movement under these two motility systems are not fully understood. M. xanthus motility is central to its predatory activity, where cells collectively secrete a pool of degradative enzymes that hydrolyze the cell envelopes of prey bacteria (23). Macromolecules released by the prey are then collectively consumed by the M. xanthus.
In response to starvation, M. xanthus sporulates after the formation of multicellular fruiting bodies. This process is social because it requires communication between different reproductive organisms, whereas development in multicellular organisms occurs within distinct reproducing individuals. After ≈4 h of starvation, M. xanthus cells begin moving toward high-density aggregation points, where fruiting structures containing ≈100,000 cells are formed within 24 h. In the fruiting bodies, rod-shaped cells differentiate into spherical spores that are resistant to heat and desiccation as well as starvation, whereas some other cells do not sporulate and thus eventually starve. This developmental process requires at least five intercellular signals specific to various stages of development (24–26).
All of these cooperative behaviors are potentially subject to “cheating” by genotypes that reap the rewards of the group activities without paying the costs (27). In a physically structured environment, proximity among related individuals tends to favor cooperation because rewards accrue to the relatives of cooperating individuals and not to relatives of cheaters, thus giving rise to kin selection (28, 29). However, in the absence of physical structure, the rewards (if any) of cooperative behavior flow equally to all genotypes, regardless of either their own contributions or the contributions of their relatives; selfish behaviors are favored.
MATERIALS AND METHODS
Bacterial Strains.
M. xanthus strain DK1622 (30) is fully motile, developmentally proficient, and has been extensively characterized. A clone of DK1622 was designated strain S and is the ancestor of all strains described in this study. Strain R is a spontaneous rifampicin-resistant mutant of strain S. Lines S1, S2, S3, S4, S5, and S6 are descendants of strain S; lines R1, R2, R3, R4, R5, and R6 derive from strain R. The rifampicin-resistance marker allows strains to be distinguished during the experimental evolution (to rule out cross-contamination) and future competition experiments (between ancestral and evolved lines).
Selection Experiment.
Six replicate clones of strain S and six of strain R were inoculated into flasks with 10 ml of CTT liquid (a rich medium composed mainly of free amino acids and short peptides) (30). These 12 lines were grown to stationary phase (32°C, 120 rpm) and diluted 100-fold into fresh medium. Such dilution transfers were performed daily, with the resulting regrowth requiring ≈6.6 (= log2 100) generations of binary fission per day. Culture samples from each line were stored frozen (−80°C in 5% glycerol) after 1,000 generations (150 days) of growth.
Growth Rates.
We compared the maximum exponential growth rates of the ancestral strains and evolved populations after 1,000 generations with 3-fold replication. All strains were acclimated to the batch regime during two 24-h growth cycles in CTT liquid prior to the experimental growth cycle. To begin the experimental cycle, 0.1 ml of each acclimated culture was transferred into 9.9 ml of fresh medium. The total biovolume of the culture, which is equal to the cell number times the average cell size (31), was measured repeatedly with a Coulter Counter (model ZM) and Channelyzer (model 256). The maximum growth rate for each strain was calculated as the slope of log-transformed biovolume vs. time during exponential growth. This parameter is equivalent to the intrinsic rate of population increase, except that biovolume is used instead of the number of individual organisms.
Motility Rates.
Motility rates were measured on CTT plates containing either 0.5% or 1.5% agar, hereafter called soft and hard agar, respectively, at a volume of 0.28 ml of medium per cm2. Plate centers were inoculated with 10 μl of stationary phase liquid culture and incubated upside down for 72 h at 32°C, at which time swarm perimeters were marked. Perimeters were marked again after 72 h of incubation at 32°C, and the distance moved by the swarm perimeter during the second 72-h period was measured along a vector radiating from the plate center. The motility rate for each strain is expressed as the mean of three replicate independent assays.
Genetic Analysis of Motility Type.
A mutation that debilitates A motility (caused by a Tn5 lac insertion that encodes kanamycin resistance) was transduced from M. xanthus strain MXH1273 (32) via the myxophage Mx4 into single clones from each descendant line and one ancestral strain. Transductants were selected on CTT agar plates containing 40 μg/ml kanamycin. Transductant swarming phenotypes were observed on soft and hard agar CTT plates.
