Abstract
Open reading frame 73 (ORF 73) is conserved among the gamma-2-herpesviruses (rhadinoviruses) and, in Kaposi's sarcoma-associated herpesvirus (KSHV) and herpesvirus saimiri (HVS), has been shown to encode a latency-associated nuclear antigen (LANA). The KSHV and HVS LANAs have also been shown to be required for maintenance of the viral genome as an episome during latency. LANA binds both the viral latency-associated origin of replication and the host cell chromosome, thereby ensuring efficient partitioning of viral genomes to daughter cells during mitosis of a latently infected cell. In gammaherpesvirus 68 (γHV68), the role of the LANA homolog in viral infection has not been analyzed. Here we report the construction of a γHV68 mutant containing a translation termination codon in the LANA ORF (73.STOP). The 73.STOP mutant virus replicated normally in vitro, in both proliferating and quiescent murine fibroblasts. In addition, there was no difference between wild-type (WT) and 73.STOP virus in the kinetics of induction of lethality in mice lacking B and T cells (Rag 1−/−) infected with 1,000 PFU of virus. However, compared to WT virus, the 73.STOP mutant exhibited delayed kinetics of replication in the lungs of immunocompetent C57BL/6 mice. In addition, the 73.STOP mutant exhibited a severe defect in the establishment of latency in the spleen of C57BL/6 mice. Increasing the inoculum of 73.STOP virus partially overcame the acute replication defected observed in the lungs at day 4 postinfection but did not ameliorate the severe defect in the establishment of splenic latency. Thus, consistent with its proposed role in replication of the latent viral episome, LANA appears to be a critical determinant in the establishment of γHV68 latency in the spleen post-intranasal infection.
The gammaherpesviruses include the human pathogens Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV, or human herpesvirus 8). These viruses establish life-long infection of the host and are associated with a number of malignancies. To better understand gammaherpesvirus pathogenesis, we and others have studied infection of mice with murine gammaherpesvirus 68 (γHV68, also referred to as MHV-68), a member of the γ2-herpesvirus family based on genome sequence (17).
The pathogenesis of γHV68 has been reviewed recently (32, 41). Briefly, γHV68 infection of inbred mice results in an acute, productive infection of multiple organs and a CD4+ T-cell-dependent splenomegaly (4, 36). Acute virus replication is largely cleared by 2 to 3 weeks postinfection (34, 42). Subsequently, γHV68 is present in its persistent, latent form, during which time the γHV68 genome is maintained in infected cells in the absence of detectable preformed infectious virus. γHV68 establishes a latent infection in B cells, macrophages, and dendritic cells and persists in lung epithelial cells (10, 33, 34, 44).
Sequence analysis of the γHV68 genome identified 80 ATG-initiated open reading frames (ORFs) predicted to encode proteins of at least 100 amino acids in length (39). The majority of these ORFs are homologous to known genes present in other gammaherpesviruses (39). ORF 73 of γHV68 is predicted to encode a latency-associated nuclear antigen (LANA) (39). Transcript analyses of infected fibroblasts have suggested that the γHV68 LANA is an immediate-early gene, as ORF 73-specific transcripts were detected in the presence of cycloheximide (27). In a reverse transcription-PCR screen for viral genes expressed during latency, the γHV68 LANA was found to be expressed preferentially in peritoneal cells following intraperitoneal infection of B-cell-deficient mice (40). Additionally, following intranasal infection with γHV68, transcription of ORF 73 was detected in the lungs but not spleens of infected animals (30).
Homologs of the putative γHV68 LANA are found in KSHV (16, 24), herpesvirus saimiri (HVS) (7), and rhesus rhadinovirus (2). In KSHV and HVS, the LANA protein has been shown to be required for maintenance of the viral genome as an episome during long-term in vitro culture (3, 7, 14). The LANA protein of KSHV was found to bind both the latency-associated origin of replication and members of the cellular origin recognition complex (3, 14, 18, 19). These findings have led to the hypothesis that LANA tethers the viral genome to host chromosomes, thus ensuring that the viral genome is passed on to daughter cells during division of the latently infected cell (3, 8, 14, 18, 29). An analogous function is encoded by the EBNA-1 protein of EBV (5, 9, 20, 25, 28, 45, 46). The putative γHV68 ORF 73 gene product is significantly smaller than the KSHV LANA (314 versus 1,162 amino acids), being closer in size to the HVS ORF 73 gene product (407 amino acids) (39). Importantly, the structure of the γHV68 ORF 73 transcript has not been determined, and it is possible that ORF 73 is extended through splicing of additional coding exons. Currently, there are no functional data demonstrating that the γHV68 ORF 73 gene product is involved in replication of the latent viral episome in proliferating cells. However, there is good evidence that the γHV68 episome is faithfully maintained in replicating cells (35, 37), indicating that γHV68 has evolved a mechanism to ensure replication of the viral episome during latent infection of proliferating cells. In addition, we have observed punctate nuclear staining of a FLAG epitope-tagged γHV68 ORF 73 in Cos-1 cells transiently transfected with an ORF 73 expression vector (N. J. Moorman and S. H. Speck, unpublished results), similar to that seen with the KSHV LANA. This is consistent with the hypothesized role of the γHV68 ORF 73 gene product in replication of the latent viral episome.
In addition to its role in viral episome maintenance, the LANA protein of KSHV has been found to play a role in transcription of both cellular and viral genes. LANA was shown to transactivate the human immunodeficiency virus long terminal repeat as well as its own promoter (15, 26), while acting as a repressor when tethered to a heterologous DNA binding domain. Cell lines constitutively expressing LANA were also found to induce a number of cellular genes by DNA microarray analysis, many of which are interferon-regulated in the normal host cell (26). In addition, expression of LANA was found to modulate expression from two EBV latency-associated promoters (13), suggesting that LANA may modulate EBV gene expression in coinfected primary effusion lymphoma (PEL) tumors.
