Abstract
The papillomavirus replicative helicase E1 and the origin recognition protein E2 are required for efficient viral DNA replication. We fused the green fluorescent protein (GFP) to the human papillomavirus type 11 E1 protein either in a plasmid with the E1 coding region alone (nucleotides [nt] 832 to 2781) (pGFP-11E1) or in a plasmid containing both the E1 and E2 regions (nt 2723 to 3826) and the viral origin of replication (ori) (p11Rc). The former supported transient replication of an ori plasmid, whereas the latter was a self-contained replicon. Unexpectedly, these plasmids produced predominantly a cytoplasmic variant GFP or a GFP-E1^E4 protein, respectively. The majority of the mRNAs had an intragenic or intergenic splice from nt 847 to nt 2622 or from nt 847 to nt 3325, corresponding to the E2 or E1^E4 messages. pGFP-11E1dm and p11Rc-E1dm, mutated at the splice donor site, abolished these splices and increased GFP-E1 protein expression. Three novel, alternatively spliced, putative E2 mRNAs were generated in higher abundance from the mutated replicon than from the wild type. Relative to pGFP-11E1, low levels of pGFP-11E1dm supported more efficient replication, but high levels had a negative effect. In contrast, elevated E2 levels always increased replication. Despite abundant GFP-E1 protein, p11Rc-E1dm replicated less efficiently than the wild type. Collectively, these observations show that the E1/E2 ratio is as important as the E1 and E2 concentrations in determining the replication efficiency. These findings suggest that alternative mRNA splicing could provide a mechanism to regulate E1 and E2 protein expression and DNA replication during different stages of the virus life cycle.
Human papillomaviruses (HPVs) comprise a family of small DNA viruses that infect human keratinocytes in stratified epithelia at mucosal or cutaneous sites. Infections are typically subclinical or benign, but certain lesions can progress to cancers at a low frequency (81, 82). The viral genome consists of a double-stranded, circular DNA of approximately 7.9 kb, and it is normally maintained stably as autonomously replicating plasmids present at a low copy number in the basal cells of the epithelium. Vegetative reproduction depends on squamous differentiation, and elevated levels of viral transcription, DNA amplification, and virion morphogenesis occur sequentially in the lower and upper spinous strata of the epithelia. Progeny virus particles are then shed with the terminally differentiated, superficial cells. Because viral DNA replication depends largely on the cellular DNA machinery and entirely on host enzymes to generate deoxyribonucleoside triphosphate substrates, the viruses encode oncoproteins to reestablish an S-phase state in the postmitotic, differentiated cells (11, 12, 25).
The HPV and bovine papillomavirus (BPV) replication origin recognition protein and transcription factor E2 bind as a dimer with high affinity and specificity to multiple copies of a consensus E2 binding site (E2BS), ACCGN4CGGT, in the viral origin of replication (ori) (4, 9, 24, 36, 42, 46, 51, 60, 62, 75, 78a). The importance of E2 to replication in transfected cells is underscored by the requirement for one or more intact E2BSs and an E2 protein capable of binding to them (8, 41, 55, 71, 74). In vitro, the BPV type 1 (BPV-1) E2 protein prevents nucleosome formation around the ori, and this is critical for efficient assembly of a preinitiation complex (34). The other essential papillomavirus replication protein is the DNA helicase E1, which binds to its cognate site in the ori with low affinity and low specificity (27, 38, 39, 63, 78, 78a, 79). Efficient targeting of E1 proteins to the ori requires their direct interaction with E2 proteins bound to the E2BS that flank the E1BS. In vitro, E1 assembles on the ori as a hexamer or dihexamer (21, 38, 40, 61), a process greatly stimulated by the heat shock/chaperone proteins Hsp70 and Hsp40 (38, 40). The E1 dihexamer functions as a replicative helicase and is required throughout initiation and elongation of DNA replication (21, 38, 39). To establish the preinitiation complex, E1 recruits DNA polymerase α/primase and the single-stranded DNA binding protein RPA (3, 14, 23, 35, 45, 50). Efficient activation of initiation requires phosphorylation of E1 by the gatekeeper of S-phase entry, cyclin E/cdk2, which is recruited to the preinitiation complex by direct interaction with a motif in the amino-terminal domain of E1 (15, 37, 43). E2 protein is absent from the active replication complexes (39). In fact, the presence of E2 inhibits the E1 helicase, but the heat shock proteins are able to promote E2 dissociation from the DNA, relieve this repression, and enable de novo DNA synthesis (38).
It is thought that E1 and E2 are expressed in the basal and parabasal cells of the squamous epithelia to support extrachromosomal viral DNA maintenance replication as these cells enter S phase periodically. In productive infections, their expression is up-regulated in differentiated spinous strata to enable viral DNA amplification. While E1 functions mainly in replication, E2 also helps regulate promoters that control viral E6-E7 oncogene expression (12, 25). Moreover, the BPV-1 E2 protein tethers the viral DNA plasmids to mitotic chromosomes, enabling viral genome segregation in dividing cells (29, 33, 67, 77). HPV E2 proteins also play a role in viral DNA partitioning, but they mediate an association with the mitotic spindles rather than with mitotic chromosomes (B. A. Van Tine, L. Dao, T. M. Sonbuchner, B. Y. Lin, T. R. Broker, and L. T. Chow, unpublished data). Thus, the role of E2 protein changes during different stages of the virus life cycle. The exact E1/E2 ratio might then be expected to change correspondingly. Indeed, differentiation induces high levels of HPV type 31 (HPV-31) E1 and E2 transcription. The E1 mRNA reaches a greater abundance than E2 mRNAs (30, 49). Another study showed that elevated levels of an HPV-1 E1 expression vector increased transient ori DNA replication, while increasing levels of an E2 expression vector reduced replication (76). A slight repression of BPV-1 DNA replication by elevated E2 expression was also reported (44).
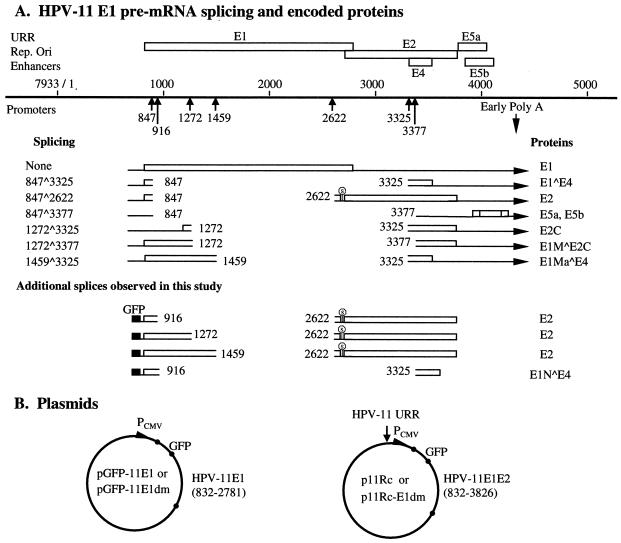
The primary transcripts for the HPV-11 E1 and E2 replication proteins extend from alternative 5′ ends to the early region polyadenylation addition sites (nucleotide [nt] 4390 or 4392) (Fig. 1A) (47). HPV-11 E1 protein is encoded by sequences spanning nt 832 to 2778 (termination codon, nt 2779 to 2781), while the E2 protein is encoded in a different reading frame from a dedicated initiation codon at nt 2723 to 2725 through nt 3823 (termination codon, nt 3824 to 3826). Thus, the E2 coding region partially overlaps the E1 gene. Embedded within the E2 gene in a different reading frame is a short E4 open reading frame (ORF) (nt 3231 to 3578; termination codon, nt 3579 to 3581). The E4 ORF is accessed through the dominant splice nt 847^3325. Translation of this message begins with the amino terminus of E1 (nt 832 to 847) and continues into E4 after the splice (Fig. 1A). In productively infected, hyperproliferative lesions, an unspliced mRNA capable of encoding the E1 protein is only rarely detected. The majority of this primary transcript is processed to generate the predominant E1^E4 mRNA species, whereas a much less abundant species containing the nt 847^2622 splice encodes the E2 protein (5, 6, 13, 47, 58, 59, 66, 69) (Fig. 1A). Spliced species that may encode other forms of E1 or E2 proteins have also been reported (7, 12, 58, 59) (Fig. 1A). Complex mRNA splicing patterns have also been observed in other mucosotropic HPVs (28, 57, 64, 65). Thus, alternative mRNA splicing might regulate the production of both the E1 and E2 proteins, thereby modulating viral DNA replication. Until now, there have been very limited investigations of this issue. One study showed that splicing within the HPV-31 upstream E6 ORF modulated E1 and E2 expression (26). Two other studies mutated the predominant splice acceptor site in the corresponding E2/E4 region or a minor donor site within E1 in genomic HPV-31. Neither mutant was able to maintain extrachromosomal viral DNA (31, 70). However, this phenotype could have been a result of indirect effects of the mutations on the production of proteins other than E1 and E2.
FIG. 1.
