Abstract
Cathepsin B (CTSB) is overexpressed in tumors of the lung, prostate, colon, breast, and stomach. However, evidence of primary genomic alterations in the CTSB gene during tumor initiation or progression has been lacking. We have found a novel amplicon at 8p22–23 that results in CTSB overexpression in esophageal adenocarcinoma. Amplified genomic NotI–HinfI fragments were identified by two-dimensional DNA electrophoresis. Two amplified fragments (D4 and D5) were cloned and yielded unique sequences. Using bacterial artificial chromosome clones containing either D4 or D5, fluorescent in situ hybridization defined a single region of amplification involving chromosome bands 8p22–23. We investigated the candidate cancer-related gene CTSB, and potential coamplified genes from this region including farnesyl-diphosphate farnesyltransferase (FDFT1), arylamine N-acetyltransferase (NAT-1), lipoprotein lipase (LPL), and an uncharacterized expressed sequence tag (D8S503). Southern blot analysis of 66 esophageal adenocarcinomas demonstrated only CTSB and FDFT1 were consistently amplified in eight (12.1%) of the tumors. Neither NAT-1 nor LPL were amplified. Northern blot analysis showed overexpression of CTSB and FDFT1 mRNA in all six of the amplified esophageal adenocarcinomas analyzed. CTSB mRNA overexpression also was present in two of six nonamplified tumors analyzed. However, FDFT1 mRNA overexpression without amplification was not observed. Western blot analysis confirmed CTSB protein overexpression in tumor specimens with CTSB mRNA overexpression compared with either normal controls or tumors without mRNA overexpression. Abundant extracellular expression of CTSB protein was found in 29 of 40 (72.5%) of esophageal adenocarcinoma specimens by using immunohistochemical analysis. The finding of an amplicon at 8p22–23 resulting in CTSB gene amplification and overexpression supports an important role for CTSB in esophageal adenocarcinoma and possibly in other tumors.
Keywords: oncogene/Barrett’s esophagus/neoplasm
Esophageal adenocarcinoma is associated with a poor prognosis with a 5-year survival rate of <5%, (1) and of importance, the incidence of this disease has increased 100% during the past 20 years (2). The major risk factor for the development of esophageal adenocarcinoma is the replacement of the squamous epithelium of the distal esophagus with a metaplastic columnar epithelium known as Barrett’s esophagus (3). This metaplastic epithelium is present in 10–12% of patients with symptomatic esophageal reflux of gastric and intestinal contents (4). Focal regions of dysplasia may develop in Barrett’s esophagus, and esophageal adenocarcinomas are often observed to arise within this epithelium, establishing Barrett’s esophagus as a premalignant lesion (3).
The cysteine protease cathepsin B (CTSB) gene, which maps to 8p22 (5), is a lysosomal enzyme that has been shown to be overexpressed or exhibit altered localization in tumors of the lung, colon, prostate, breast, and stomach (reviewed in refs. 6 and 7). Overexpression and/or altered localization of CTSB is thought to result in degradation of the basement membrane facilitating tumor invasion and metastasis (8). Other studies have shown altered expression of CTSB is an independent predictor of poor prognosis in tumors of the lung, colon, and breast (9–11). However, there has been a lack of evidence for primary genomic alterations involving CTSB.
In the present study, two-dimensional (2D) DNA electrophoresis was used to identify amplified restriction-fragments in specimens of esophageal adenocarcinoma. Two such fragments were cloned from an early stage tumor and were found to map to chromosome bands 8p22–23, thus identifying a previously undescribed amplicon. Amplification and expression of CTSB and additional known genes from this region were examined to determine potential associations of overexpression of these genes with esophageal cancer. We demonstrate the occurrence of CTSB gene amplification in a subset of esophageal adenocarcinomas, which overexpress CTSB.
MATERIALS AND METHODS
Patients and Tissues.