In light of the above data, we undertook to determine what role the γHV68 LANA plays in the life cycle of γHV68, both in vitro and in vivo, employing a null mutant containing a premature translation stop codon in ORF 73.
MATERIALS AND METHODS
Viruses and tissue culture.
Viral stocks were generated by transfecting γHV68 bacterial artificial chromosome (BAC) clones into Vero-Cre cells. Following the appearance of cytopathic effect, wells were harvested and used to infect Vero-Cre cells to generate high-titer viral stocks. NIH 3T12 and MEF (mouse embryonic fibroblast) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 U of penicillin per ml, 100 mg of streptomycin per ml, and 2 mM l-glutamine (complete DMEM). Cells were maintained in a 5% CO2 tissue culture incubator at 37°C. MEFs were obtained from either BALB/c or C57BL/6 mouse embryos as described previously (42). Vero-Cre cells were a gift from David Leib. Cells were passaged in DMEM supplemented with 10% FCS and 300 μg of hygromycin B/ml.
Construction of γHV68 73.KAN and 73.STOP viruses.
The γHV68 genome cloned as a BAC was a kind gift of Ulrich Koszinowski (1). For the generation of the 73.KAN virus, a kanamycin resistance cassette (KanR) was inserted into ORF 73. The KanR cassette was amplified from the vector pCP15 with primers containing BstEII sites. The products were purified and then ligated into the unique BstEII site of the vector pL3700 to create the vector pL73.KAN. pL3700 contains nucleotides 101654 to 105377 of the γHV68 genome. The γHV68 sequence of pL73.KAN was liberated with a BamHI, XbaI digest, purified, and transformed into DH10B Escherichia coli cells containing the γHV68 BAC and a temperature-sensitive RecA expression plasmid, p2650. This plasmid contains a temperature-sensitive origin of replication and is lost at temperatures greater than 42°C. Following transformation, the cells were allowed to recover at 37°C for 2 h and then plated on Luria-Bertani (LB) medium plus chloramphenicol (Cam) and Kan and incubated overnight at 42°C. The γHV68 Bac provides Camr in this selection process, while only genomes which have also recombined the KanR cassette into ORF 73 will be doubly resistant. Clones were streaked onto LB plus ampicillin (Amp) plates to confirm the loss of p2650 to ensure no further recombination took place. Individual clones were then grown in LB plus Cam and Kan, and DNA was isolated by using the modified Qiagen Midi Prep kit as described in the manufacturer's protocol. Correct insertion of the KanR cassette was confirmed by Southern blotting.
The 73.STOP virus was generated by allelic exchange in E. coli, essentially as described by Smith and Enquist (31). Briefly, the region of the γHV68 genome from genomic coordinates 101654 to 105377 was cloned into the suicide vector pGS284. The pGS284 vector contains an ampicillin cassette and the levansucrase gene for positive and negative selection, respectively. Nucleotides 104779 to 104773 were modified from 5′-ATGCCT-3′ to 5′-GTCTAGA-3′, resulting in the introduction of a stop codon and a frameshift mutation. The resulting plasmid was named pGS73STOP. pGS73STOP was transfected into S17λpir E. coli cells. These bacteria were then mated to GS500 E. coli (RecA+) cells. Cointegrants were selected on LB plus Cam and Amp plates and allowed to resolve by overnight growth in LB plus Cam only. Following resolution, bacteria were plated on LB plus Cam and 7% sucrose plates to select for recombinants which had lost plasmid sequence. Individual colonies from these plates were grown in LB plus Cam, and the incorporation of the STOP mutation was determined by Southern blotting.
Mice, infections, and organ harvests.
C57BL/6 mice were purchased from Jackson Laboratory and used between 8 and 12 weeks of age. Mice were placed under isofluorane anesthesia prior to infection as well as before sacrifice by cervical dislocation. Intranasal infections were performed with 103 PFU of virus in a volume of 20 μl of complete DMEM. Upon sacrifice, spleens were placed in 0.5 ml of complete DMEM on ice and frozen at −80°C. Sentinel mice were assayed every 3 months and were negative for adventitious mouse pathogens by serology.
Plaque assays.
Plaque assays were performed using NIH 3T12 monolayers under noble agar overlay as described previously (42), with the following alterations. NIH 3T12 cells were plated in six-well plates at 2 × 105 cells per well the day prior to infection. Organs for which titers were to be determined were thawed and homogenized by mechanical disruption in a Mini-Beadbeater 8 (Biospec Products, Bartlesville, Okla.), and then 10-fold dilutions were made in complete DMEM and plated onto NIH 3T12 cell monolayers. Infections were performed in a 100-μl volume, and plates were rocked every 15 min for 1 h at 37°C. Samples were overlaid with 3 ml of a 1:1 mixture of 3% noble agar and 2× MEM supplemented with 20% FCS and a 2× concentration of antibiotic and l-glutamine. An additional 2 ml of noble agar, 2× MEM mixture was added 3 days postplating. Monolayers were stained between day 6 and 8 by the addition of 2 ml of neutral red overlay (0.01% neutral red in serum-free DMEM). After 18 to 24 h, plaques were counted. The limit of detection for this assay is 10 PFU per organ.
Limiting dilution ex vivo reactivation assay.