HPV-11 mRNA and plasmid constructs. (A) E region mRNAs identified in previous and present work and proteins encoded or inferred. A portion of the HPV-11 genome spanning the regulatory and E regions is represented as a solid line. For simplicity, only ORFs E1 through E5b are shown; they are depicted as open boxes, with splice sites marked by vertical arrows below the line. Numbers are nucleotide positions of the splice sites. Only the E region poly(A) site is shown. Below the solid line, mRNAs derived from the E1 promoter are diagrammed, with ORFs shown as open boxes. E5a and E5b, present in all E region mRNAs, are shown once. Proteins demonstrated or inferred are given to the right (except for E5a and E5b). Additional mRNAs with distinct 5′ ends but identical splices are not illustrated. Stop signs upstream of the E2 ORF indicate the termination of the presumptive native or GFP fusion peptide. The bottom four splicing species are identified in this investigation. The proteins inferred from these cDNA sequences are also given to the right. The GFP coding sequence is depicted as a filled box upstream. The transcription initiation site and poly(A) site of the expression vector are not shown. (B) Schematic representation of expression plasmids or replicons in this study. Circles represent the pEGFP-C1 vector backbone with the GFP coding region fused in frame with the HPV-11 E1 ORF. The expression clone contains either the HPV-11 E1 sequence (nt 832 to 2781) or the overlapping E1/E2 sequence (nt 832 to 3826) as indicated. The HPV-11 URR was inserted at the AseI site of the vector (arrow), which lies upstream of the CMV promoter (PCMV). The E1 donor mutation (E1dm) abolishes the consensus AGGT splice donor sequence at nt 847 without affecting the E1 amino acids encoded.
In this study, we fused green fluorescent protein (GFP) to the N terminus of HPV-11 E1 in a plasmid expressing E1 alone (pGFP-11E1) and in a replicon (p11Rc) that also expressed E2 from the native sequence context of the viral genome. We found that the majority of the transcripts spanning E1 were intragenically spliced within nt 847^2622 in the former and intergenically spliced within nt 847^3325 in the latter. Very little E1 mRNA or protein was produced. These conclusions were supported by Northern blotting for RNA, reverse transcription-PCR (RT-PCR) and sequence determination for cDNAs, Western blotting for proteins, and microscopic examination of the distribution of GFP. A mutation at the predominant splice donor site at nt 847 (E1dm) in the two plasmids abolished the respective dominant spliced species and greatly increased the expression of the nuclear GFP-11E1 protein. Low levels of pGFP-11E1dm supported highly efficient ori replication. But a negative effect was observed when high levels were transfected. In contrast, increasing the amount of the E2 expression plasmid always supported more-efficient replication. Relative to the wild-type replicon, the mutated replicon p11Rc-E1dm replicated less efficiently, perhaps due to an imbalanced E1/E2 ratio. We identified, by RT-PCR, several new but minor spliced transcripts that apparently can encode E2. Interestingly, replication of either of the replicons increased when complemented by an E2 expression vector, indicating that E2 protein was limiting. These results demonstrate that the E1/E2 ratio is as important as the absolute concentrations of the E1 and E2 proteins in determining the efficiency of viral DNA replication. We suggest that papillomaviruses use alternative mRNA splicing to regulate E1 and E2 protein expression and, through their coordinated production, DNA replication.
MATERIALS AND METHODS
Plasmids.
HPV-11 E1 and E2 expression plasmids pMT2-E1, pMT2-E2, and the origin-containing plasmid p7730-99 have been described previously (9, 32, 39). To construct pGFP-11E1, which expresses a GFP-E1 fusion protein (Fig. 1B), the HPV-11 E1 sequence (nt 832 to 2781) was amplified by Pfu Turbo DNA polymerase (Stratagene, La Jolla, Calif.) by using primers containing restriction enzyme recognition sequences at their ends and was inserted between KpnI and BamHI cloning sites of the pEGFP-C1 vector (Clontech Laboratories, Inc., Palo Alto, Calif.). The resulting fusion has 17 extra amino acids linking the two protein moieties. pGFP-11E1dm was constructed in the same way except that splicing donor site mutations spanning nt 847 were introduced into the 5′ primer (from nt 844 to 849, TCAGGT to TCTGGC) without affecting the coding capacity. The p11Rc replicon was obtained as follows. First, the upstream regulatory region (URR) of HPV-11 (nt 7070 to 7933/1 to 99) was inserted into the AseI site of the pEGFP-C1 vector upstream of the cytomegalovirus (CMV) promoter. The E1 and E2 coding sequences (nt 832 to 3826) were then inserted at the KpnI site in frame and downstream of GFP (Fig. 1B). Both the URR and E1-E2 DNA segments were amplified by Pfu Turbo DNA polymerase using primers containing the corresponding restriction enzyme recognition sequences. To generate the replicon p11Rc-E1dm, splice donor site mutations were introduced as described for pGFP-11E1dm. All PCR-amplified fragments were confirmed by DNA sequencing.
Cell cultures and transfections.
The monkey kidney epithelial cell line COS7, which expresses the simian virus 40 large T antigen, and human kidney epithelial cell line 293 transformed by adenovirus E1A and E1B regions were maintained and transfected by electroporation as described previously (9, 80). 293 cells were used for transient replication assays and Western blotting, whereas COS7 cells were transfected for direct fluorescence microscopy, Western blot analysis, and mRNA isolation.
Transient replication assays.
Transient replication assays were performed in 293 cells as described previously (9, 80). In assays using individual E1 and E2 expression plasmids, 0.5 μg of the HPV-11 origin-containing plasmid p7730-99 was cotransfected together with the indicated amounts of E1 and E2 plasmids. In assays using replicons, only the indicated amounts of replicons were transfected. In some experiments, an E1 or E2 expression plasmid was cotransfected with the replicon as indicated. Low-molecular-weight DNA was harvested 48 h posttransfection. One-half of the recovered DNA was digested with HindIII, which linearized the ori plasmid, while the other half was digested together with DpnI, which cut unreplicated input plasmids into small fragments. The digested DNAs were resolved in 0.8% agarose gels, Southern-blotted, and detected after hybridization with [32P]dCTP-labeled probes using the Megaprime DNA Labeling system (Amersham Biosciences Corp., Piscataway, N.J.). The probes were generated from either the ori plasmid p7730-99 or the pEGFP-C1 plasmid, the backbone of the replicons. The results were analyzed by PhosphorImager (Molecular Dynamics Inc., Sunnyvale, Calif.).
Direct fluorescence microscopy.
After electroporation, 105 COS7 cells were grown on two-well chamber slides (Nalge Nunc International, Naperville, Ill.) for 24 h, fixed at room temperature with 4% paraformaldehyde in phosphate-buffered saline (PBS), washed five times with PBS, and overlaid with coverslips by using mounting medium with DAPI (4′,6-diamidino-2-phenylindole) (Vector Laboratories, Inc., Burlingame, Calif.). Images were captured and processed with an Olympus AX70 fluorescence microscope with fluorescein isothiocyanate (FITC)/DAPI filters using an AxioCam digital camera (Carl Zeiss, Inc., Thornwood, N.Y.).
Antibodies and Western blotting.
Rabbit polyclonal antibodies against HPV-11 E1, E2, and E1^E4 were raised in our lab (7, 9). Mouse monoclonal antibody JL-8 against GFP was purchased from Clontech, Inc. For Western blots, COS7 cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [2-amino-2-hydroxymethyl-1,3-propanediol] [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride, and 1% protease inhibitor cocktail [Sigma-Aldrich Co., St. Louis, Mo.]) 24 h posttransfection. 293 cells were lysed in the same way 48 h posttransfection. Protein concentrations were determined by a Bradford assay using Protein Assay Dye Reagent (Bio-Rad Laboratories, Inc., Hercules, Calif.). Proteins from each sample (in the amounts given in figure legends) were resolved by electrophoresis through SDS-12% polyacrylamide gels, followed by immunoblotting with individual antibodies, and were detected by enhanced chemiluminescence (ECL) reagents (Amersham Biosciences Corp.).
mRNA isolation, Northern blotting, and RT-PCR.
COS-7 cells were harvested 24 h posttransfection. Total and cytoplasmic poly(A) RNAs were isolated using an Oligotex Direct mRNA Mini Kit (Qiagen Inc., Valencia, Calif.). Five hundred nanograms of mRNA from each sample was resolved in formaldehyde gels under conditions recommended by the supplier and transferred to a Hybond N+ nylon membrane (Amersham Biosciences Corp.) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [pH 7.2]). The membrane was washed with 5× SSC, cross-linked using a Bio-Rad GS Gene Linker, incubated at 42°C overnight in prehybridization buffer containing 50% formamide, 5× Denhardt's solution, 0.5% SDS, 200 μg of single-stranded DNA/ml, and 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), and hybridized with [α-32P]dCTP-labeled probes. The 32P-labeled probes specific for GFP or for E1 (nt 1072 to 2556) were prepared by nick translation. mRNA bands were detected by PhosphorImager.
To perform RT-PCR, mRNA samples were reverse transcribed with the Invitrogen SuperScript First-Strand Synthesis system. The cDNAs then served as templates for PCR amplification using Taq DNA polymerase (Invitrogen Co.) with different primer pairs, as indicated in each figure. Briefly, sense-strand primers were K (nt 5′ 613 to 634 of pEGFP-C1), M (nt 5′ 1333 to 1355 of pEGFP-C1), and P (nt 5′ 848 to 870 of HPV-11 E1), and antisense primers were N (nt 3′ 2757 to 2781 of HPV-11 E1), O (nt 3′ 1467 to 1488 of pEGFP-C1), Q (nt 3′ 3826 to 3803 of HPV-11 E2), and R (nt 3′ 3383 to 3400 of HPV-11 E2).
RESULTS
Multiple intragenic splices within the E1 transcript reduce the expression of E1 mRNA and protein.
Previously, we detected a very low level of native HPV-11 E1 or an epitope-tagged E1 protein in COS cells when E1 was expressed from the adenovirus major late promoter in the pMT2 vector (9, 32). To facilitate tracking of the expression and localization of E1, we constructed an expression plasmid for a GFP-E1 fusion protein under the control of the CMV immediately-early promoter (pGFP-11E1) (Fig. 1B). This protein was competent in supporting the transient replication of an HPV-11 ori plasmid at a level similar to that of pMT2-E1 when cotransfected with an HPV-11 E2 expression plasmid (see Fig. 4; also data not shown). However, to our surprise, the green fluorescence was localized throughout the cell, similar to that with GFP alone (Fig. 2A), despite the fact that E1 is known to be a nuclear protein (72).