After obtaining written consent, specimens of normal squamous esophagus and esophageal adenocarcinoma from 66 patients including the Barrett’s metaplasia from 20 of these patients were obtained after esophagectomy at the University of Michigan Medical Center from 1992 to 1997. A portion of each tissue was immediately frozen in liquid nitrogen, and a second portion was embedded in OCT compound (Miles Scientific, Naperville, IL), frozen in isopentane cooled to the temperature of liquid nitrogen, and then stored at −70°C.
2D DNA Electrophoresis.
High molecular weight DNA for 2D gel electrophoresis was extracted as described (12). 2D electrophoresis was performed as described (13). In brief, DNA from control and tumor tissues was digested with the methylation sensitive restriction enzyme NotI, end-labeled with [α-32P]dCTP and [α-32P]dGTP (DuPont/NEN), and further digested with EcoRV. The resulting fragments were size-fractionated in the first dimension in 32-cm 0.9% disc-agarose-gels. The separated DNA fragments were further digested in situ with the HinfI restriction enzyme, and the DNA was further size-fractionated in the second dimension in large format (25 × 43 cm) 5.25% polyacrylamide gels. The gels were either dried and autoradiographic images obtained by phosphor storage technology (Molecular Dynamics) or left hydrated and placed on Hyperfilm (Amersham) for preparative gels. Digitized images were analyzed by using software previously developed (13). The level of DNA amplification for specific fragments was estimated by densitometry using nonamplified, two-copy spot level intensities from normal tissue DNA as standards.
Isolation and Cloning of Amplified Fragments from 2D Gels.
Amplified fragments were cloned directly from preparative gels as described (14). In brief, the DNA fragment of interest was recovered by elution with a high-salt buffer (50 mM Tris⋅HCl pH 8.0, 1 M NaCl, 10 mM EDTA) and ligated in the presence of DNA ligase at 16°C for 40 h into a modified NotI–HinfI fragment compatible Bluescript SK+ vector digested with SapI and NotI. Transformation of competent cells was performed by electroporation. DNA sequence determination of the NotI–HinfI fragments was performed by automated sequence analysis (Applied Biosystems) at the DNA Sequencing Core Facility of The University of Michigan.
Identification of Bacterial Artificial Chromosome (BAC) Clones.
BAC clones that contained the amplified fragment sequences were identified by PCR-based probing of a human BAC library as specified by the supplier (Research Genetics, Huntsville, AL). The fragment-specific primers for the D4 fragment were 5′-AGGGGAGCGGAGAGAAG-3′ and 5′-AGGGGACGAGCAGGAAC-3′, and for the D5 fragment, the specific primers were 5′-TCAGCACTCAACACCTTC-3′ and 5′-GGGTTGTTCGATTTCAGG-3′. Amplifications were performed in a 25 μl of reaction volume by using Taq polymerase (Promega) and 2 μl of pooled BAC library DNA in a mixture containing 0.75 pM of each primer, 0.15 mM each of dATP, dCTP, dGTP, and dTTP (Perkin–Elmer), 2.0 mM MgCl2, and 1.2% formamide. The mixtures were denatured at 94°C for 2 min, followed by 30 thermal cycles with each cycle representing 30 sec at 94°C and 30 sec at 62°C for D4-specific primers and 56°C for D5-specific primers and 30 sec at 72°C. The presence of an amplification product was determined by size fractionation in a 2% agarose gels containing ethidium bromide.
Fluorescent in Situ Hybridization (FISH).
Individual BAC and YAC clones were initially mapped by using procedures previously described (15). In brief, total DNA from BACs 393H12 and 538D6 containing the D4 and D5 fragments, respectively, and YAC 725C12 containing CTSB (Research Genetics) were labeled with biotin and hybridized to normal human metaphase chromosome spreads in the presence of human Cot-1 DNA. Probes were detected with FITC-conjugated avidin and FITC-conjugated anti-avidin (Vector Laboratories).