Detection of γHV68 reactivation from latency was performed as previously described (42, 43). Briefly, splenocytes were harvested either on day 16 or 42 postinfection and single cell suspensions were generated. Erythrocytes were lysed with ammonium chloride, resuspended in complete DMEM, and plated in a series of twofold dilutions, starting at 105 cells per well, onto MEF monolayers in 96-well tissue culture plates. After 21 days, wells were scored microscopically for the presence of cytopathic effect. In some cases, samples were replated on fresh MEF monolayers to confirm the presence of infectious virus. Twenty-four wells were plated per dilution with a total of 12 dilutions per sample per experiment. To detect preformed infectious virus, parallel samples were resuspended in 1/3× DMEM in the presence of 0.5-mm silica beads and subjected to four rounds of 1-min mechanical disruption in a Mini-Beadbeater 8 as previously described. This disruption procedure kills >99% of cells, with at most a twofold effect on the titer of preformed infectious virus (42). Disrupted cells were plated in a similar series of twofold dilutions.
Real-time PCR for the quantitation of viral genomes present in infected tissues.
To determine the relative number of viral genomes present in tissues, DNA was extracted from tissue using the Qiagen DNeasy kit. DNA was quantitated in a fluorometer, and 0.5 μg of DNA was included in each PCR. The PCR mix consisted of 5 mM MgCl2, a 200 μM concentration each of dATP, dGTP, and dCTP, 400 μM dUTP, a 900 nM concentration of each primer, 250 nM probe, 0.5 U of AmpErase UNG, and 2.25 U of AmpliTaq. The total reaction volume was 50 μl. PCR primers for ORF 50 were 5′-GGC CGC AGA CAT TTA ATG AC-3′ and 5′-GCC TCA ACT TCT CTG GAT ATG CC-3′. The sequence of the Taqman probe used was 5′-(6-carboxyfluorescein) ATT TGG GCG CAA TGT GTT GGA TGA A-3′. To control for variation in input DNA, glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific PCR was also performed on each sample. The same PCR mixture was used for the GAPDH-specific PCR with the primers 5′-CCT GCA CCA CCA ACT GCT TAG-3′ and 5′-GTG GAT GCA GGG ATG ATG TTC-3′. The Taqman probe for the GAPDH reactions was 5′-(6-carboxyfluorescein) CAC TCA GAA GAC TGT TGA TGG CCC CTC-3′. A two-step PCR was used which consisted of the following steps: 2 min at 50°C, 10 min at 95°C, and 50 cycles of 95°C for 15 s, followed by 60°C for 30 s. The same PCR conditions were used for both the ORF50 and GAPDH reactions. The number of copies of ORF50 and GAPDH in each sample was determined by comparison to a series of standard curve reactions using a plasmid control containing appropriate sequences. The standard curve dilutions used represented a range from 108 to 101, in serial 10-fold dilutions, and were performed in a background of 0.5 μg of splenic DNA from naive mice. All real-time PCRs were performed on a Becton Dickinson iCycler. All samples were analyzed in a minimum of two reactions. Data points represent the log square means of the replicate samples.
Statistical analyses.
All data were analyzed using GraphPad Prism (GraphPad Software, San Diego, Calif.). Titer data were statistically analyzed using the nonparametric Mann-Whitney test. The frequencies of reactivation and genome-positive cells were statistically analyzed using the paired t test. To accurately obtain the frequency for each limiting dilution, data were subjected to nonlinear regression (using a sigmoidal dose curve with nonvariable slope to fit the data). Frequencies of reactivation and genome-positive cells were obtained by calculating the cell density at which 63% of the wells scored positive for reactivating virus based on a Poisson distribution.
RESULTS
Disruption of ORF 73.
Traditionally, homologous recombination in mammalian cells has been used for the generation of herpesvirus mutants. With the introduction of the γHV68 genome cloned as a BAC (1), the process of generating mutant viruses has been greatly simplified. We have taken advantage of two systems, one described by Cherepanov and Wackernagel (6) and the other by Smith and Enquist (31), to introduce mutations into ORF 73. We initially inserted a kanamycin selection cassette into ORF 73 by using targeted homologous recombination in an E. coli DH10B strain containing the γHV68-BAC and a RecA expression plasmid containing a temperature-sensitive origin. Thus, at 37°C RecA is expressed and the plasmid is maintained, thereby allowing recombination to take place between homologous sequences. Following transformation the bacteria were allowed to recover for 2 h at 37°C and then plated on LB plates containing chloramphenicol and kanamycin overnight at 42°C. At 42°C the RecA expression plasmid is incapable of replication and thus is lost from the bacteria. Chloramphenicol resistance was provided by the γHV68 BAC. Chloramphenicol- and kanamycin-resistant colonies were individually grown and analyzed by Southern blot analyses for appropriate insertion of the kanamycin resistance cassette to generate γHV68 LANA.KAN (see Discussion, below).