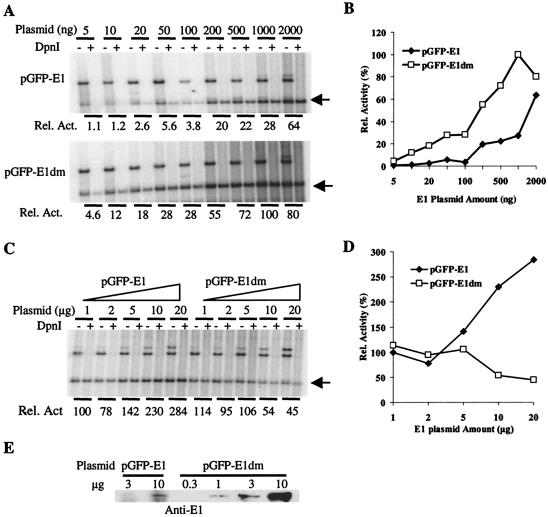
FIG. 4.
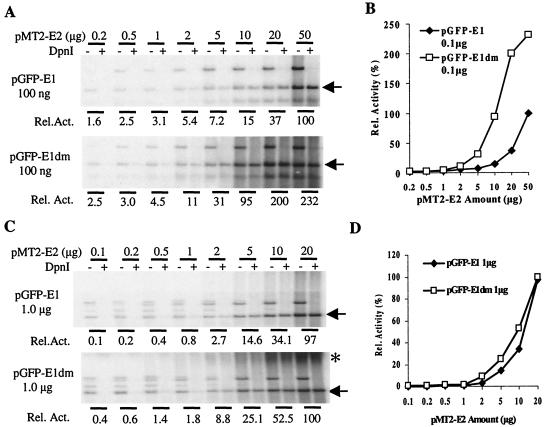
Effects on HPV-11 origin plasmid replication of increasing amounts of pGFP-11E1 or pGFP-11E1dm. Transient replication assays were performed in 293 cells as described in references 9 and 80. Five micrograms of plasmid pMT2-E2 was cotransfected with increasing amounts of E1 plasmid. (A and C) The low-molecular-weight DNA was digested either with HindIII alone (− lanes) or with both HindIII and DpnI (+ lanes), resolved in 0.8% agarose gels, Southern blotted, and then hybridized with 32P-labeled DNA probes to reveal the replicated ori plasmid (arrow). Bands above the arrow represent E1 and E2 expression plasmids, which were removed upon DpnI treatment. The replicated, linearized ori plasmids (arrow) in the DpnI + lanes were quantified and normalized by using ImageQuant (version 1.1; Molecular Dynamics) to obtain the relative replication activities. (B and D) The relative replication activities in panels A and C were plotted against the amount of input E1 or E2 plasmid, respectively. (E) E1 protein expression in 293 cells transfected with a range of expression plasmid amount. Western blotting was performed as described in the legend to Fig. 2B. Cells were harvested 48 h posttransfection, and 50 μg of the cell lysate was loaded in each lane.
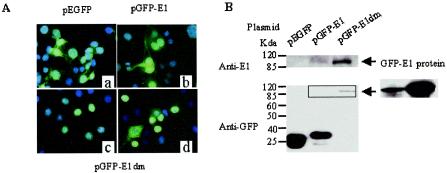
FIG. 2.
Detection of GFP and GFP fusion proteins in COS cells transfected with the vector pEGFP-C1, pGFP-11E1, or pGFP-11E1dm. (A) Localization of proteins by direct fluorescence microscopy of GFP. (a) pEGFP-C1; (b) pGFP11E1; (c and d) majority (c) and minority (d) of cells transfected with pGFP-11E1dm. Twenty-four hours posttransfection, cells were fixed with paraformaldehyde and stained with DAPI before mounting. Images were taken with a FITC/DAPI filter. (B) Western blots showing relative protein expression levels. The right panel represents the result of GFP-E1 detection after a long exposure time. Ten micrograms of each plasmid was separately transfected into 5 × 106 COS cells. Twenty-four hours posttransfection, cells were lysed with RIPA buffer. Portions (30 or 10 μg each) of the lysates were resolved by SDS-polyacrylamide gel electrophoresis and probed with an anti-E1 or anti-GFP antibody, respectively.
To discover the reason for the unexpected GFP distribution, we conducted Western blotting using either a polyclonal antibody to HPV-11 E1 or a monoclonal antibody to GFP. The E1 antibody detected a protein with a molecular mass of about 110 kDa, corresponding to full-length GFP-E1. Its level was very low but similar to that expressed from pMT2-E1 (Fig. 2B and data not shown). The anti-GFP antibody detected a dominant band of approximately 33 kDa, slightly bigger than GFP. Only after a prolonged exposure did we detect the full-length GFP-E1 (110 kDa) (Fig. 2B). What might be the nature of the 33-kDa variant GFP protein? There are three known mRNA splicing donor sites within the E1 region, at nt 847, 1272, and 1459, and one splicing acceptor site at nt 2622 (7, 12, 58, 59) (Fig. 1A). The E1 intragenic splice nt 847^2622 would generate a peptide of 29 amino acids which would start from the E1 AUG at nt 832 and terminate shortly after the splice at a position upstream of the E2 AUG. All but 5 of the 29 residues are encoded by sequences downstream of the splice in a frame different from E1. From the recombinant plasmid, this splice could generate a GFP fused to a 17-residue linker peptide plus the peptide of 29 residues. The size of this putative fusion protein corresponds exactly to the 33-kDa variant GFP observed. A much weaker band corresponding to GFP was also observed. It probably represents GFP generated upon proteolytic cleavage in the linker region of the fusion protein (Fig. 2B).
To substantiate our hypothesis, Northern blot hybridization was conducted using total or cytoplasmic poly(A) RNAs isolated from COS7 cells transfected with pGFP-11E1 or the pEGFP-C1 vector. When a GFP-specific probe was used, a single, highly abundant GFP mRNA of 1.0 kb (Fig. 3A, band 1) was observed in pEGFP-C1-transfected cells. In cells transfected with pGFP-11E1, we detected a rare full-length 3.0-kb GFP-11E1 transcript (Fig. 3A, band 2) and several more-abundant but shorter species: a minor 1.6- to 1.8-kb doublet (band 3) and a major 1.2-kb species (band 4), containing E1 sequences of about 600 to 800 and 200 bases, respectively. This would explain why the probe to a portion of the E1 region detected only the full-length transcript (Fig. 3A, right panel). These data suggest that alternative E1 intragenic mRNA splicing events occurred. For comparison, the full-length E1 mRNA generated from the pMT2-E1 plasmid was also of very low abundance, presumably due to the same intragenic splices (Fig. 3A).
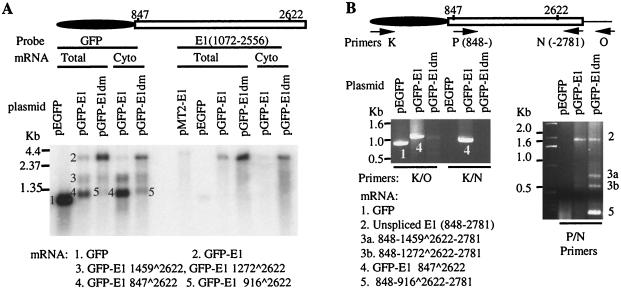
FIG. 3.
Spliced mRNA generated from pGFP-11E1 and pGFP-11E1dm in transfected COS7 cells. (A) Northern blots showing different spliced transcripts (bands 1 to 5). Twenty-four hours posttransfection, total or cytoplasmic mRNAs were isolated, and 0.5 μg of each sample was resolved in formaldehyde-agarose gels and hybridized with a 32P-labeled DNA probe specific either for the GFP sequence or for the E1 sequence (nt 1072 to 2556). The E1 probe detected only the full-length E1 ORF. The spliced species either were too rare or contained E1-specific sequences too short to be detected with this probe. The identity of each mRNA species in bands 1 to 5 was determined by RT-PCR (panel B) and is shown below. (B) Identification of splicing sites by RT-PCR. mRNAs were reverse transcribed and amplified with the indicated primer pairs. The 5′ nucleotide positions of the forward or reverse primers are indicated with arrows pointing in the 5′-to-3′ direction (see Materials and Methods). The splicing junctions were determined by direct sequencing of the products, designated as bands 1 to 5, each corresponding to one of the mRNA species detected in panel A.
To verify our interpretation, we performed RT-PCR. The bands recovered were given numbers corresponding to mRNA species detected in the Northern blots (Fig. 3A). The forward primer was specific for GFP (Fig. 3B, left panel, primer K), and the reverse primer was placed at the 3′ end of the E1 sequence (primer N). Only a strong, very short band was obtained (band 4). When the reverse primer was moved downstream in the vector (primer O), the product obtained was correspondingly longer (band 4). Sequence determination confirmed the same splice of nt 847^2622 in both RT-PCR products. In the control, only the combination of primers K and O, not primers K and N, generated the expected RT-PCR product from the cells transfected with the pEGFP-C1 vector (Fig. 3B, band 1, left panel). But we could not amplify either the full-length or the other spliced transcripts observed in the Northern blots. This is undoubtedly due to the preferential amplification of the shortest and predominant spliced species over the longer, rare ones. Additional transcripts were, however, recovered by using a forward primer placed immediately downstream of the dominant splice donor (Fig. 3B, primer P, right panel). Band 2, which was not always detected, had the predicted length of an unspliced species. Bands 3a, 3b, and 5 were new spliced species, and their identities will be discussed later. Based on these findings, we conclude that the primary GFP-11E1 transcript is alternatively spliced, that the most abundant, 33-kDa protein is a variant GFP protein encoded by a predominant mRNA resulting from an intragenic splice of nt 847^2622, and that a full-length GFP-11E1 mRNA and protein are very rare.