Developmental Morphology and Sporulation Frequency.
To examine the developmental morphology of the evolved lines and their ancestors, two replicate cultures per strain were centrifuged for 10 min at 12,000 × g and 4°C and resuspended in TPM buffer (33) at ≈5 × 109 cells/ml. From each resuspended culture, a 20-μl sample was inoculated onto 1.5% agar TPM developmental plates (34) and allowed to starve for 72 h, at which time photographs were taken at ×15 magnification through a dissecting microscope.
Sporulation frequencies were measured by using the same protocol as above, except that 100-μl aliquots of resuspended cultures were spotted onto TPM plates; the viable spore number was estimated after heat selection and dispersion of spores by sonication. After 72 or 168 h of starvation, cells were harvested into 0.5 ml of TPM liquid and subjected to heat (2 h at 50°C) and sonication (5 min in a horn sonicator at maximum output) to select for viable spores. These spore cultures were then diluted onto CTT hard agar plates. The plates were immediately overlaid with CTT soft agar to prevent motile colonies from swarming into one another after spore germination, thereby allowing accurate counts of distinct colonies. Sporulation frequencies reflect the proportion of cells in starving populations that had formed heat-resistant spores after 72 and 168 h of development. Estimates are the mean of two or three independent replicates.
RESULTS
Growth Rate in Liquid Batch Culture.
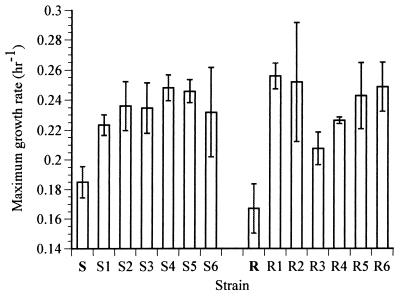
Six populations (S1–S6) derived from ancestral M. xanthus strain S and six populations (R1–R6) from strain R were inoculated into liquid growth medium and transferred daily for 150 days (see Materials and Methods), yielding 1,000 generations of evolution in the liquid environment. This environment was expected to select for increases in the maximum growth rate of cells (35). As shown in Fig. 1, all 12 evolved lines show significantly faster growth in the batch environment than do their common ancestors. This finding demonstrates that heritable adaptation to the batch regime occurred during the experimental evolution. The average improvement was ≈37%, with values ranging from 21% for line S1 all the way to 57% for line R1. This variation in growth rates among the evolved lines was statistically significant (see below), suggesting that they followed different adaptive pathways in the same selective regime.
Figure 1.
Maximum exponential growth rates of ancestral and evolved strains. Error bars indicate 95% confidence intervals.
The growth rate data suggest that the rifampicin resistance of ancestral strain R may impose a fitness cost (although this cost was not statistically significant). If so, then the increases in growth rate of the R-derived populations may be partially caused by evolutionary compensation for the cost of rifampicin resistance. The mean growth rate of the S-derived populations (0.240 h−1) is almost identical to that of the R-derived strains (0.239 h−1), suggesting that the R-derived lines improved more relative to their ancestor than did the S-derived lines. These results are consistent with the interpretation that R-derived lines underwent adaptation via both compensation for resistance and additional pathways for improvement that were also available to the S-derived lines.
Phenotypic Motility Pattern.
Shi and Zusman (36) have shown that A+S− and A−S+ motility mutants exhibit distinct motility patterns on hard and soft agar. A+S− mutants swarm relatively fast on hard agar but are effectively nonmotile on soft agar. A−S+ mutants show the opposite pattern, being swift on soft agar but very poor at swarming on hard agar. A+S+ cells swarm faster than either mutant type on both soft and hard agar. We measured the motility rates of our ancestral and derived lines (by using mixed population samples obtained at generation 1,000) on hard and soft agar as a phenotypic screen for motility mutants that may have swept through evolving populations.