To generate a more subtle mutation in ORF 73, we utilized the allelic exchange method described by Smith and Enquist (31). This system utilizes a suicide donor vector, pGS284, to incorporate a mutation and a counterselection marker to drive removal of vector sequences. Thus, pGS284 contains an ampicillin resistance cassette for positive selection and the levansucrase gene, sacB, for negative selection (31). In addition, the pGS284 vector is derived from a vector based on the RP4oriT and the R6K origin of replication of pGP704 (22) and will not replicate in E. coli strain GS500. Bacteria containing the pGS284 suicide vector, into which the desired ORF 73 targeting sequences had been cloned, were mated to the RecA+ E. coli strain (GS500) containing the γHV68 BAC. GS500 cells, in which the pGS284 targeting plasmid had recombined by a single crossover event with the γHV68 BAC, were ampicillin and chloramphenicol resistant. Resolution of the single crossover was allowed to occur by overnight growth in LB medium containing only chloramphenicol to ensure maintenance of the γHV68 BAC. Cultures were then plated onto LB plates containing chloramphenicol and 7% sucrose to select for γHV68-BAC recombinants that had lost the pGS284 vector sequences and then were further screened for sensitivity to ampicillin. Ampicillin-sensitive, sucrose-resistant colonies were then grown and analyzed by Southern blot analyses for incorporation of the translation stop codon into ORF 73. This approach was used to replace the kanamycin cassette in the γHV68 73.KAN virus with a mutant ORF 73 in which the methionine codon at amino acid 30 was mutated to a translation stop codon. In addition to the introduction of a translation stop codon, an extra nucleotide was inserted following the stop codon to introduce a reading frameshift, thus preventing expression of a functional LANA protein via either translational read-through or reversion of the stop codon. The introduced changes in ORF 73 also created a novel XbaI site which was used as a genetic marker in the Southern blotting shown in Fig. 1. The γHV68 73.KAN virus was also rescued to wild-type (WT) sequence using allelic exchange to ensure no spurious mutations were incorporated during the recombination process. This virus is referred to as γHV68 73.MR.
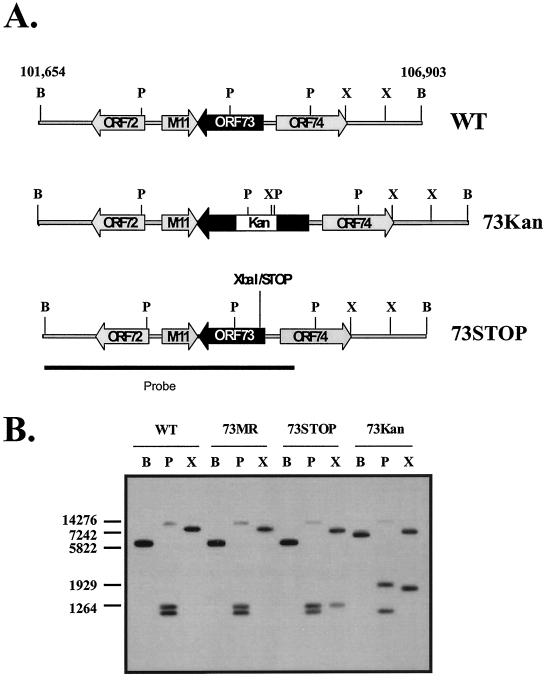
FIG. 1.
Construction of 73.KAN, 73.STOP, and 73.MR viruses. (A) γHV68-BAC (1) was used to generate the 73.KAN, 73.STOP, and 73.MR mutants. The WT BAC was mutagenized by RecA-mediated recombination to include a kanamycin resistance cassette in ORF 73. The Kan cassette was replaced by allelic exchange with the ORF 73 coding sequence containing a translation stop codon 30 amino acids into the coding frame. In addition, immediately following the introduced stop codon an extra base was added, resulting in a reading frameshift. The sum of these two mutations generates a unique XbaI site in ORF 73. To control for spurious mutations generated during insertion of the Kan cassette, 73.MR was created by replacing the Kan cassette in ORF 73 in the 73.KAN virus with the WT ORF 73 sequence. (B) Southern blot analyses of 73.STOP, 73.MR, and WT virus. Insertion of the STOP/XbaI cassette resulted in a unique band of 1,708 bp in the 73.STOP lane. The rescue of the kanamycin resistance cassette insertion is demonstrated by comparison of the PstI digests of 73.STOP and 73.MR. The probe contains sequence from genomic coordinates bp 101,654 to 105,377. B, BamHI; P, PstI; and X, XbaI.
As shown in Fig. 1B, both the WT and marker rescue viruses gave the expected pattern of hybridizing bands in the BamHI (5,249-bp), PstI (13,561-, 1,296-, and 1,164-bp) and XbaI (9,314-bp) digests. Notably, the insertion of the kanamycin cassette in the 73.Kan virus was readily apparent in both the PstI (increase in size of the 1,296-bp fragment to 1,803 bp) and XbaI (decrease in size of the 9,314-bp fragment to 8,959 bp plus the addition of a 1,708-bp fragment) digests (Fig. 1B). In addition, in the 73.Kan virus, the expected increase in the size of the BamHI fragment was apparent (from 5,250 bp to 6,601 bp). In the 73.STOP virus, which was generated from the 73.KAN virus, the WT banding pattern was restored for the BamHI and PstI digests; however, the introduction of the translation stop codon resulted in the introduction of a novel XbaI fragment (9,314-bp fragment replaced by 8,017- and 1,298-bp fragments) (Fig. 1B).
To generate viral stocks for subsequent infections, BAC DNA prepared from 73.STOP, 73.MR, and WT bacterial cultures was transfected into a Vero cell line that constitutively expresses Cre recombinase (Vero-Cre). By virtue of the loxP sites surrounding the BAC vector sequences, Cre-mediated excision of the BAC vector occurred during growth in this cell line. The virus from these transfections was then used to prepare large, high-titer viral stocks in Vero-Cre cells. The viral stocks were screened for the presence of BAC vector sequences by both PCR and Southern blotting, and both assays confirmed the complete removal of all BAC vector sequences (data not shown).
γHV68 73.STOP replicates comparably to WT γHV68 in vitro.