Two color FISH mapping was performed by using a modification of the procedure of Wilke et al. (16). The D4 and D5 BACs labeled with biotin and the CTSB YAC labeled with digoxygenin were simultaneously hybridized to normal human metaphase chromosomes cultured in the presence of BrdUrd for enhancement of chromosome banding (17). The biotin-labeled probes were detected with FITC-conjugated avidin and FITC-conjugated anti-avidin, and the digoxygenin-labeled probe was detected with rhodamine-conjugated anti-digoxygenin and Texas-red conjugated anti-sheep IgG (Vector Laboratories). The chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Analysis was performed by using a Zeiss Axioscope epifluorescence microscope equipped with a Zeiss triple band set, allowing simultaneous visualization of FITC, Texas Red, and DAPI. Images for publication were captured digitally with a Photometrics CCD camera, processed on a MAC Quadra 900 computer with photoshop software (Adobe Systems, San Jose, CA), and printed on a Kodak XLS 8600 PS Printer.
Probes.
The CTSB, farnesyl-diphosphate farnesyltransferase (FDFT1), and arylamine N-acetyltransferase (NAT-1) probes were generated by reverse transcription–PCR using primer pairs designed from the published cDNA sequences (18–20). The CTSB primers were: 5′-GGGGACGGCTGTAATGGT-3′ and 5′-AGAAGCCATTGTCACCCCAG-3′, the FDFT1 primers were: 5′-GCGGAAGGTGATGCCCAAGA-3′ and 5′-TCCGACCAGCCCAGCAACAT-3′, and the NAT-1 primers were: 5′-TTTCGTTTTGTTTTCCTTGCTT-3′ and 5′GTTGGGTTCTGATTTGGTCT-3′. Ten micrograms of total RNA from esophageal tissues was reverse transcribed as described (21). PCR conditions for both CTSB and FDFT1 cDNA amplification were 35 cycles of 94°C for 30 sec, 57°C for 30 sec, and 72°C for 30 sec. Products were size-fractionated in 1% agarose gels with ethidium bromide. Lipoprotein lipase (22), (LPL) (Consortium clone number 612741, Research Genetics) and the expressed sequence tag (EST) clone number yn69e08.sl (GenBank accession no. H22460) were used as probes after inserts were released by restriction digestion and purified by electro-elution. The EST was previously mapped to the chromosomal marker D8S503. Identity of probes was confirmed by automated DNA sequence analysis. A cDNA representing the single-copy gene chemokine-like receptor 1 (CMKLR1) (generous gift of Dr. Ira Gantz; ref. 23) was used to probe membranes as a control for loading and transfer of DNA. Probes were radiolabeled with [α-32P]dCTP (Dupont/NEN) by using a random primer kit (Life Technologies, Gaithersburg, MD).
Southern and Northern Blot Analysis.
Genomic DNA was isolated from tissues and purified as described (12), digested with EcoRI (Promega), size-fractionated on 0.9% agarose gels containing ethidium bromide, and vacuum-transferred to nylon membranes. Total RNA was extracted by using the Trizol reagent (Life Technologies) as recommended by the supplier. Sample RNA (10 μg) was size-fractionated in 1.2% agarose/2.2 M formaldehyde gels and vacuum-transferred to nylon membranes.
Membranes were hybridized and washed as described (21). Phosphor storage technology (Molecular Dynamics) was used for signal detection. Normalization for DNA loading and transfer was obtained by using a ratio of each individual probe/CMKLR1 obtained by computerized densitometry (Molecular Dynamics). An increase in signal intensity ratio of 3-fold or greater in the tumor compared with that in the normal control tissue was considered as representing gene amplification. Normalization for RNA loading and transfer was obtained by using a 32P-labeled ribosomal 28S oligomer described (24).
Western Blot Analysis.
Total tissue protein was extracted in a buffer containing Nonidet P-40 as described (21). Ten micrograms of total protein extract was fractionated by 12% SDS/PAGE and transferred to nylon membranes. The CTSB protein was detected with a anti-CTSB polyclonal antibody (K90049C, Biodesign International, Kennebunk, ME) at a 1:750 dilution and followed by incubation with a peroxidase-conjugated secondary antibody at a 1:20,000 dilution (ICN Biomedicals, Aurora, OH). Enhanced chemilumnescence (Pierce) and Hyperfilm (Amersham) were used for detection. After stripping the membranes as previously described (21), the membranes were probed with an anti-β-actin antibody (Sigma), which served as a control for loading and transfer of total protein.