Table 1 shows that 7 of the 12 derived lines possess the motility pattern expected for mutants that have lost social motility (A+S− phenotype). These seven lines (S2, S3, S4, S6, R2, R3, and R5) all showed minimal movement (attributable to normal colony expansion) on soft agar; in contrast, they continued to swarm significantly on hard agar, although at a slower rate than the ancestral strains. Three other lines (S1, R1, and R4) exhibited motile sectors on soft agar that extended outward from the main swarm perimeter, which was nonmotile. Motility rates for these three lines were measured along a vector radiating out through the nonmotile swarm perimeter rather than through the motile sector. The entire perimeter of line S5 swarmed on soft agar, but the expanding swarm was subdivided into sectors. The sectors in these four lines were presumably caused either by A+S+ cells present at low frequencies in the evolved populations prior to the motility test or by spontaneous mutation back to the A+S+ phenotype after placing cells on agar. If individual clones isolated from the evolved populations show motile sectors, then spontaneous mutation is the most likely cause of sectoring in the mixed populations. To test this possibility, the soft agar motility phenotype was observed for three clones from each of these four derived populations. In one case (S5), all three clones exhibited the same sectoring pattern as the mixed population, supporting the hypothesis of spontaneous mutation. However, none of the clones from the remaining three lines showed sectoring, suggesting that these populations still contained a low frequency of A+S+ cells at 1,000 generations.
Table 1.
Motility rates of ancestral and evolved lines on soft and hard agar
| Strain | Motility rate (mm/day) and significance level
|
Strain | Motility rate (mm/day) and significance level
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Soft agar | Hard agar | Soft agar | Hard agar | ||||||
| S | 5.94 | 1.98 | R | 5.33 | 2.02 | ||||
| S1 | 0.31 | ** | 1.14 | ** | R1 | 0.21 | ** | 1.43 | ** |
| S2 | 0.12 | ** | 0.88 | ** | R2 | 0.24 | ** | 1.41 | ** |
| S3 | 0.24 | ** | 1.07 | ** | R3 | 0.64 | ** | 1.12 | ** |
| S4 | 0.29 | ** | 0.95 | ** | R4 | 0.28 | ** | 1.43 | ** |
| S5 | 2.10 | ** | 1.52 | * | R5 | 0.24 | ** | 1.66 | NS |
| S6 | 0.24 | ** | 1.57 | NS | R6 | 1.05 | ** | 1.71 | NS |
Asterisks (or NS) indicate significance levels of differences between ancestral and evolved strains based on the Dunnett multiple comparisons test. ∗, P < 0.05; ∗∗, P < 0.01; NS, P > 0.05.
The remaining line (R6) showed significant although reduced swarming on soft agar by the entire colony perimeter without any obvious sectoring. However, the majority (518/560 = 93%) of clones observed from line R6 were not motile on soft agar. The remaining 7% were evidently sufficient to account for the soft agar motility exhibited by the mixed population sample.
All 12 descendant lines were motile on hard agar but, in most cases, at rates significantly slower than the two ancestors (Table 1). It has been shown that both A and S motility tend to be slower when the alternative motility system is nonfunctional (36), indicating that each system affects the function of the other. Therefore, it is likely that the reductions in hard agar motility observed in the evolved lines are pleiotropic effects of lost S motility rather than indicating a high cost of maintaining high-level A motility. If maintenance of functional A motility in the batch culture environment is detrimental to fitness, then there should be further reductions in A motility rate during continued evolution of lines that have already lost S motility.
Genetic Analysis of Motility Types.
Clones from each derived line were tested to determine whether their motility genotypes were indeed A+S−, as their inability to spread on soft agar suggested. A−S+ and A+S− mutant strains exist in which the motility defect is caused by a Tn5 lac insertion (37). To test for motility genotype, these Tn5 lac insertions can be transduced into a strain of interest. If an A− Tn5 lac insertion abolishes all motility in the recipient strain, then that strain is A+S−; if an S− insertion stops all motility on transduction, then the strain is A−S+. If neither insertion completely eliminates motility, then the recipient strain is A+S+, and some other genetic defect must be responsible for its motility phenotype.