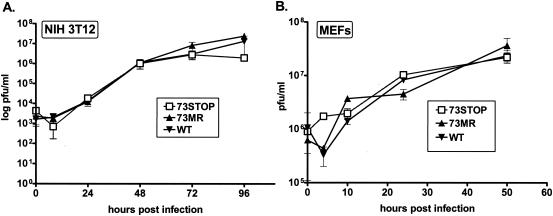
The ability to generate 73.STOP virus stocks of comparable titer to WT and 73.MR virus stocks from transfection of infectious γHV68-BAC DNA into Vero-Cre cells demonstrated that ORF 73 is not essential for viral replication. To quantitatively assess whether the γHV68 LANA plays a role in γHV68 replication in vitro, 73.STOP, 73.MR, and WT viruses were compared in a multistep growth assay in vitro on murine fibroblast NIH 3T12 monolayers. Cells were infected at a multiplicity of infection (MOI) of 0.05. Wells were harvested at the indicated time points, and the titer was determined by plaque assay on NIH 3T12 cells. As shown in Fig. 2A, all viruses replicated equivalently at all time points measured. Thus, the γHV68 LANA protein is not required for efficient virus replication in vitro.
FIG. 2.
γHV68 LANA is not required for virus replication in vitro. (A) NIH 3T12 monolayers were infected at an MOI of 0.05 with the indicated virus. Wells were harvested at the indicated time points, and virus titers were determined on NIH 3T12 cells as described in Materials and Methods. The data were compiled from two independent experiments. (B) MEF monolayers were infected at an MOI of 5.0 with the indicated virus. Wells were harvested at the indicated time points, and virus titers were determined on NIH 3T12 cells as described in Materials and Methods. The data were compiled from two independent experiments. The standard error of the mean is shown for both panels A and B.
To further assess 73.STOP virus growth in vitro, we infected growth-arrested MEF cultures. Confluent (contact-inhibited) MEFs were rested for 2 weeks prior to infection with 73.STOP, 73.MR, or WT virus. MEFs were infected for a single-step growth assay at an MOI of 5, and virus growth was assessed over a 48-h time period postinfection (Fig. 2B). Notably, no difference in the growth of the 73.STOP mutant compared to that of either WT or 73.MR virus was observed.
γHV68 73.STOP is compromised for viral replication in vivo.
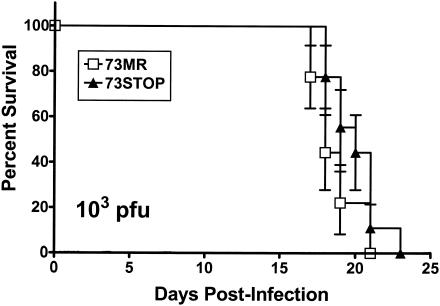
While not required for in vitro replication, it is possible that the γHV68 LANA may be required for efficient virus replication in vivo. To examine in vivo replication of the 73.STOP mutant, we initially examined induction of lethality in severely immunocompromised Rag 1 knockout mice (Rag 1−/−) which lack B and T lymphocytes. Rag 1−/− mice were infected with 1,000 PFU of either 73.STOP or 73.MR virus via intranasal inoculation and monitored daily thereafter for survival (Fig. 3). No statistically significant difference in the induction of lethality was observed between 73.STOP and 73.MR under these experimental conditions. To further examine 73.STOP mutant replication in vivo, we infected normal immunocompetent C57BL/6 mice intranasally with 1,000 PFU of 73.STOP, 73.MR, or WT virus and examined acute virus replication in the lungs at days 4 and 9 postinfection (Fig. 4A). Surprisingly, while virus replication could readily be detected at day 4 in the lungs of mice infected with either 73.MR or WT virus, no virus was detected in the lungs of 73.STOP virus-infected mice (Fig. 4A). However, at day 9 73.STOP virus replication could readily be detected in the lungs of infected mice, although at a three- to fourfold-lower level than in WT- and 73.MR-infected animals (Fig. 4A). Increasing the inoculating dose of virus from 1,000 PFU to 106 PFU revealed acute replication of the 73.STOP virus in the lungs of infected mice at day 4 postinfection (Fig. 4B). However, the observed levels of 73.STOP virus replication at day 4 were still reduced >10-fold compared to that with 73.MR. It is unclear why the observed defect in acute virus replication in C57BL/6 mice is not reflected in a delayed kinetics in lethality in Rag 1−/− mice. It is possible that the restriction in 73.STOP virus replication in C57BL/6 mice is not present in Rag 1−/− mice. Alternatively, the replication of the 73.STOP mutant may not be impaired in all tissues. Since we do not know precisely why the Rag 1−/− mice die from γHV68 infection, it is difficult to further assess this issue.
FIG. 3.
γHV68 LANA is not required for virus-induced lethality in Rag1−/− mice. The 73.STOP mutant kills B6.Rag1-deficient mice with the same kinetics as 73.MR virus. B6.Rag1-deficient mice were infected with 1,000 PFU of the indicated virus via intranasal inoculation and assessed daily for survival. The data were compiled from two independent experiments each containing four to five mice per group. The standard error of the mean is shown.
FIG. 4.
γHV68 LANA is required for efficient acute virus replication in the lungs following intranasal inoculation. (A) Mice were infected with 1,000 PFU of the indicated virus via intranasal inoculation. Lung tissue was harvested at the indicated time points and titers were determined on NIH 3T12 cells as described in Materials and Methods. The data were compiled from two independent experiments containing four to five mice per group. (B) Mice were infected with 106 PFU of the indicated virus via intranasal inoculation. Lung tissue was harvested at day 4, and titers were determined on NIH 3T12 cells as described in Materials and Methods. The data were compiled from a single experiment containing five mice per group.