Mutation at the dominant splicing donor site at nt 847 abolishes the major spliced RNA and greatly increases E1 mRNA and protein expression
To construct an efficient expression vector for the GFP-11E1 protein, we mutated the splicing donor site at nt 847 without changing the coding sequence. pGFP-11E1dm was then transfected into COS7 cells. Northern and Western blotting, fluorescent microscopy, and RT-PCR were performed, and the results were compared with expression from the wild type pGFP-11E1 vector.
Northern blotting with a GFP probe showed that the 3-kb unspliced transcript was the predominant RNA species (Fig. 3A, band 2). There were also some minor shorter species of approximately 1.6 to 1.8 kb (band 3) and 1.25 kb (band 5). In particular, band 5 appeared to migrate slightly more slowly than band 4, which was generated by the wild type pGFP-11 clone. Hybridization with the E1-specific probe (Fig. 3A, right panel) confirmed a significant increase in levels of the full-length GFP-E1 mRNA from the vector containing the mutation.
RT-PCR was then performed using primers (K-O and K-N) (Fig. 3B, left panel) to span the whole GFP-E1 sequence. The result showed that the nt 847^2622 splicing was abolished in the pGFP-11E1dm mRNA preparation, because band 4, which was abundant in pGFP-11E1 mRNA, was no longer detected. With primers P and N, we obtained an unspliced, nearly full length E1 product (nt 848 to 2781) (band 2) as well as several shorter products (Fig. 3B, right panel). By DNA sequencing, three alternative intragenic splices were identified. The splices of nt 1459^2622 (band 3a) and of nt 1272^2622 (band 3b) were identical to those recovered from the wild-type vector, but the band intensities were stronger. In addition, band 5, which was the shortest and had the highest intensity, contained a splice of nt 916^2622. This splice was not detected in mRNA derived from the wild-type vector. Collectively, these results showed that the loss of the major spliced species of nt 847^2622 increased the unspliced transcript level and also potentiated the formation of minor or new alternatively spliced species relative to the RNA species generated from the pGFP-11E1 plasmid containing the wild-type E1 sequence.
Consistent with the above analyses on RNAs, Western blotting (Fig. 2B) using an anti-E1 antibody clearly showed that pGFP-11E1dm expressed a much higher level of GFP-E1 fusion protein than pGFP-11E1. When blots were probed with the anti-GFP antibody, full-length GFP-E1 was the predominant protein expressed from pGFP-11E1dm. In contrast, the 33-kDa variant GFP observed with pGFP-11E1 was no longer detected. These results indicate that intragenic splicing plays a significant role in regulating the level of E1 protein expression.
The GFP distribution as revealed by fluorescent microscopy was consistent with the Western blotting results. When cells were transfected with pGFP-11E1dm, GFP was localized exclusively to the nucleus in the majority of the cells (Fig. 2A). However, in about 20 to 30% of GFP-positive cells, there was still some cytoplasmic GFP, with or without nuclear GFP as well. This might be a result of GFP-E1 overproduction. Alternatively, the subcellular localization of E1 could be cell cycle regulated, but the cytoplasmic E1 could have escaped detection in the past because of the low level of native E1 protein expression. When cells were cotransfected with an HPV-11 E2 expression vector, bright green dots were observed in the nuclei (data not shown), consistent with a previous report showing colocalization of E1 and E2 in distinct nuclear foci (72).
HPV ori replication is differentially regulated by the relative amounts of E1 and E2 in transient replication.
We proceeded to examine the effects of the absolute and relative amounts of E1 and E2 proteins on the level of HPV ori replication in 293 cells by cotransfecting different amounts of E1 and E2 expression vectors. With small amounts of pGFP-11E1 and a fixed 5 μg of pMT2-E2, replication increased with increasing amounts of the E1 expression plasmid (Fig. 4A and B). In parallel, the pGFP-11E1dm plasmid was much more efficient than the wild-type pGFP-11E1 plasmid. Only 5 to 10 ng of pGFP-11E1dm was able to promote a detectable level of replication. In comparison, about 100 ng of the wild-type GFP-E1 plasmid was required to achieve a similar level of replication. Unexpectedly, with further increases in the amount of the pGFP-11E1dm plasmid to 0.5 to 1.0 μg or more, the extent of replication progressively decreased (Fig. 4A and B). These results were reproducible in several experiments (data not shown). To investigate this phenomenon, we repeated the experiments by using as much as 20 μg of the E1 plasmids in the presence of 5.0 μg of the E2 vector. Replication reached a plateau at approximately 1.0 μg of pGFP-11E1dm. Much beyond this input, replication decreased. In contrast, this decline was not observed with pGFP-11E1. The results of one of several such experiments are shown in Fig. 4C and D. To correlate the extent of ori replication with the level of E1 protein expression, Western blotting was performed with equal amounts of lysates of 293 cells transfected with a range of either E1 plasmid. Figure 4E shows that GFP-E1 protein was detected only when 10 μg of pGFP-E1 was transfected, but not with smaller amounts. In agreement with the results for COS cells, pGFP-E1dm generated higher levels of E1 protein, and 1 μg of plasmid expressed detectable GFP-E1 protein.
We then performed replication assays using increasing amounts of the E2 expression vector in the presence of 100 ng input of either pGFP-11E1 or pGFP-11E1dm, a level at which no negative effect of the pGFP-11E1dm was observed (Fig. 5A and B). Replication consistently increased with increasing levels of the E2 plasmid. Even at 50 μg of the E2 plasmid, we observed no repression. In addition, GFP-E1dm was always more efficient than GFP-E1 because of the higher levels of E1 protein produced by the mutated vector. These results were reproducibly observed. We also examined the effects of increasing the amount of the E2 expression plasmid in the presence of 1 μg of either of the E1 plasmids, a level at which replication by pGFP-E1dm peaked in the previous experiments (Fig. 4). The results showed that replication increased with E2 input (Fig. 5C and D). Although pGPF-E1dm was still more effective than pGFP-E1, the difference was not as dramatic as that in experiments in which 0.1 μg of the E1 expression vector was used. At the highest input level of E2 plasmid tested, the two E1 expression vectors supported comparable replication (Fig. 5C and D). However, as the amount of the E2 plasmid increased, a significant amount of slow-migrating, DpnI-resistant ori DNA was detected in cells transfected with pGFP-E1dm, but not in cells transfected with pGFP-E1. This material cannot be attributed to incomplete digestion of restriction enzymes, because an input of expression plasmids as high as 20 μg was completely digested.
FIG. 5.
Effects on HPV-11 origin plasmid replication of increasing amounts of pMT2-E2 plasmids. Transient replication assays were performed and data were processed as described in the legend to Fig. 4. One hundred nanograms (A and B) or 1 μg (C and D) of pGFP-11E1 or pGFP-11E1dm was contransfected with increasing amounts of the E2 plasmid. Asterisk marks the high-molecular-weight, newly replicated DNA of unknown structures.
Collectively, these results demonstrate that the absolute amounts of E1 and E2 proteins and their ratios are both important in determining the efficiency of HPV ori replication. Beyond certain optimal E1/E2 ratios, high levels of E1 protein do not generate more completely replicated ori DNA. In contrast, increasing E2 levels always leads to higher replication when E1 levels are low, suggesting that E2, rather than E1, is limiting in these replication assays.
Multiple spliced RNA species are generated from a replicon containing the genomic E1 and E2 sequences.
It is possible that, with the separate expression vectors, the replication experiments described above did not reflect the true relative E1/E2 ratio normally achieved by the virus. To simulate the viral genome, we constructed a replicon, p11Rc (Fig. 1B), which contained the overlapping genomic E1 and E2 coding sequences (nt 832 to 3826). GFP was fused in frame with E1 as in pGFP-11E1. Into the plasmid, the 1-kb HPV-11 URR region (nt 7070 to 7933/1 to 99) spanning the ori was inserted upstream of its CMV promoter, which controls the expression of the viral proteins. In productively infected HPV-11 lesions, the primary transcript ending at the E region poly(A) site gives rise to E1, E2, E1^E4, E2C (i.e., E8^E2C), and possibly other proteins by alternative splicing. The p11Rc replicon contained all known splicing donor sites (nt 847, 1272, and 1459) and acceptor sites (nt 2622, 3325, and 3377) in the E region. We anticipated that the replicon would encode a GFP-E1 protein from an unspliced mRNA and a native E2 protein from a transcript with a splice of nt 847^2622.