Three clones from each derived line were chosen at random, all of which were nonmotile on soft agar except for one clone from line R6. We then attempted to transduce the Tn5 lac insertion that eliminates A motility from the A−S+ strain MXH1273 into these clones and their two ancestors. Several transductants were obtained from all 35 clones that were nonmotile on soft agar and from ancestral strain R. Further attempts to obtain transductants of ancestor S and the one other evolved clone were unsuccessful. Motility was abolished in all of the derived transductants, whereas transductants of the ancestral strain lost motility on hard agar but retained their motility on soft agar. These results imply that all of the evolved clones that are nonmotile on soft agar (i.e., all except one clone from line R6) are indeed A+S− genotypes.
Developmental Morphology and Sporulation Frequency.
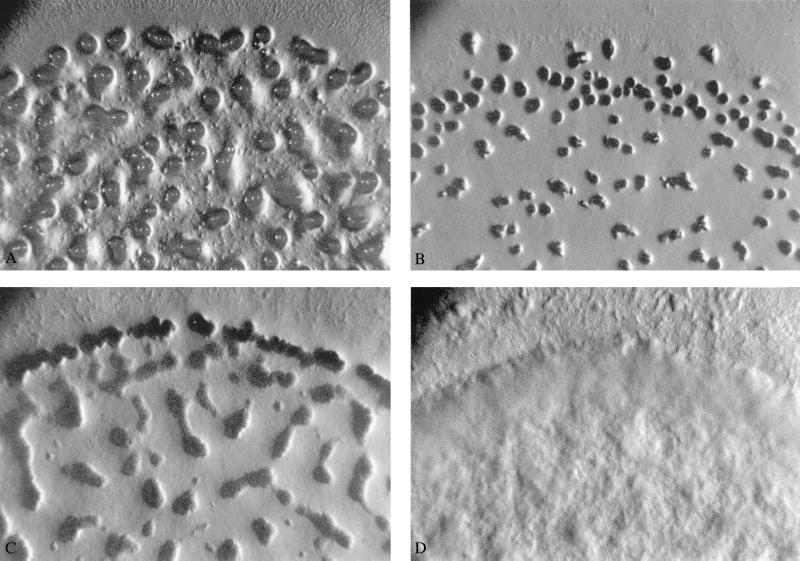
Table 2 indicates that only 3 of the 12 derived lines (S3, R4, and R6) showed formation of fruiting body structures after 72 h of starvation. Moreover, as exemplified in Fig. 2, the fruiting bodies of these three lines were small and irregular compared with those of the ancestors (Fig. 2 A–C), suggesting defective development. Fig. 2D shows the developmental morphology of line R3, which is representative of the 9 lines unable to form fruiting bodies.
Table 2.
Fruiting body phenotypes and sporulation frequencies (log-transformed) of ancestral and evolved lines
| Strain | Fruiting phenotype | Log (sporulation frequency)
|
Strain | Fruiting phenotype | Log (sporulation frequency)
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 72 h | 168 h | 72 h | 168 h | ||||||||
| S | + | −2.17 | −2.21 | R | + | −2.76 | −2.21 | ||||
| S1 | − | <−7.70 | ** | <−7.70 | ** | R1 | − | <−7.70 | ** | <−7.70 | ** |
| S2 | − | −6.53 | ** | −2.63 | * | R2 | − | <−7.70 | ** | <−7.70 | ** |
| S3 | + | −2.36 | NS | −2.20 | NS | R3 | − | <−7.70 | ** | <−7.70 | ** |
| S4 | − | −4.38 | * | −5.60 | ** | R4 | + | −4.29 | * | −2.14 | NS |
| S5 | − | −7.35 | ** | <−7.70 | ** | R5 | − | <−7.70 | ** | <−7.70 | ** |
| S6 | − | <−7.70 | ** | <−7.70 | ** | R6 | + | −3.39 | NS | −2.18 | NS |
+, Forms fruiting bodies; −, does not form fruiting bodies. Asterisks (or NS) indicate significance levels of differences between ancestral evolved strains based on the Dunnett multiple comparisons test. ∗, P < 0.05; ∗∗, P < 0.01; NS, P > 0.05.
Figure 2.
Developmental phenotypes of ancestral strain S (A) and evolved lines R4, S3, and R3 (B–D), respectively, at ×15 magnification.