Immunohistochemistry.
Frozen specimens were sectioned at 5 μm, placed on 0.1% poly-l-lysine-coated slides, and fixed in 100% acetone at −20°C for 10 min. Endogenous peroxidase activity was quenched with two changes of 0.5% hydrogen peroxide for 45 min each. Nonspecific binding was blocked using a 1:20 dilution of rabbit serum (Vector Laboratories) in PBS-1% BSA. The CTSB protein was detected by using the anti-CTSB antibody (Biodesign International) at a 1:1500 dilution in PBS-1% BSA. On each slide, a negative control of either no primary antibody or sheep serum diluted 1:1500 in PBS-1% BSA was used to identify potential nonspecific staining. Immunoreactivity was detected by using the Vectastain avidin/biotin complex kit (Vector Laboratories) according to the manufacturers specifications by using 3,3′-diamino-benzadine (Sigma) as the chromogen. The slides were lightly counterstained with Harris-modified hematoxylin and permanently mounted. Staining was analyzed by two independent observers by using light microscopy.
RESULTS AND DISCUSSION
Identification and Cloning of Amplified Restriction Fragments.
DNA from 66 patients’ normal esophagus and esophageal adenocarcinomas were analyzed by Southern blot analysis for amplification of the erbB2, epidermal growth factor receptor (EGFR), c-myc, and K-ras genes (unpublished data). Adenocarcinoma specimens not demonstrating amplifications of these known proto-oncogenes (23 patients) were selected for 2D DNA electrophoresis, so as to improve the probability that identified amplified restriction fragments would represent sequences of previously undescribed amplicons.
Amplified restriction-digest fragments observed in the 2D patterns of tumor DNA were identified and their intensity determined by computer-assisted densitometry. Amplified fragments were cataloged by using both the estimated NotI–EcoRV and NotI–HinfI fragment lengths. One stage I esophageal adenocarcinoma, of particular interest because this tumor resulted in a worse-than-expected prognosis, demonstrated five amplified restriction fragments. Two NotI–HinfI fragments (D4 and D5), each amplified ≈20-fold in comparison with control tissue, were extracted from a preparative gel by using DNA from this tumor, and cloned into a NotI–HinfI fragment compatible, Bluescript SK+ vector (Fig. 1). Southern blot analysis of DNA from this patient by using the D4 and D5 fragments as probes confirmed both fragments to be amplified 20-fold.
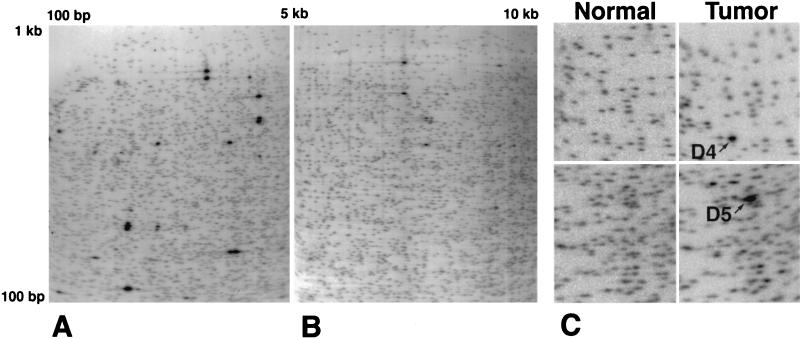
Figure 1.
2D DNA electrophoresis gels from esophageal squamous epithelium of NotI–EcoRV fragments 100–5,000 bp in size (A) and NotI–EcoRV fragments 5,000–10,000 bp in size (B). In normal controls, spots of increased intensity as compared with the intensity of spots from two-copy fragments represent multiple copy ribosomal DNA fragments. This technique allows analysis of over 6,000 genomic fragments. (C) Enlargement of a portion of two regions of larger fragment images comparing spot patterns from normal control tissue and esophageal adenocarcinoma DNA from the same patient. The fragments labeled D4 and D5 demonstrate an ≈20-fold amplification in the tumor and were subsequently extracted from a preparative gel and cloned.