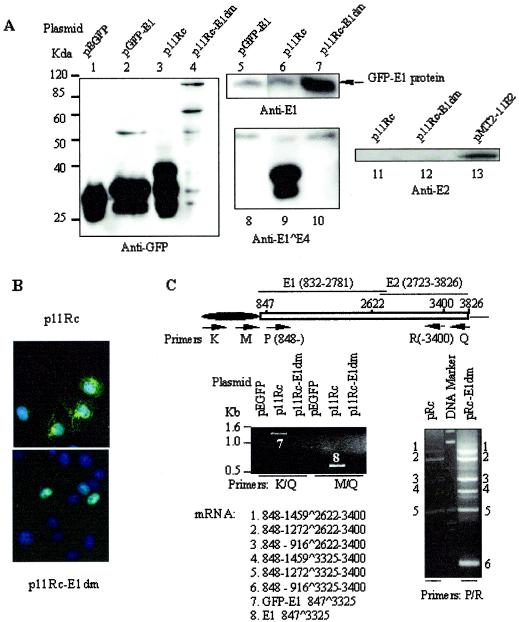
The p11Rc replicon, pEGFP-C1, and pGFP-11E1 were separately transfected into COS7 cells. Lysates were analyzed by Western blotting in parallel. No GFP-E1 protein was detected from the replicon with the anti-GFP antibody except after prolonged exposure (Fig. 6A, lane 3; also data not shown). Rather, the antibody revealed three lower-molecular-mass bands. The fastest-migrating band had a mobility corresponding to that of GFP (Fig. 6A; compare lanes 1 and 3); it was much weaker than the other two at a shorter exposure and could be a result of cleavage in the linker between the GPF and fusion protein moieties (data not shown; but see Fig. 2B, lower panel). The middle band had a mobility similar to that of the variant GFP detected in pGFP-11E1-transfected cells (Fig. 6A; compare lanes 2 and 3), but it was not this protein (see below). The slowest and by far the most abundant of the three bands had the mobility of a GFP-E1^E4 protein (40 kDa) predicted from an mRNA with a splice between the dominant splice donor and acceptor sites, nt 847^3325.
FIG. 6.
Protein expression and mRNA splicing from replicons p11Rc and p11Rc-E1dm in COS7 cells. (A) Western blots showing the relative protein expression levels of GFP-E1, GFP-E1^E4, and native E2. A total of 5 × 106 cells were transfected with 10 μg of each plasmid as indicated above each lane. Lysates from cells transfected with pEGFP, pGFP-E1, or pMT2-E2 were used as controls. Cells were lysed with RIPA buffer after 24 h. Portions (30 μg each) of the lysates were resolved by SDS-polyacrylamide gel electrophoresis, Western blotted, and then probed with antibodies to E1 or E2. Portions (10 μg each) of the lysates were probed with an anti-GFP or anti-E1^E4 antibody. (B) Distribution of GFP-tagged proteins as detected by direct fluorescence microscopy, as described for Fig. 2A. (C) Identification of mRNA splices by RT-PCR as described for Fig. 3B. The primer pairs used are given below the gels.
To identify this trio of low-molecular-weight GFP proteins, the lysates were Western blotted and probed with a polyclonal antibody to the HPV-11 E1^E4 protein (Fig. 6A, lanes 8, 9, and 10). The antibody detected the middle and upper bands of the three revealed by the anti-GFP antibody. In a shorter exposure, the faster band was much weaker than the slower GFP-E1^E4 band (data not shown). We suggest that the faster band was primarily derived from the GFP-E1^E4 protein, not the 33-kDa variant GFP resulting from the nt 847^2622 splice detected in pGFP-11E1-transfected cells (Fig. 6A, lane 2), because the variant GFP could not be detected by the anti-E1^E4 antibody (Fig. 6A; compare lanes 2 and 8). Furthermore, fluorescence microscopy showed that GFP in the p11Rc-transfected cells formed a filamentous network exclusively in the cytoplasm (Fig. 6B), typical of the E1^E4 proteins of mucosal HPVs (5, 6, 52, 56). In contrast, the 33-kDa variant GFP was diffuse in appearance and was observed in both the cytoplasm and the nucleus, like GFP (compare Fig. 6B, top panel, with Fig. 2Aa and b).
Consistent with these interpretations, RT-PCR with primer pair K-Q or M-Q detected only one product, the spliced species of nt 847^3325 (Fig. 6C, bands 7 and 8). However, the predominance of this splice in the shortest of all the messages prevented the detection of any longer but much less abundant spliced mRNAs, such as E2 mRNA with the splice of nt 847^2622. To detect minor spliced species, we used a forward primer (primer P) immediately downstream of the splice donor site at nt 847. When forward primer P was used in conjunction with reverse primer R, five faint products were detected. The identities of these species are discussed in the following section.
A replicon with a mutation at the dominant splice donor site at nt 847 abolishes the E1^E4 mRNA and increases E1 mRNA production as well as that of alternatively spliced species.
To test how a donor mutation would affect the production of both the E1 and E2 proteins and hence DNA replication, we introduced the same donor mutation at nt 847 as that present in pGFP-11E1dm into the replicon (p11Rc-E1dm). RT-PCR of transcripts from p11Rc-E1dm-transfected COS7 cells revealed the successful elimination of the dominant splice of nt 847^3325 (Fig. 6C, left panel). Western blotting showed that the major GFP fusion protein was full-length GFP-E1, which was much more abundant when generated by p11Rc-E1dm than when generated by p11Rc (Fig. 6A; compare lanes 3 and 6 with 4 and 7). The trio of low-molecular-weight GFP fusion proteins was below detection (Fig. 6A, lanes 4 and 10). In agreement with these findings, fluorescence microscopy revealed that GFP was primarily nuclear, as expected of the nuclear GFP-E1 protein (Fig. 6B).
We then performed RT-PCR to determine the spliced species generated by the p11Rc-E1dm replicon which could encode E2, in parallel with those generated by p11Rc. The sense-strand primer P (nt 5′ 848 to 870) (Fig. 3B and 6C) and the reverse primer R (nt 3′ 3383 to 3400) were used. This reverse primer is located downstream of all known splice acceptors, but it reduces the lengths of PCR products, facilitating their recovery. A number of bands from p11Rc-E1dm transcripts were detected (Fig. 6C). The sequences of six of the more prominent bands were determined. Each was a spliced species: 1459^2622, 1272^2622, 916^2622, 1459^3325, 1272^3325, and 916^3325. The first five spliced species were also observed in RNA from p11Rc, but the bands were much weaker (Fig. 6C). This is because the majority of the transcripts from p11Rc were the E1^E4 species (nt 847^3325), which was not amplified by the primer pair used. Sequence analysis showed that translation from all three spliced species using the nt 2622 acceptor site terminates upstream of the E2 initiation codon at nt 2723. Ribosome reinitiation could then give rise to a native E2 protein, as it does in the known E2 mRNA containing the nt 847^2622 splice. At present, we do not know which of the three alternative species encodes E2 in vivo. The nt 916^3325 splice fuses the amino-terminal 28 amino acids of E1 to the E4 frame; we call this product E1N^E4 (Fig. 1A). We attribute the high relative abundance of this RNA species in RT-PCR to preferential amplification of this shortest template. In actuality, GFP-E1N^E4 must be present in very small amounts, since the E4 antibody did not detect any band of the expected size (Fig. 6A, lane 10).
Interestingly, we could not detect E2 protein from either p11Rc-transfected or p11Rc-E1dm-transfected COS7 cells. For comparison, E2 from lysates of cells transfected with pMT2-11E2 was readily detected (Fig. 6A; compare lanes 11 and 12 with lane 13). Nevertheless, we infer that E2 was expressed from the wild-type and mutated vectors, because both replicated in 293 cells in the absence of a separate E2 expression vector, as described below. Collectively, these results suggest that the E2 messages generated from the genomic context are of reduced abundance or are translated less efficiently than a dedicated E2 mRNA from pMT2-E2. But part of the distinction might also be attributable to a difference in promoter strength between these vectors.
p11Rc replicates more efficiently than p11Rc-E1dm.
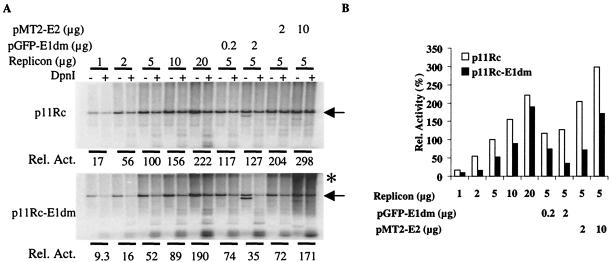
Transient replication assays with both p11Rc plasmids were performed in 293 cells (Fig. 7). With a wide range of transfected DNA amounts, the full-length, linearized product generated from p11Rc was always more abundant than that from p11Rc-E1dm. These results suggest that E2 might be expressed at a reduced level from the mutated replicon relative to expression from the wild-type vector. To test this hypothesis, we complemented the replicon with either an E1 or an E2 expression vector. When 5 μg of either replicon was cotransfected with 0.2 μg of pGFP-11E1dm, replication increased only by a moderate amount, indicating that the E1 protein was not limiting (Fig. 7). However, when p11Rc-E1dm was cotransfected with 2.0 μg of the GFP-E1dm plasmid, a depression in replication was observed, consistent with the negative effects of elevated E1 protein levels or of an elevated E1/E2 ratio observed with individual expression vectors (Fig. 4). Cotransfection of 2.0 μg of the GFP-E1dm plasmid had very little effect on p11Rc (Fig. 7). We speculate that replication by p11Rc may have declined after peaking between the addition of 0.2 and 2 μg of p11Rc-E1dm, as described for pGFP-11E1dm (Fig. 4). In contrast, when 5 μg of either replicon was cotransfected with the pMT2-11E2 plasmid, replication increased greatly, up to about threefold with 10.0 μg of the E2 plasmid (Fig. 7), indicating that E2 was limiting for both replicons in the genomic context. However, the wild-type replicon still yielded more of the linearized full-length replication product than the mutated replicon. Interestingly, at a moderate to high input of p11Rc-E1dm, there was slower-migrating, newly replicated DNA (Fig. 7). Levels of this material increased significantly when the E2 plasmid was cotransfected, a finding similar to those observed for transfections with large amounts of pGFP-E1dm and E2 expression plasmids (Fig. 5C).
FIG. 7.