Table 2 also shows that seven derived lines completely lost their ability to sporulate, whereas another line (S4) sporulated at a frequency several orders of magnitude below its ancestor. Only two strains (S3 and R6) retained sporulation frequencies comparable with the ancestral level at both 72 and 168 h, whereas two additional strains (S2 and R4) showed reduced sporulation at 72 h but attained near ancestral levels after 168 h.
In seven of the derived lines, the absence of fruiting bodies corresponded with sporulation frequencies at or near 0 (<10−7). Interestingly, two lines (S2 and S4) that were unable to form fruiting bodies nonetheless exhibited intermediate sporulation frequencies. The three lines that did develop fruiting structures (albeit irregular, see Fig. 2) sporulated at frequencies within an order of magnitude of their ancestors.
Variation Among Lines and Correlations Across Traits.
As shown in Table 3, each of the five quantitative traits that we measured in this study exhibited significant variation among the independently derived lines. Therefore, these data clearly indicate that the populations adapted to the asocial regime by different genetic and phenotypic pathways. This conclusion is further supported by the diverse combinations of trait values. In all 12 cases, the increased growth rates of evolved lines were associated with significant reductions in (or complete loss of) at least one of the major M. xanthus social behaviors. However, the degree of improvement in the liquid batch selective regime did not correlate with the extent of reduction in social motility, fruiting body formation ability, or sporulation frequency. For instance, lines S1 and R3 had the smallest growth rate gains among the 12 derived lines, yet both completely lost all three of these social traits. Alternatively, line R6 had one of the largest gains in batch performance as it kept some social motility, retained fruiting ability, and sporulated at ancestral levels. Furthermore, loss of social motility was not always associated with loss of fruiting ability or a major drop in sporulation frequency, as exemplified by lines S3 and R4. Finally, loss of fruiting ability did not necessarily entail a complete loss of sporulation, as seen in lines S2 and S4.
Table 3.
Summary of ANOVAs to assess heritable variation among replicate populations in five quantitative traits
| Trait | Ancestor | df | F | P |
|---|---|---|---|---|
| Maximum growth rate | S | 5, 12 | 9.128 | 0.0009 |
| R | 5, 12 | 15.504 | <0.0001 | |
| Motility rate: soft agar | S | 5, 12 | 57.613 | <0.0001 |
| R | 5, 12 | 21.867 | <0.0001 | |
| Motility rate: hard agar | S | 5, 12 | 13.762 | 0.0001 |
| R | 5, 12 | 3.574 | 0.0328 | |
| Sporulation frequency at 72 h | S | 5, 10 | 18.326 | <0.0001 |
| R | 5, 12 | 100.94 | <0.0001 | |
| Sporulation frequency at 168 h | S | 5, 11 | 970.21 | <0.0001 |
| R | 5, 12 | 412.38 | <0.0001 |
The numerator and denominator df depend on number of populations (six per ancestor in all cases) and the number of assays per population (usually three), respectively. A significant effect (P < 0.05) indicates heterogeneity among the populations above and beyond that which is caused by variability in the quantitative trait assays.
DISCUSSION
The importance of social interaction to the evolutionary fitness of many higher organisms is clear (1). However, the costs, and hence the fate, of such interactions in the absence of positive selection are difficult to determine experimentally with such organisms because of their longevity and size. By using a social bacterium, M. xanthus, we have demonstrated that genetically based cooperation rapidly degrades in an asocial selective environment. Indeed, performance gains in asocial selective conditions were often associated with the complete loss of one or more sophisticated social phenotypes, which strongly suggests that sociality is very costly to fitness under such conditions.