Sequence analysis of the 716-bp NotI–HinfI D4 fragment showed it to be GC-rich and to contain an Alu repetitive element. The 598-bp NotI–HinfI D5 fragment contained a long terminal repeat repetitive sequence. A computer-based query of the D4 and D5 sequences against genomic and EST sequence databases showed that neither fragment shared homology with previously described sequences. For further sequence analysis, and because the D4 and D5 fragments were of sub-optimal length as probes for FISH analysis to determine the chromosomal location of the fragments, larger genomic clones containing the D4 and D5 sequences were isolated.
Identification of BAC Clones and the Chromosomal Location of the D4 and D5 Fragments.
A BAC library was used as a source of genomic DNA clones. PCR-based screening of the BAC library with primers specific for the D4 and the D5 fragments identified representative BAC clones for each fragment. The BAC clone 393H12 was confirmed to contain the D4 fragment, and the BAC clone 538D6 was confirmed to contain the D5 fragment by Southern blot analysis of NotI–EcoRV digested BAC DNA. Reciprocal hybridization of the BAC Southern blots indicated no homology between the BAC clones.
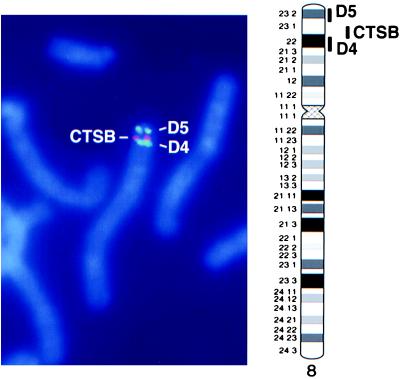
The BAC clones were used as probes for FISH to normal chromosomes. The D4 BAC mapped at chromosome band 8p22 and the D5 BAC mapped at chromosome band 8p23 (Fig. 2). This finding and the similar copy number for both fragments suggested the D4 and D5 fragments were part of a single amplicon at 8p22–23. An amplicon in this chromosomal region has not been previously reported.
Figure 2.
FISH to normal human chromosomes by using a BAC representative of the D4 fragment (centromeric green band) and a BAC representative of the D5 fragment (telomeric green band) defines the location of a previously undescribed amplicon at 8p22–23. Cohybridization with a YAC containing the CTSB gene (red band) demonstrates the CTSB locus in relation to the D4 and D5 loci.
Identification of Genes Encompassed in the 8p22–23 Amplicon.
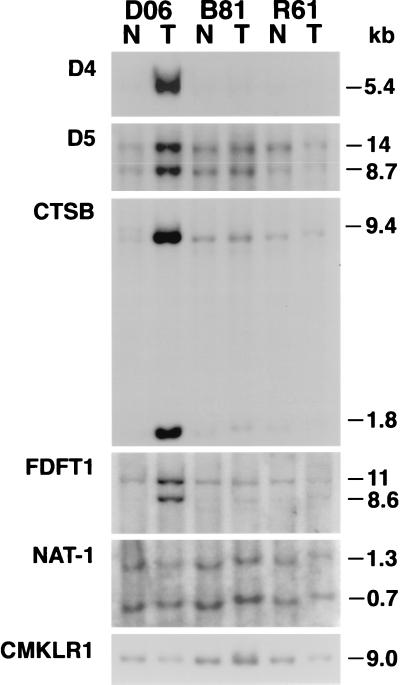
To determine whether this amplification event was unique to this single tumor or present in other tumors, Southern blot analysis of DNA from 66 patients specimens of normal and tumor tissues was performed by using the D4 and D5 fragments as probes. Amplification of the D4 fragment was identified in 1 of 66 (1.5%), and amplification of the D5 fragment was identified in 2 of 66 (3.0%) of the esophageal adenocarcinomas (Fig. 3).
Figure 3.
Southern blot analysis of DNA from three patients specimens of normal squamous esophagus (N) and esophageal adenocarcinoma (T) hybridized with CTSB, FDFT1, and NAT-1 cDNA probes demonstrating 20-fold amplification of CTSB and FDFT1 genes in tumor (D06). The NAT-1 gene was not amplified in any of the 66 esophageal adenocarcinomas analyzed.