Replication activities of the p11Rc and p11Rc-E1dm replicons. Transient replication assays were performed in 293 cells by transfecting the indicated amounts of replicons. (A) Southern blotting to reveal the replicated DNA, as described for Fig. 4. The replicated, linearized replicons (arrows) in the DpnI+ lanes were quantified and normalized to obtain the relative replication activities. The asterisk marks the high-molecular-weight, newly replicated DNA of unknown structures. This band was more prominent in the presence of large amounts of E1 and E2. Bands below the arrow represent the cotransfected expression plasmids for E1 or E2. (B) Relative replication activities in panel A were plotted against the amounts of input replicons.
DISCUSSION
In this study, we have used pGFP-11E1 expression plasmids or replicons that express both GFP-E1 and native E2 proteins to examine how alternative mRNA splicing affects the expression of E1 and E2 proteins and also how the changing E1/E2 ratios may modulate the efficiency of transient HPV ori-specific replication. These studies are possible because the addition of the GFP moiety at the amino terminus of E1 does not compromise its biological function, since it supports transient replication as well as does the expression vector for the wild-type protein (Fig. 4, 5, and 7; also data not shown). Using these plasmids, we have shown that production of E1 mRNA, and hence E1 protein production, is highly depressed by intragenic and intergenic mRNA splicing involving primarily the dominant splice donor at nt 847, an observation consistent with the mRNAs present in productive lesions (13). We have further shown that a mutation in this dominant splice donor site, E1dm, significantly increases E1 protein production from both plasmids. By comparing wild-type and mutated plasmids or replicons, we have demonstrated that the efficiency of replication is determined by the absolute amounts of E1 and E2 proteins as well as by the ratio of these two proteins. A large amount of E2 expression plasmid invariably increases ori plasmid replication. In contrast, an elevated ratio of E1 over E2 beyond a certain point has a negative effect. Consequently, the mutated replicon replicated less efficiently than the wild-type replicon despite its high levels of GFP-E1 protein. Interestingly, our data also show that the E2 protein is limiting in reaching the capacity of the E1 helicase even for the wild-type replicon. Finally, we have identified three new transcripts, all of which have the potential to encode the E2 protein.
In the context of the E1 expression plasmid, the majority of the transcripts were a spliced species of nt 847^2622, and very little unspliced mRNA remained to encode the E1 protein (Fig. 2 and 3). Viral mRNA with this splice encodes the E2 protein (58). The small peptide of 29 residues resulting from the splice has not been detected previously from E1 expression vectors due to its small size. It was revealed in this study only because of the fusion to GFP (Fig. 2). We believe that this splice allows ribosome reinitiation to access the E2 ORF, after termination of the short upstream peptide, as is also hypothesized for the effect of E6 intragenic splices on the translation of E7 in the oncogenic HPVs (68). In contrast, in the context of the replicon, the predominant splice was nt 847^3325, generating the highly abundant GFP-E1^E4 mRNA and protein (Fig. 6). Consequently, only very low levels of E1 and E2 mRNA and protein are generated from the replicon (Fig. 6). It is interesting that, despite the rarity of the unspliced E1 mRNA, the E2 protein, but not the E1 protein, is limiting, as replication of the replicon is greatly enhanced when the E2 expression vector is cotransfected. In comparison, the effect of cotransfection of an E1 expression vector was less significant (Fig. 7). We speculate that E1 is efficiently translated, since it is the first protein encoded in the message, whereas E2 synthesis relies on translation reinitiation after the termination of an upstream peptide regardless of which of the splice donors is used (see below).
Since the splicing donor and acceptor sites and their various combinations in multiple spliced messages are highly conserved (13), our results concerning the regulation of E1 and E2 expression by alternative mRNA splicing are most likely applicable to other HPVs. Interestingly, although many comparable splice sites are also conserved in BPV-1, the corresponding E1 intragenic splice (nt 864^2558) has not been reported. Rather, the BPV-1 E2 mRNA is initiated from a separate promoter immediately upstream of the E2 ORF (2). Consequently, Rangasamy and colleagues (53, 54) were able to express a nuclear BPV-1 E1 protein tagged at the amino terminus with GFP and observed no abnormal distribution of GFP. Thus, this splice, if it indeed exists in BPV-1 mRNA, must occur at a very low frequency.
We have shown that, when the dominant splice donor at nt 847 was mutated, most of the E1 transcript was unspliced, accounting for the highly elevated production of E1 protein either from the E1 expression vector or from the replicon. Lysate dilutions showed that the level of E1 protein expressed in COS7 cells was 50- to 100-fold that detected from the wild-type expression vector (Fig. 2B and data not shown). In 293 cells, in which the replication assays were conducted and the expression plasmids did not replicate, a significant difference was also observed (Fig. 4E). These results validate our interpretation that the differences in the E1 protein levels and the ratios of E1/E2 proteins account for the differences in their replication activities (Fig. 4A through D). In addition, RT-PCR showed that the usage of previously known alternative minor splice donors at nt 1272 or nt 1459 increased relative to that observed in wild-type plasmids (Fig. 3B). A cryptic donor site at nt 916 was uncovered. The splice of nt 916^2622 was more frequently used by p11Rc-E1dm than by p11Rc (Fig. 6), and the nt 916^3325 splice was observed only with the mutated replicon. This novel donor site at nt 916 has not been reported previously in RNA preparations from lesions. We speculate that it is scarcely used because of its proximity to the dominant donor site at nt 847. Only when the dominant donor site is mutated is this cryptic site potentiated. Even then, the predicted E1N^E4 protein was not detected by Western blotting (Fig. 6).
As is observed in natural infections, there is also a hierarchy in the usage of the splice acceptor sites. The predominant spliced species from the replicon was nt 847^3325. E1^E4 is also the most abundant viral mRNA and protein in HPV-infected cells (5, 6, 13, 18, 52, 56, 66). Notably, the splice nt 847^2622 was recovered by RT-PCR only from the E1 expression vector, not from the wild-type replicon (data not shown). In fact, because this nt 847^3325 species was highly abundant and also because it was the shortest product, we recovered the minor alternative splices only when the forward primer was placed downstream of this donor site (Fig. 3 and 6). We also note that the splices of nt 916^2622, 1272^2622, and 1459^2622 have not been described previously. Rather, nt 1272 or 1459 is spliced to nt 3325 or 3377, generating either E2C or fusion peptides between portions of the E1 and E2 coding regions (7, 59) (Fig. 1A). We believe that at least one of these new species encodes the E2 protein, as p11Rc-E1dm supported its own replication. Indeed, a group of alternatively spliced HPV-16 mRNAs containing the E2 ORF has also been identified (64), and all of these can be translated to E2 in vivo and in vitro (1).
Our data show that E1/E2 ratios can significantly influence the level of replication. This was evident in three-plasmid cotransfection experiments (Fig. 4), where a high input of pGFP-11E1dm depressed rather than increased replication at a fixed input of the E2 expression vector. We believe that, for the same reason, the mutated replicon, which produced high levels of GFP-E1 protein, had a reduced efficiency of replication relative to the wild-type replicon (Fig. 6 and 7). It is possible that the negative effect of a high level of E1 protein is further exacerbated by less efficient alternative mRNA splicing or translation reinitiation of these transcripts from the mutated replicon relative to that of the primary E2 mRNA with the splice of nt 847^2622 produced by the wild-type replicon. This repression of replication by a high HPV E1/E2 ratio has not been observed previously because the wild-type E1 expression vector produces mostly spliced transcripts, so that it does not express an amount of E1 protein comparable to that generated by the E1dm mutant examined in this study.
What may have caused replication depression in the presence of high E1 concentrations? It is not likely due to a titration of critical interacting host proteins, as these proteins should be highly abundant and sufficient to support host DNA replication. A more plausible possibility is that multiple initiations from the template DNA could have occurred, resulting in disruption of orderly replication. DNA breakage and recombination might then lead to the formation of slow-migrating products (Fig. 5 and 7). Männik et al. (44) reported a smear of products, consisting primarily of small fragments shorter than the BPV-1 replicons, when the replicons produced large amounts of BPV-1 E1 protein. These abnormal products were attributed to an onion-skin mode of replication reinitiation from the ori. However, we observed only significantly longer products. The precise origin and nature of these products remain to be investigated.
In contrast to the negative effect of a high E1/E2 ratio, increasing the amount of the E2 expression vector invariably increases replication, indicating that E2 is limiting (Fig. 5). This result agrees with the previous observation in cell-free replication (14) but is in contrast to the report on HPV-1 (76). The reason for the discrepancy is unclear, but we note that, unlike the situation with the mucosotropic HPVs, HPV-1 E1 alone can initiate replication in the absence of E2 in transfected cells (22). What is unexpected is that E2 is limiting even for the wild-type replicon (Fig. 7). One might ask why the virus would want to limit E2 protein so as not to be able to achieve the highest level of replication possible for the amounts of E1 protein available. The answer may lie in the facts that the viral origin and E6 promoter overlap and that there are different modes of viral DNA replication as the keratinocytes differentiate. HPVs need E7 and E6 proteins to induce an S-phase environment in the differentiated, stratified epithelial cells in order to support productive viral DNA amplification. However, large amounts of E2 would inhibit transcription from the E6 promoter (10, 16, 17, 19, 20, 48, 73). Thus, a delicate balance must be struck between transcription and replication regulation.