The γHV68 LANA is required for efficient establishment and reactivation from latency.
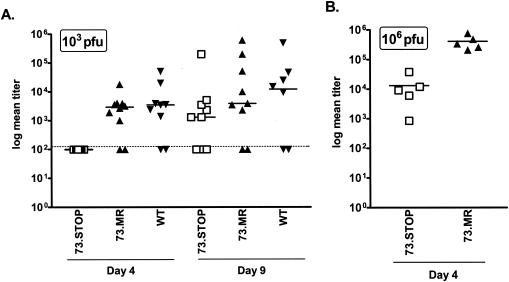
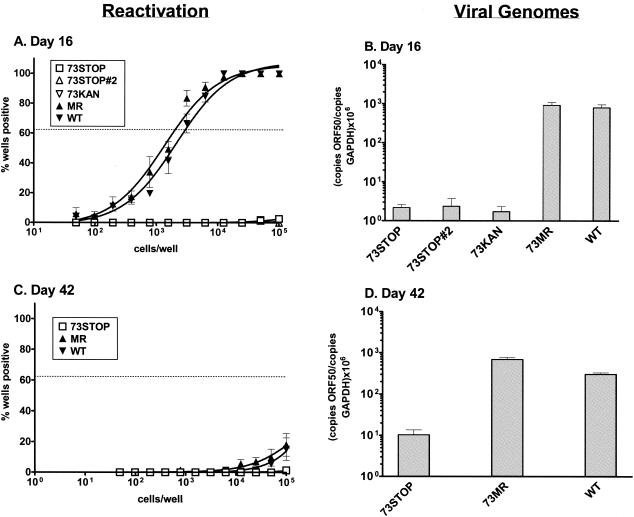
To assess the ability of the 73.STOP mutant to establish latency, we examined splenic latency in C57BL/6 mice following intranasal inoculation with 1,000 PFU of 73.STOP, 73.MR, or WT γHV68. At early times (day 16) postinfection, no reactivation of 73.STOP virus was observed from splenocytes (Fig. 5A). In contrast, at day 16 postinfection ca. 1 in 3,000 splenocytes reactivated 73.MR or WT virus. To determine whether the failure to detect reactivation of the 73.STOP mutant virus reflected a defect in the establishment of latency or a defect in reactivation, we determined by real-time PCR the amount of viral genome present in the spleens of mice infected with 73.STOP versus either 73.MR or WT virus (Fig. 5B). This analysis demonstrated that there was >200-fold less viral DNA present in the spleens of mice infected with the 73.STOP mutant, demonstrating that LANA function is required for the efficient establishment of splenic latency. Because the 73.MR virus was directly generated from the 73.KAN mutant (see Materials and Methods) (Fig. 1) and not by rescuing the mutation in 73.STOP, it is possible that the phenotypes exhibited by 73.STOP arise from a spurious distal mutation rather than the mutation introduced into ORF 73. To assess this possibility, we assayed the ability of the 73.KAN mutant, as well as an independent 73.STOP mutant virus (73.STOP#2) to establish latency. As shown in Fig. 5A and B, the results obtained with the 73.KAN and 73.STOP#2 mutants were indistinguishable from those observed with the 73.STOP mutant. Thus, these results conclusively map the failure to efficiently establish latency in the spleen to the mutations introduced into ORF 73.
FIG. 5.
γHV68 LANA is required for efficient establishment or maintenance of latency following intranasal infection. Mice were infected with 1,000 PFU of the indicated virus in 20 μl of complete DMEM. (A and C) Limiting dilution assays were performed on splenocytes plated on MEF indicator monolayers as described in Materials and Methods. (B and D) Real-time PCR analysis was performed to determine the number of viral genomes present per 0.5 μg of splenic DNA. Input DNA was standardized using a separate real-time PCR assay specific for the cellular GAPDH gene on the same DNA sample. The data were compiled from at least two independent experiments, each containing four or five mice per group. The standard error of the mean is shown.
By day 42 postinfection little reactivation could be detected from either WT-, 73.MR-, or 73.STOP-infected splenocytes (Fig. 5C). Analysis of viral DNA in the spleens of 73.STOP-infected mice revealed a slight increase compared to day 16 levels, while the amount of WT and 73.MR DNA slightly decreased over this time period (Fig. 5D). However, there remained ≥30-fold less 73.STOP DNA in the spleens of these mice compared to the WT or 73.MR groups. Whether the 73.STOP genomes detected at day 42 reflect latently infected splenocytes or the remnants of lytic virus replication in the spleen is unclear. The latter is an important consideration, since even at the peak of viral latency only a small percentage (≤1.0%) of viral genome-positive splenocytes reactivate in the ex vivo reactivation assay. In addition, it is important that the number of viral genomes present in 73.STOP-infected samples was at or near the limit of detection for this assay and, thus, the observed levels of 73.STOP viral genome in the spleens of these animals may be overestimated by the real-time PCR analyses.