Genes previously mapped to the 8p22–23 region were examined as candidate genes encompassed in the amplicon (ref. 5 and National Center for Biotechnology Information Gene Map: http://www.ncbi.nlm.nih.gov/genemap). The cysteine protease CTSB, a gene that has been associated with malignancy, was considered to be an excellent candidate cancer-related gene encompassed in the 8p22–23 amplicon. It also was likely that the 8p22–23 amplicon contained multiple genes. The erbB2 amplicon is known to contain several genes, most of which are overexpressed as a result of amplification (25, 26). Other previously characterized gene loci from the 8p22–23 region were evaluated for amplification and overexpression and included: farnesyl-diphosphate farnesyltransferase (FDFT1), an enzyme involved in cholesterol synthesis (19); NAT-1, an enzyme involved in the metabolism of aromatic amines, hydrazines, and N-hydroxylamines (27, 28); and LPL, an enzyme that catalyzes the hydrolysis of triglycerides (22). For the initial characterization of the 8p22–23 amplicon, investigation was limited to the evaluation of these characterized genes and one uncharacterized EST mapped to this region.
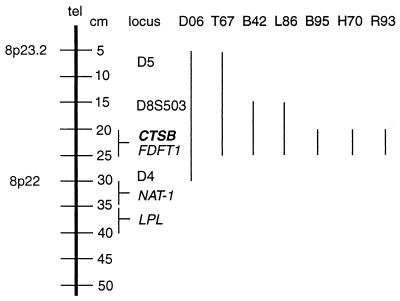
Southern blot analysis of 66 esophageal adenocarcinomas using the above described probes demonstrated amplification of both CTSB and FDFT1 in eight (12.1%) of the tumors (Fig. 3). The level of amplification ranged from 4- to 20-fold for both genes. Twenty cases of Barrett’s esophagus also were examined from these patients. Low-level (4-fold) CTSB and FDFT1 amplification was only observed in one specimen of dysplastic Barrett’s esophagus from whose adenocarcinoma DNA the D4 and D5 fragments were cloned (D06). Neither NAT-1 nor LPL were amplified in any of the tumors, suggesting these genes are not part of this amplicon. An uncharacterized EST (D8S503) mapped to a region telomeric to CTSB was found to be amplified in only three of seven tumors showing CTSB amplification. These analyses narrowed the region of most frequent amplification to an ≈5–14 cM region flanking the CTSB and FDFT1 loci (Fig. 4). Given the established role of CTSB in tumorigenesis in other tissues, these data suggested that CTSB is a candidate gene whose amplification may play a role in esophageal adenocarcinoma tumorigenesis.
Figure 4.
Summary of Southern blot data demonstrating amplification of 8p loci in esophageal adenocarcinomas. Bars at right indicate the loci amplified in the seven tumors studied with all probes. Mapping data for CTSB, FDFT1, LPL, NAT-1, and D8S503 were taken from the human chromosome 8 Gene Map of the National Center for Biotechnology Information (http://www.ncbi.nih.gov/genemap). The D4 locus was mapped by FISH and by radiation hybrid panel analysis to 29 cM. The D5 locus was mapped by FISH to 8p23.2.
CTSB Is Overexpressed in Tumors with 8p22–23 Amplification.
To determine whether gene amplification is associated with overexpression of CTSB and FDFT1 mRNA, Northern blot analysis of 12 patients’ specimens (six amplified and six without amplification) was performed. Overexpression of CTSB and FDFT1 mRNA was detected in six of six (100%) of the amplified esophageal adenocarcinomas (Fig. 5). Importantly, CTSB mRNA overexpression was present in two of six (33%) of nonamplified tumors; however, FDFT1 mRNA overexpression without amplification was not observed. CTSB and FDFT1 mRNA overexpression also was observed in 1 of 20 (5%) of specimens of Barrett’s esophagus. This specimen showed low-level amplification of the D4 and D5 fragments and suggests amplification of the 8p22 region occurs infrequently in Barrett’s metaplasia. Amplification in a stage I esophageal adenocarcinoma suggests that amplification may occur early in tumorigenesis. This data also showed that amplification of the CTSB and FDFT1 gene loci was associated with overexpression of CTSB and FDFT1 mRNA. Furthermore, overexpression of CTSB mRNA, but not FDFT1 mRNA, may occur by a mechanism(s) other than amplification. This finding indicates that overexpression of the CTSB candidate gene may drive selection for the 8p22–23 amplicon.