The question of why so many spliced species are observed in transfected cells and in natural lesions is intriguing. There are several possible explanations. First, some of the alternatively spliced minor mRNAs indeed code for functional proteins. However, once the splice sites exist, the virus many not have a way to, or may not care to, preclude other, unintended splice combinations. Since most of the alternative spliced species tend to be rather minor or rare, the presence of the minor proteins encoded may not interfere with the more abundant functional proteins. Second, the less dominant splice sites might be beneficial for the virus because the same protein could be generated from a variety of spliced transcripts, as demonstrated in our study of p11Rc-E1dm. The third possibility is that the use of different splice sites could be conditional and might in fact be employed as a mechanism to regulate gene expression posttranscriptionally. Our data demonstrate that, by alternative mRNA splicing, the virus can control the expression of multiple proteins, their absolute levels, and their relative ratios without resorting to regulating separate promoters. This is a very flexible and efficient way to regulate its gene expression. In the case of the replication proteins, little or no E1 and E2 protein expression is needed in the quiescent basal cells until the cells enter S phase. More is needed in the rapidly cycling parabasal keratinocytes to ensure proper partitioning of viral DNA into the daughter cells. Still higher levels of E1 and E2 are needed to promote vegetative DNA amplification for progeny virion assembly in the differentiating spinous cells. Additional experiments will be needed to address this last possibility in vivo. Finally, our evidence that an E1dm mutant has high protein expression and authentic protein localization will facilitate future research on the E1 protein.
Acknowledgments
This work was supported by Public Health Service grant CA83679. The Digital Imaging Microscopy Facility was established with funds provided in large measure by the UAB Health Services Foundation and by Public Health Service grant DE/CA11910.
DNA sequencing was performed by the UAB CFAR DNA sequencing core facility.
REFERENCES
- 1.Alloul, N., and L. Sherman. 1999. The E2 protein of human papillomavirus type 16 is translated from a variety of differentially spliced polycistronic mRNAs. J. Gen. Virol. 80:29-37. [DOI] [PubMed] [Google Scholar]
- 2.Baker, C. C., and C. Calef. 1996. Maps of papillomavirus mRNA transcripts, section III, p. 1-20. In G. Myers, C. C. Baker, C. Wheeler, A. Halpern, A. A. McBride, and J. Doorbar (ed.), Human papillomaviruses 1996: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics, Los Alamos, N.Mex.
- 3.Bonne-Andrea, C., S. Santucci, P. Clertant, and F. Tillier. 1995. Bovine papillomavirus E1 protein binds specifically DNA polymerase α but not replication protein A. J. Virol. 69:2341-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonne-Andrea, C., F. Tillier, G. D. McShan, V. G. Wilson, and P. Clertant. 1997. Bovine papillomavirus type 1 DNA replication: the transcriptional activator E2 acts in vitro as a specificity factor. J. Virol. 71:6805-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, D. R., J. Bryan, M. Rodriguez, R. C. Rose, and D. G. Strike. 1991. Detection of human papillomavirus types 6 and 11 E4 gene products in condylomata acuminatum. J. Med. Virol. 34:20-28. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. R., M. T. Chin, and D. G. Strike. 1988. Identification of human papillomavirus type 11 E4 gene products in human tissue implants from athymic mice. Virology 165:262-267. [DOI] [PubMed] [Google Scholar]
- 7.Chiang, C.-M., T. R. Broker, and L. T. Chow. 1991. An E1M∧E2C fusion protein encoded by human papillomavirus type 11 is a sequence-specific transcription repressor. J. Virol. 65:3317-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang, C.-M., G. Dong, T. R. Broker, and L. T. Chow. 1992. Control of human papillomavirus type 11 origin of replication by the E2 family of transcription regulatory proteins. J. Virol. 66:5224-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang, C.-M., M. Ustav, A. Stenlund, T. F. Ho, T. R. Broker, and L. T. Chow. 1992. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc. Natl. Acad. Sci. USA 89:5799-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin, M. T., T. R. Broker, and L. T. Chow. 1989. Identification of a novel constitutive enhancer element and an associated binding protein: implications for human papillomavirus type 11 enhancer regulation. J. Virol. 63:2967-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow, L. T., and T. R. Broker. 1994. Papillomavirus DNA replication. Intervirology 37:150-158. [DOI] [PubMed] [Google Scholar]
- 12.Chow, L. T., and T. R. Broker. 1997. Small DNA tumor viruses, p. 267-301. In N. Nathanson (ed.), Viral pathogenesis. Lippincott-Raven Publishers, Philadelphia, Pa.
- 13.Chow, L. T., M. Nasseri, S. M. Wolinsky, and T. R. Broker. 1987. Human papillomavirus types 6 and 11 mRNAs from genital condylomata acuminata. J. Virol. 61:2581-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conger, K. L., J.-S. Liu, S.-R. Kuo, L. T. Chow, and T. S.-F. Wang. 1999. Human papillomavirus DNA replication. Interactions between the viral E1 protein and two subunits of human DNA polymerase α /primase. J. Biol. Chem. 274:2696-2705. [DOI] [PubMed] [Google Scholar]
- 15.Cueille, N., R. Nougarede, F. Mechali, M. Philippe, and C. Bonne-Andrea. 1998. Functional interaction between the bovine papillomavirus virus type 1 replicative helicase E1 and cyclin E-cdk2. J. Virol. 72:7255-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demeret, C., C. Desaintes, M. Yaniv, and F. Thierry. 1997. Different mechanisms contribute to the E2-mediated transcriptional repression of human papillomavirus type 18 viral oncogenes. J. Virol. 71:9343-9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong, G., T. R. Broker, and L. T. Chow. 1994. Human papillomavirus type 11 E2 proteins repress the homologous E6 promoter by interfering with the binding of host transcription factors to adjacent elements. J. Virol. 68:1115-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doorbar, J., C. Foo, N. Coleman, L. Medcalf, O. Hartley, T. Prospero, S. Napthine, J. Sterling, G. Winter, and H. Griffin. 1997. Characterization of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology 238:40-52. [DOI] [PubMed] [Google Scholar]
- 19.Dostatni, N., P. F. Lambert, R. Sousa, J. Ham, P. M. Howley, and M. Yaniv. 1991. The functional BPV-1 E2 trans-activating protein can act as a repressor by preventing formation of the initiation complex. Genes Dev. 5:1657-1671. [DOI] [PubMed] [Google Scholar]
- 20.Dowhanick, J. J., A. A. McBride, and P. M. Howley. 1995. Suppression of cellular proliferation by the papillomavirus E2 protein. J. Virol. 69:7791-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fouts, E. T., X. Yu, E. H. Egelman, and M. R. Botchan. 1999. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 274:4447-4458. [DOI] [PubMed] [Google Scholar]
- 22.Gopalakrishnan, V., and S. A. Khan. 1994. E1 protein of human papillomavirus type 1a is sufficient for initiation of viral DNA replication. Proc. Natl. Acad. Sci. USA 91:9597-9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han, Y., Y.-M. Loo, K. T. Militello, and T. Melendy. 1999. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J. Virol. 73:4899-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirochika, H., R. Hirochika, T. R. Broker, and L. T. Chow. 1988. Functional mapping of the human papillomavirus type 11 transcriptional enhancer and its interaction with the trans-acting E2 proteins. Genes Dev. 2:54-67. [DOI] [PubMed] [Google Scholar]
- 25.Howley, P. M. 1996. Papillomavirinae: the viruses and their replication, p. 2045-2076. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 26.Hubert, W. G., and L. A. Laimins. 2002. Human papillomavirus type 31 replication modes during the early phases of the viral life cycle depend on transcriptional and posttranscriptional regulation of E1 and E2 expression. J. Virol. 76:2263-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes, F. J., and M. A. Romanos. 1993. E1 protein of human papillomavirus is a DNA helicase/ATPase. Nucleic Acids Res. 21:5817-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hummel, M., J. B. Hudson, and L. A. Laimins. 1992. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J. Virol. 66:6070-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klumpp, D. J., and L. A. Laimins. 1999. Differentiation-induced changes in promoter usage for transcripts encoding the human papillomavirus type 31 replication protein E1. Virology 257:239-246. [DOI] [PubMed] [Google Scholar]
- 31.Klumpp, D. J., F. Stubenrauch, and L. A. Laimins. 1997. Differential effects of the splice acceptor at nucleotide 3295 of human papillomavirus type 31 on stable and transient viral replication. J. Virol. 71:8186-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo, S.-R., J.-S. Liu, T. R. Broker, and L. T. Chow. 1994. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J. Biol. Chem. 269:24058-24065. [PubMed] [Google Scholar]
- 33.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, R., and M. R. Botchan. 1994. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc. Natl. Acad. Sci. USA 91:7051-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, R., and M. R. Botchan. 1993. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell 73:1207-1221. [DOI] [PubMed] [Google Scholar]
- 36.Li, R., J. Knight, G. Bream, A. Stenlund, and M. R. Botchan. 1989. Specific recognition nucleotides and their DNA context determine the affinity of E2 protein for 17 binding sites in the BPV-1 genome. Genes Dev. 3:510-526. [DOI] [PubMed] [Google Scholar]
- 37.Lin, B. Y., T. Ma, J.-S. Liu, S.-R. Kuo, G. Jin, T. R. Broker, J. W. Harper, and L. T. Chow. 2000. HeLa cells are phenotypically limiting in cyclin E/CDK2 for efficient human papillomavirus DNA replication. J. Biol. Chem. 275:6167-6174. [DOI] [PubMed] [Google Scholar]