In principle, two distinct population genetic processes could explain the losses of social traits. One hypothesis is that, in the absence of selection for these social functions, they simply decayed by random drift of mutations that were strictly neutral in the asocial regime (38). The second hypothesis is that social functions are costly to maintain and perform, and so the bacteria gained a selective advantage under the asocial regime by getting rid of such functions. Under the first hypothesis, therefore, different mutations were responsible for improving growth in the liquid medium from those that caused the loss of social functions; under the second hypothesis, the gains and losses were pleiotropic effects of the same mutations. Random drift seems unlikely to be a sufficient explanation for our results, however, given the rapidity with which the losses occurred (1,000 generations) and the several independent traits that were repeatedly lost in the replicate populations. The more likely explanation is that these losses were caused by natural selection to rid the bacteria of functions that were both costly and unnecessary. Accordingly, the variation among the derived lines reflects the stochastic appearance of several different mutations that all improve fitness in the asocial regime while having heterogeneous effects on social functions (depending on the underlying genes). This interpretation is consistent with the substantial variability among the derived lines with regard to their fitness in the selective regime itself (Fig. 1 and Table 3). The drift and pleiotropy hypotheses are not mutually exclusive, so both may have contributed to these results. In future work, we will attempt to identify the specific mutations responsible for the loss of social functions, move them into the ancestral strain, and thereby test directly whether they are neutral or beneficial in the asocial batch culture environment. Preliminary experiments have already implicated a particular set of loci (involved in the production of pili) as responsible for the loss of social motility in several of our evolved strains (G.V., unpublished data).
Unfortunately, the ecology of M. xanthus in the wild is not well characterized. What are the precise contributions that M. xanthus social behaviors make to evolutionary fitness under various natural conditions? We know that, in the laboratory, social motility allows faster swarming on soft surfaces than does adventurous motility (36), but under what specific natural conditions does such gregarious motility confer an advantage (or disadvantage)? Also, what is the evolutionary benefit of maintaining a complex process of fruiting body formation when mutants exist (as we have found) that can sporulate without undergoing such development? Although these questions are beyond the scope of this study, our results do have some ecological implications. It is reasonable to assume that different natural populations of M. xanthus experience environments that differ in their growth regimes and physical structure. Our results suggest that natural populations which frequently encounter abundant resources or physically unstructured conditions are likely to exhibit lower levels of social cooperation than do populations that have adapted to resource scarcity or more structured habitats. Indeed, it would be very interesting to survey natural populations of M. xanthus for this variation and, moreover, to apply modern comparative methods (39) to examine the patterns of emergence and loss of social behaviors within the phylogeny of the myxobacteria and their relatives.
Additional evolution experiments are currently underway in our laboratories to investigate adaptation to selective conditions designed to favor the enhancement of M. xanthus social motility and developmental sporulation, respectively. In the future, we intend also to examine the evolutionary reversibility of the losses in social functions that were seen in this study. Finally, we hope that other groups will choose to pursue a direct experimental approach to study the evolution of social interactions by using other suitable organisms, such as the slime mold Dictyostelium discoideum (40, 41), and perhaps even certain social insects.
Acknowledgments
This research was funded by the National Science Foundation Center for Microbial Ecology (DEB-912006), by National Science Foundation Grant DEB-9421237 to R.E.L., by the Michigan State University Biotechnology Research Center, and by the Michigan Agricultural Experiment Station.
References
- 1. Krebs J R, Davies N B, editors. Behavioral Ecology: An Evolutionary Approach. 4th Ed. Cambridge, MA: Blackwell; 1997. [Google Scholar]