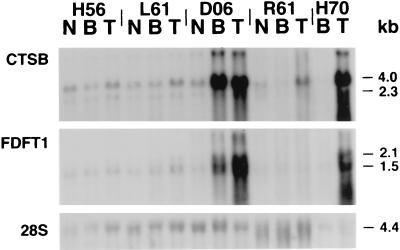
Figure 5.
Northern blot analysis of total RNA from normal squamous esophagus (N), Barrett’s esophagus (B), and esophageal adenocarcinoma (T) specimens hybridized with CTSB and FDFT1 cDNA probes. CTSB mRNA is overexpressed in tumors identified to contain the 8p22–23 amplicon (D06 and H70) as well as a tumor not containing the amplicon (R61). FDFT1 mRNA is only overexpressed in tumors containing the 8p22–23 amplicon.
Overexpression of FDFT1 appeared to be associated only with gene amplification and therefore its overexpression is less likely to be causally associated with malignancy. The FDFT1 promoter contains two potential nuclear factor (NF)-1-binding sites, two steroid receptor element-1 elements, two Sp1 promoter elements, and one CCAAT box (29). The presence of both housekeeping-promoter activity and inducible-promoter activity suggests that gene amplification could be expected to result in mRNA overexpression. The overexpression of FDFT1 likely results from inclusion within the CTSB amplicon.
Analysis of CTSB Protein Expression.
We next evaluated CTSB protein levels because elevated mRNA levels do not necessarily correlate with high protein levels. The regulation of CTSB expression in other tumor types is poorly understood but may include alternative mRNA splicing, the use of different promoter regions, post-translational protein modification resulting in overexpression, and altered trafficking of the protein resulting in increased protease activity and extra-lysosomal expression (6). Evaluation of CTSB protein levels in esophageal tissues was performed by using Western blot analysis of total protein extracts using a commercially available polyclonal anti-CTSB antibody. The specificity of this antibody and any potential quantitative differences in CTSB protein levels in the normal squamous esophagus, Barrett’s metaplasia, and tumors from the same patients were evaluated. Western blot analysis identified CTSB-specific bands similar to those previously reported (10) with a doublet band of ≈27 kDa in size observed in all esophageal tissue types (Fig. 6). The smaller of the two bands represents the single chain form of CTSB (10) and was found to be increased in tumor specimens compared with normal controls. A larger band of ≈33 kDa in size was observed only in tumors containing the 8p22–23 amplicon. Higher Mr forms of CTSB have been shown to be associated with extracellular expression of CTSB (8). These studies demonstrated that CTSB gene amplification and mRNA overexpression result in a substantial increase in CTSB protein, including the occurrence of higher Mr forms that may be associated with extracellular expression of CTSB in tumors.
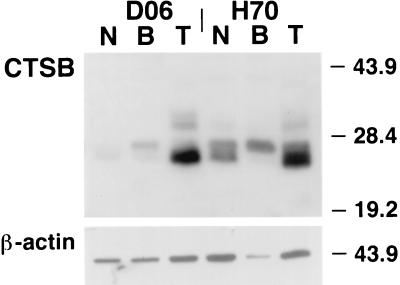
Figure 6.
Western blot analysis of total protein extract from normal squamous esophagus (N), Barrett’s esophagus (B), and esophageal adenocarcinoma (T) specimens probed with a polyclonal anti-CTSB antibody. A 27-kDa doublet band is identified in all specimens. The lower Mr band comprising this doublet is more abundant in tumor specimens. A higher Mr band, associated with extracellular expression of CTSB protein, also is identified in the tumor specimens.