- 38.Lin, B. Y., A. M. Makhov, J. D. Griffith, T. R. Broker, and L. T. Chow. 2002. Chaperone proteins abrogate inhibition of the human papillomavirus (HPV) E1 replicative helicase by the HPV E2 protein. Mol. Cell. Biol. 22:6592-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, J.-S., S.-R. Kuo, T. R. Broker, and L. T. Chow. 1995. The functions of human papillomavirus type 11 E1, E2, and E2C proteins in cell-free DNA replication. J. Biol. Chem. 270:27283-27291. [DOI] [PubMed] [Google Scholar]
- 40.Liu, J.-S., S.-R. Kuo, A. M. Makhov, D. M. Cyr, J. D. Griffith, T. R. Broker, and L. T. Chow. 1998. Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J. Biol. Chem. 273:30704-30712. [DOI] [PubMed] [Google Scholar]
- 41.Lu, J.-Z., Y.-N. Sun, R. C. Rose, W. Bonnez, and D. J. McCance. 1993. Two E2 binding sites (E2BS) alone or one E2BS plus an A/T-rich region are minimal requirements for the replication of the human papillomavirus type 11 origin. J. Virol. 67:7131-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lusky, M., J. Hurwitz, and Y. S. Seo. 1993. Cooperative assembly of the bovine papilloma virus E1 and E2 proteins on the replication origin requires an intact E2 binding site. J. Biol. Chem. 268:15795-15803. [PubMed] [Google Scholar]
- 43.Ma, T., N. Zou, B. Y. Lin, L. T. Chow, and J. W. Harper. 1999. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc. Natl. Acad. Sci. USA 96:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Männik, A., K. Runkorg, N. Jaanson, M. Ustav, and E. Ustav. 2002. Induction of the bovine papillomavirus origin “onion skin”-type DNA replication at high E1 protein concentrations in vivo. J. Virol. 76:5835-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masterson, P. J., M. A. Stanley, A. P. Lewis, and M. A. Romanos. 1998. A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase α-primase p68 subunit. J. Virol. 72:7407-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohr, I. J., R. Clark, S. Sun, E. J. Androphy, P. MacPherson, and M. R. Botchan. 1990. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694-1699. [DOI] [PubMed] [Google Scholar]
- 47.Nasseri, M., R. Hirochika, T. R. Broker, and L. T. Chow. 1987. A human papilloma virus type 11 transcript encoding an E1∧E4 protein. Virology 159:433-439. [DOI] [PubMed] [Google Scholar]
- 48.Nishimura, A., T. Ono, A. Ishimoto, J. J. Dowhanick, M. A. Frizzell, P. M. Howley, and H. Sakai. 2000. Mechanisms of human papillomavirus E2-mediated repression of viral oncogene expression and cervical cancer cell growth inhibition. J. Virol. 74:3752-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ozbun, M. A., and C. Meyers. 1998. Human papillomavirus type 31b E1 and E2 transcript expression correlates with vegetative viral genome amplification. Virology 248:218-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park, P., W. Copeland, L. Yang, T. Wang, M. R. Botchan, and I. J. Mohr. 1994. The cellular DNA polymerase α-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc. Natl. Acad. Sci. USA 91:8700-8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prakash, S. S., S. R. Grossman, R. B. Pepinsky, L. A. Laimins, and E. J. Androphy. 1992. Amino acids necessary for DNA contact and dimerization imply novel motifs in the papillomavirus E2 trans-activator. Genes Dev. 6:105-116. [DOI] [PubMed] [Google Scholar]
- 52.Pray, T. R., and L. A. Laimins. 1995. Differentiation-dependent expression of E1∧E4 proteins in cell lines maintaining episomes of human papillomavirus type 31b. Virology 206:679-685. [DOI] [PubMed] [Google Scholar]
- 53.Rangasamy, D., and V. G. Wilson. 2000. Bovine papillomavirus E1 protein is sumoylated by the host cell Ubc9 protein. J. Biol. Chem. 275:30487-30495. [DOI] [PubMed] [Google Scholar]
- 54.Rangasamy, D., K. Woytek, S. A. Khan, and V. G. Wilson. 2000. SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J. Biol. Chem. 275:37999-38004. [DOI] [PubMed] [Google Scholar]
- 55.Remm, M., R. Brain, and J. R. Jenkins. 1992. The E2 binding sites determine the efficiency of replication for the origin of human papillomavirus type 18. Nucleic Acids Res. 20:6015-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roberts, S., I. Ashmole, G. D. Johnson, J. W. Kreider, and P. H. Gallimore. 1993. Cutaneous and mucosal human papillomavirus E4 proteins form intermediate filament-like structures in epithelial cells. Virology 197:176-187. [DOI] [PubMed] [Google Scholar]
- 57.Rohlfs, M., S. Winkenbach, S. Meyer, T. Rupp, and M. Dürst. 1991. Viral transcription in human keratinocyte cell lines immortalized by human papillomavirus type-16. Virology 183:331-342. [DOI] [PubMed] [Google Scholar]
- 58.Rotenberg, M. O., C.-M. Chiang, M. L. Ho, T. R. Broker, and L. T. Chow. 1989. Characterization of cDNAs of spliced HPV-11 E2 mRNA and other HPV mRNAs recovered via retrovirus-mediated gene transfer. Virology 172:468-477. [DOI] [PubMed] [Google Scholar]
- 59.Rotenberg, M. O., L. T. Chow, and T. R. Broker. 1989. Characterization of rare human papillomavirus type 11 mRNAs coding for regulatory and structural proteins, using the polymerase chain reaction. Virology 172:489-497. [DOI] [PubMed] [Google Scholar]
- 60.Sedman, J., and A. Stenlund. 1995. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 14:6218-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sedman, J., and A. Stenlund. 1998. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J. Virol. 72:6893-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seo, Y.-S., F. Müller, M. Lusky, E. Gibbs, H.-Y. Kim, B. Phillips, and J. Hurwitz. 1993. Bovine papilloma virus (BPV)-encoded E2 protein enhances binding of E1 protein to the BPV replication origin. Proc. Natl. Acad. Sci. USA 90:2865-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seo, Y.-S., F. Müller, M. Lusky, and J. Hurwitz. 1993. Bovine papilloma virus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc. Natl. Acad. Sci. USA 90:702-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sherman, L., and N. Alloul. 1992. Human papillomavirus type 16 expresses a variety of alternatively spliced mRNAs putatively encoding the E2 protein. Virology 191:953-959. [DOI] [PubMed] [Google Scholar]
- 65.Sherman, L., N. Alloul, I. Golan, M. Dürst, and A. Baram. 1992. Expression and splicing patterns of human papillomavirus type-16 mRNAs in pre-cancerous lesions and carcinomas of the cervix, in human keratinocytes immortalized by HPV 16, and in cell lines established from cervical cancers. Int. J. Cancer 50:356-364. [DOI] [PubMed] [Google Scholar]
- 66.Sherman, L., Y. Golan, S. Mitrani-Rosenbaum, and A. Baram. 1992. Differential expression of HPV types 6 and 11 in condylomas and cervical preneoplastic lesions. Virus Res. 25:23-36. [DOI] [PubMed] [Google Scholar]
- 67.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smotkin, D., H. Prokoph, and F. O. Wettstein. 1989. Oncogenic and nononcogenic human genital papillomaviruses generate the E7 mRNA by different mechanisms. J. Virol. 63:1441-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stoler, M. H., S. M. Wolinsky, A. Whitbeck, T. R. Broker, and L. T. Chow. 1989. Differentiation-linked human papillomavirus types 6 and 11 transcription in genital condylomata revealed by in situ hybridization with message-specific RNA probes. Virology 172:331-340. [DOI] [PubMed] [Google Scholar]
- 70.Stubenrauch, F., M. Hummel, T. Iftner, and L. A. Laimins. 2000. The E8∧E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J. Virol. 74:1178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sverdrup, F., and S. A. Khan. 1995. Two E2 binding sites alone are sufficient to function as the minimal origin of replication of human papillomavirus type 18 DNA. J. Virol. 69:1319-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swindle, C. S., N. Zou, B. A. Van Tine, G. M. Shaw, J. A. Engler, and L. T. Chow. 1999. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tan, S. H., L. E. Leong, P. A. Walker, and H. U. Bernard. 1994. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J. Virol. 68:6411-6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ustav, E., M. Ustav, P. Szymanski, and A. Stenlund. 1993. The bovine papillomavirus origin of replication requires a binding site for the E2 transcriptional activator. Proc. Natl. Acad. Sci. USA 90:898-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Horn, G., S. Sheikh, and S. A. Khan. 2001. Regulation of human papillomavirus type 1 replication by the viral E2 protein. Virology 287:214-224. [DOI] [PubMed] [Google Scholar]
- 77.Voitenleitner, C., and M. R. Botchan. 2002. E1 protein of bovine papillomavirus type 1 interferes with E2 protein-mediated tethering of the viral DNA to mitotic chromosomes. J. Virol. 76:3440-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.White, P. W., A. Pelletier, K. Brault, S. Titolo, E. Welchner, L. Thauvette, M. Fazekas, M. G. Cordingley, and J. Archambault. 2001. Characterization of recombinant HPV6 and 11 E1 helicases. Effect of ATP on the interaction of E1 with E2 and mapping of a minimal helicase domain. J. Biol. Chem. 276:22426-22438. [DOI] [PubMed] [Google Scholar]
- 78a.Yang, L., R. Li, I. J. Mohr, R. Clark, and M. R. Botchan. 1991. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature 353:628-632. [DOI] [PubMed] [Google Scholar]
- 79.Yang, L., I. Mohr, E. Fouts, D. A. Lim, M. Nohaile, and M. R. Botchan. 1993. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 90:5086-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zou, N., B. Y. Lin, F. Duan, K.-Y. Lee, G. Jin, R. Guan, G. Yao, E. J. Lefkowitz, T. R. Broker, and L. T. Chow. 2000. The hinge of the human papillomavirus type 11 E2 protein contains major determinants for nuclear localization and nuclear matrix association. J. Virol. 74:3761-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.zur Hausen, H. 2000. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 92:690-698. [DOI] [PubMed] [Google Scholar]
- 82.zur Hausen, H., and E.-M. de Villiers. 1994. Human papillomaviruses. Annu. Rev. Microbiol. 48:427-447. [DOI] [PubMed] [Google Scholar]