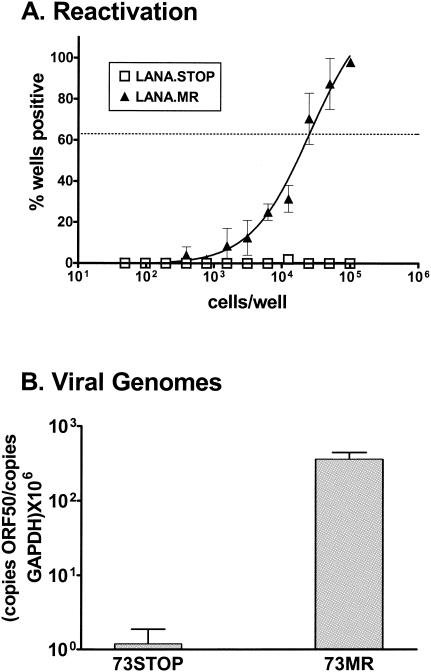
To assess whether the failure to efficiently establish splenic latency with the 73.STOP mutant could be accounted for by the observed defect in acute virus replication (see above), we infected C57BL/6 mice with 106 PFU of virus. As discussed above, we have shown that the defect in virus replication at day 4 postinfection in the lungs can be partially overcome by inoculating the mice with a higher dose. Notably, increasing virus dose did not lead to a detectable increase in the frequency of splenocytes reactivating 73.STOP virus (Fig. 6A) or the frequency of cells harboring viral genome (Fig. 6B). Thus, although 106 PFU of 73.STOP virus leads to substantial acute virus in the lungs at day 4 postinfection (ca. fivefold higher than WT virus replication following inoculation with 1,000 PFU [Fig. 4]), the 73.STOP mutant still failed to establish detectable latency in the spleen.
FIG. 6.
γHV68 LANA is required for efficient establishment or maintenance of latency following high-dose intranasal infection. Mice were infected with 106 PFU of the indicated virus in 20 μl of complete DMEM. (A) Limiting dilution assays were performed on splenocytes plated on MEF indicator monolayers as described in Materials and Methods. (B) Real-time PCR analysis was performed to determine the number of viral genomes present per 0.5 μg of splenic DNA. Input DNA was standardized using a separate real-time PCR assay specific for the cellular GAPDH gene on the same DNA sample. The data were compiled from at least two independent experiments, each containing four or five mice per group. The standard error of the mean is shown.
DISCUSSION
The function of γHV68 LANA homologs in other gammherpesviridae has been extensively studied in vitro. As discussed above, these functions include episomal maintenance of the viral genome during latent infection, modulation of transcription of both cellular and viral genes, and interaction with the cellular origin recognition complex (3, 8, 12, 13, 15, 18, 19, 21, 26). The studies presented here were designed to assess the role of the putative γHV68 LANA homolog during lytic and latent infection in vivo. We determined that the putative γHV68 LANA is not required for efficient lytic viral replication in vitro but does appear to play a role in acute virus replication in vivo. The latter was a surprising result and suggests that either (i) some aspect of LANA function which is not required for virus replication in vitro augments virus replication in vivo (e.g., a function which alters the innate immune response to γHV68 infection); or (ii) establishment of latency in some cell type(s) followed by reactivation contributes to acute virus replication in vivo. Distinguishing between these possibilities will likely be difficult.
As expected, γHV68 LANA function appears to be critical for establishment of latency in vivo. Following intranasal infection, the γHV68 73.STOP virus was found to establish or maintain latency at a reduced level compared to WT to 73.MR virus at both early (day 16) and late (day 42) times postinfection. The 73.STOP virus reactivated at a much lower frequency than either WT or 73.MR at early times postinfection, presumably as a result of the decreased number of latently infected splenocytes. It was not possible to determine differences in reactivation at late times postinfection, as splenocytes do not reactivate to sufficient levels to accurately assess the frequency of cells reactivating virus in this assay. The detection of low levels of viral genome at day 42 in the spleens of mice infected with the 73.STOP mutant could reflect either (i) the remnants of virus replication in the spleen or (ii) latent infection of a noncycling cell population (e.g., splenic macrophages). In the latter case, if the sole role of LANA is to maintain the latent viral genome in dividing cells, then it may be dispensable in latency reservoirs that never divide post-establishment of γHV68 latency. Overall, these data are consistent with the proposed role of the γHV68 LANA in latent viral genome replication. Studies are currently under way to elucidate the role of the γHV68 LANA in episomal maintenance.
The impact of mutating the other known genes (M11 [v-bcl2], gene 72 [v-cyclin], and gene 74 [v-GPCR]) in this region of the viral genome on establishment and reactivation from latency has been well characterized. Notably, the phenotype of the ORF 73 mutant does not overlap with the phenotype observed with null mutations in any of these genes (11, 23, 38). Indeed, unlike ORF 73, none of these genes appears to be required for efficient establishment of viral latency (11, 23, 38).
Finally, in addition to its role in episomal maintenance, the KSHV LANA has been shown to modulate transcription from a number of promoters. Both cellular and viral promoters have been found to be modulated by KSHV LANA expression. KSHV LANA was shown to upregulate transcription from its own promoter and from a number of cellular promoters, many of which drive expression of interferon-inducible genes (26). In addition, KSHV LANA was also found to activate transcription from the human immunodeficiency virus long terminal repeat and from the EBV latency-associated promoters Cp and Qp (13, 15). The ability of the KSHV LANA to modulate transcription is dependent on its ability to bind DNA sequences in these promoters. In light of this information, the loss of transcriptional regulatory functions that may be provided by the γHV68 LANA could also contribute to the severe latency defect observed with the 73.STOP mutant. Perhaps changes in cellular gene expression induced by γHV68 LANA are required for the establishment or maintenance of latent viral genomes in infected cells. Clearly, further analyses of the impact of the γHV68 LANA on viral and cellular gene expression are required to address this issue.
Acknowledgments
This research was supported by National Institutes of Health grants CA43143, CA52004, and CA58524 to S.H.S.