- 2.Wilson E O. The Insect Societies. Cambridge, MA: Harvard Univ. Press; 1971. [Google Scholar]
- 3.Scheel D, Packer C. Anim Behav. 1991;41:697–710. [Google Scholar]
- 4.Creel S, Creel N M. Anim Behav. 1995;50:1325–1339. [Google Scholar]
- 5.Bednarz J C. Science. 1988;239:1525–1527. doi: 10.1126/science.239.4847.1525. [DOI] [PubMed] [Google Scholar]
- 6.Shoemaker D D, Ross K G, Arnold M L. Evolution. 1996;50:1958–1976. doi: 10.1111/j.1558-5646.1996.tb03583.x. [DOI] [PubMed] [Google Scholar]
- 7.Ross K G, Vargo E L, Keller L. Proc Natl Acad Sci USA. 1996;93:3021–3025. doi: 10.1073/pnas.93.7.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross K G. Genetics. 1997;145:961–974. doi: 10.1093/genetics/145.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mech L D. The Wolf: The Ecology and Behavior of an Endangered Species. New York: Natural History Press; 1970. [Google Scholar]
- 10.Bouma J E, Lenski R E. Nature (London) 1988;335:351–352. doi: 10.1038/335351a0. [DOI] [PubMed] [Google Scholar]
- 11.Modi R I, Adams J. Evolution. 1991;45:656–667. doi: 10.1111/j.1558-5646.1991.tb04336.x. [DOI] [PubMed] [Google Scholar]
- 12.Lenski R E, Simpson S C, Nguyen T T. J Bacteriol. 1994;176:3140–3147. doi: 10.1128/jb.176.11.3140-3147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrag S J, Perrot V. Nature (London) 1996;381:120–121. doi: 10.1038/381120b0. [DOI] [PubMed] [Google Scholar]
- 14.Schrag S J, Perrot V, Levin B R. Proc R Soc London B. 1997;264:1287–1291. doi: 10.1098/rspb.1997.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Björkman J, Hughes D, Andersson D I. Proc Natl Acad Sci USA. 1998;95:3949–3953. doi: 10.1073/pnas.95.7.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, DC: Am. Soc. Microbiol.; 1993. [Google Scholar]
- 17.Zahavi A, Ralt D. In: Myxobacteria: Development and Cell Interactions. Rosenberg E, editor. New York: Springer; 1984. pp. 215–220. [Google Scholar]
- 18.Hodgkin J, Kaiser D. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 19.Wu S S, Kaiser D. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 20.Behmlander R M, Dworkin M. J Bacteriol. 1991;173:7810–7821. doi: 10.1128/jb.173.24.7810-7820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodgkin J, Kaiser D. Proc Natl Acad Sci USA. 1977;74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser D, Crosby C. Cell Motil. 1983;3:227–245. [Google Scholar]
- 23.Rosenberg E, Varon M. In: Myxobacteria: Development and Cell Interactions. Rosenberg E, editor. New York: Springer; 1984. pp. 109–123. [Google Scholar]
- 24.Hagen D C, Bretscher A P, Kaiser D. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 25.Downard J, Ramaswamy S V, Kil K-S. J Bacteriol. 1993;175:7762–7770. doi: 10.1128/jb.175.24.7762-7770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser D, Kroos L. In: Myxobacteria II. Dworkin M, Kaiser D, editors. Washington, DC: Am. Soc. Microbiol.; 1993. pp. 257–283. [Google Scholar]
- 27.Maynard Smith J. Evolution and the Theory of Games. Cambridge, U.K.: Cambridge Univ. Press; 1982. [Google Scholar]
- 28.Hamilton W D. J Theor Biol. 1964;7:1–52. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 29.Chao L, Levin B R. Proc Natl Acad Sci USA. 1981;78:6324–6328. doi: 10.1073/pnas.78.10.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaiser D. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenski R E, Bennett A F. Am Nat. 1993;142:S47–S64. doi: 10.1086/285522. [DOI] [PubMed] [Google Scholar]
- 32.MacNeil S D, Calara F, Hartzell P L. Mol Microbiol. 1994;14:785–796. doi: 10.1111/j.1365-2958.1994.tb01267.x. [DOI] [PubMed] [Google Scholar]
- 33.Bretscher A P, Kaiser D. J Bacteriol. 1978;133:763–768. doi: 10.1128/jb.133.2.763-768.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroos L, Kuspa A, Kaiser D. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 35.Vasi F, Travisano M, Lenski R E. Am Nat. 1994;144:432–456. [Google Scholar]
- 36.Shi W, Zusman D R. Proc Natl Acad Sci USA. 1993;90:3378–3382. doi: 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacNeil S D, Calara F, Hartzell P L. Mol Microbiol. 1994;14:61–71. doi: 10.1111/j.1365-2958.1994.tb01267.x. [DOI] [PubMed] [Google Scholar]
- 38.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]
- 39.Harvey P H, Pagel M D. The Comparative Method in Evolutionary Biology. Oxford, U.K.: Oxford Univ. Press; 1991. [Google Scholar]
- 40.Devreotes P N. Science. 1989;245:1054–1058. doi: 10.1126/science.2672337. [DOI] [PubMed] [Google Scholar]
- 41.Bonner J T. The Cellular Slime Molds. 2nd Ed. Princeton, NJ: Princeton Univ. Press; 1967. [Google Scholar]