We also acknowledge helpful discussions with Skip Virgin and David Leib, as well as members of the Speck lab. Special thanks go to Greg Smith for technical advice regarding allelic exchange protocols.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 4.Brooks, J. W., A. M. Hamilton-Easton, J. P. Christensen, R. D. Cardin, C. L. Hardy, and P. C. Doherty. 1999. Requirement for CD40 ligand, CD4+ T cells, and B cells in an infectious mononucleosis-like syndrome. J. Virol. 73:9650-9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 7.Collins, C. M., M. M. Medveczky, T. Lund, and P. G. Medveczky. 2002. The terminal repeats and latency-associated nuclear antigen of herpesvirus saimiri are essential for episomal persistence of the viral genome. J. Gen. Virol. 83:2269-2278. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, M. A., C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 9.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 10.Flano, E., S. M. Husain, J. T. Sample, D. L. Woodland, and M. A. Blackman. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J. Immunol. 165:1074-1081. [DOI] [PubMed] [Google Scholar]
- 11.Gangappa, S., L. F. van Dyk, T. J. Jewett, S. H. Speck, and H. W. Virgin. 2002. Identification of the in vivo role of a viral bcl-2. J. Exp. Med. 195:931-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groves, A. K., M. A. Cotter, C. Subramanian, and E. S. Robertson. 2001. The latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus activates two major essential Epstein-Barr virus latent promoters. J. Virol. 75:9446-9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu, J., A. C. Garber, and R. Renne. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76:11677-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyun, T. S., C. Subramanian, M. A. Cotter, R. A. Thomas, and E. S. Robertson. 2001. Latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus interacts with Tat and activates the long terminal repeat of human immunodeficiency virus type 1 in human cells. J. Virol. 75:8761-8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein, G. 1981. The relative role of viral transformation and specific cytogenetic changes in the development of murine and human lymphomas. Hamatol. Bluttransfus. 26:3-10. [DOI] [PubMed] [Google Scholar]
- 18.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leight, E. R., and B. Sugden. 2000. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev. Med. Virol. 10:83-100. [DOI] [PubMed] [Google Scholar]
- 21.Lim, C., Y. Gwack, S. Hwang, S. Kim, and J. Choe. 2001. The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 276:31016-31022. [DOI] [PubMed] [Google Scholar]
- 22.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moorman, N. J., H. W. Virgin IV, and S. H. Speck. 2003. Disruption of the gene encoding the γHV68 v-GPCR leads to decreased efficiency of reactivation from latency. Virology 307:179-190. [DOI] [PubMed] [Google Scholar]
- 24.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by ORF 73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reisman, D., J. Yates, and B. Sugden. 1985. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 5:1822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rochford, R., M. L. Lutzke, R. S. Alfinito, A. Clavo, and R. D. Cardin. 2001. Kinetics of murine gammaherpesvirus 68 gene expression following infection of murine cells in culture and in mice. J. Virol. 75:4955-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shinohara, H., M. Fukushi, M. Higuchi, M. Oie, O. Hoshi, T. Ushiki, J. Hayashi, and M. Fujii. 2002. Chromosome binding site of latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus is essential for persistent episome maintenance and is functionally replaced by histone H1. J. Virol. 76:12917-12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simas, J. P., D. Swann, R. Bowden, and S. Efstathiou. 1999. Analysis of murine gammaherpesvirus-68 transcription during lytic and latent infection. J. Gen. Virol. 80:75-82. [DOI] [PubMed] [Google Scholar]
- 31.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speck, S. H., and H. W. Virgin. 1999. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr. Opin. Microbiol. 2:403-409. [DOI] [PubMed] [Google Scholar]
- 33.Stewart, J. P., E. J. Usherwood, A. Ross, H. Dyson, and T. Nash. 1998. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J. Exp. Med. 187:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 35.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1993. Interactions of murine gammaherpesvirus 68 with B and T cell lines. Virology 193:825-833. [DOI] [PubMed] [Google Scholar]
- 36.Usherwood, E. J., A. J. Ross, D. J. Allen, and A. A. Nash. 1996. Murine gammaherpesvirus-induced splenomegaly: a critical role for CD4 T cells. J. Gen. Virol. 77:627-630. [DOI] [PubMed] [Google Scholar]
- 37.Usherwood, E. J., J. P. Stewart, and A. A. Nash. 1996. Characterization of tumor cell lines derived from murine gammaherpesvirus-68-infected mice. J. Virol. 70:6516-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Dyk, L. F., H. W. Virgin, and S. H. Speck. 2000. The murine gammaherpesvirus 68 v-cyclin is a critical regulator of reactivation from latency. J. Virol. 74:7451-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Virgin, H. W., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virgin, H. W., R. M. Presti, X. Y. Li, C. Liu, and S. H. Speck. 1999. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J. Virol. 73:2321-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Virgin, H. W., and S. H. Speck. 1999. Unraveling immunity to gamma-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr. Opin. Immunol. 11:371-379. [DOI] [PubMed] [Google Scholar]
- 42.Weck, K. E., M. L. Barkon, L. I. Yoo, S. H. Speck, and H. W. Virgin IV. 1996. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J. Virol. 70:6775-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weck, K. E., S. S. Kim, H. W. Virgin IV, and S. H. Speck. 1999. B cells regulate murine gammaherpesvirus 68 latency. J. Virol. 73:4651-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weck, K. E., S. S. Kim, H. W. Virgin IV, and S. H. Speck. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol. 73:3273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wysokenski, D. A., and J. L. Yates. 1989. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J. Virol. 63:2657-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yates, J. L., S. M. Camiolo, and J. M. Bashaw. 2000. The minimal replicator of Epstein-Barr virus oriP. J. Virol. 74:4512-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]