We evaluated CTSB protein localization by immunohistochemical analysis of 40 esophageal adenocarcinoma, 14 Barrett’s metaplasia, and 4 squamous esophageal epithelium specimens. The absence of staining in control sections confirmed specificity of staining with anti-CTSB antibody. Low-level, but detectable CTSB protein was observed throughout the cytoplasm of esophageal squamous epithelium (Fig. 7). A pattern of expression similar to that observed in normal colon epithelium (10) was seen in the columnar cells of Barrett’s metaplasia with CTSB protein located in the apical region of the cells. In regions of Barrett’s esophagus determined to be dysplastic due to loss of goblet cell formation, and the presence of stratified and atypic nuclei, immunostaining of CTSB was observed throughout the cytoplasm. It is unclear whether this staining represents extra-lysosomal expression of the protein or a loss of normal cell architecture and altered localization of lysosomes in dysplastic cells.
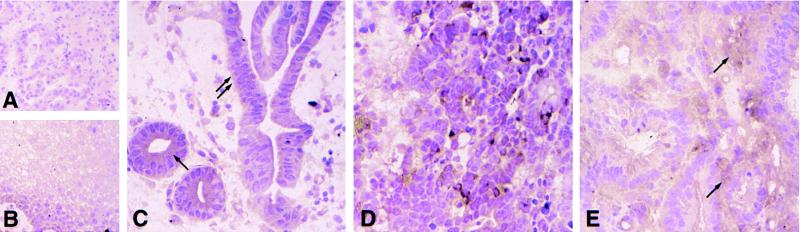
Figure 7.
Immunohistochemical analysis for CTSB protein expression. (A) Esophageal adenocarcinoma specimen incubated with sheep serum served as a negative control. (B) The squamous epithelium of the esophagus demonstrates faint cytoplasmic staining for CTSB protein expression predominantly in the basal proliferative zone of the epithelium. (C) The columnar cells of Barrett’s esophagus demonstrate CTSB protein primarily localized to the apical region of the columnar cells (arrow). In contrast, CTSB protein is identified diffusely throughout the cytoplasm in regions of dysplasia (double arrows) within Barrett’s esophagus specimens. (D) Esophageal adenocarcinoma specimens stain heterogeneously for CTSB protein in a diffuse cytoplasmic pattern. (E) Extracellular CTSB expression (arrows) was identified in 29 of 40 (72.5%) of the esophageal adenocarcinoma specimens. (original magnifications, ×200).
The pattern of diffuse cytoplasmic staining also was identified in all the specimens of esophageal adenocarcinoma, and staining also was identified in the extracellular matrix of 29 of 40 (72.5%) of the tumor specimens. The finding of extracellular expression of CTSB correlated with the identification of higher Mr forms of the protein by Western blot analysis. CTSB staining was heterogeneous throughout the tumor specimens, similar to that observed in adenocarcinomas of the prostate and lung (30, 31). CTSB protein did not localize consistently along the basement membrane as described in colon and prostate cancer (10, 30). Further studies are necessary to determine how the pattern of CTSB expression in tumors with gene amplification may differ from tumors overexpressing this protease due to other mechanisms.
All of the patients with CTSB amplification and/or expression had poor clinical outcome. However, because most patients with esophageal adenocarcinoma also do poorly and the small number of patients in this study, a definitive association between CTSB amplification and survival at the present time could not be established.
In summary, we have identified a previously undescribed amplicon at chromosome bands 8p22–23 in esophageal adenocarcinoma. We evaluated four characterized and one uncharacterized genes previously localized to this region for their potential occurrence in the amplicon. Importantly, the cysteine protease CTSB for which altered expression has been associated with malignancy in other tissues was found to be amplified and overexpressed in esophageal adenocarcinoma, demonstrating genomic alteration involving CTSB. These data provide the first evidence of amplification as a mechanism for overexpression of CTSB. These findings provide strong support for an important role for the CTSB protease in malignant transformation and/or progression in esophageal adenocarcinoma. Our studies suggest amplification of the CTSB gene may be a mechanism of overexpression in other malignancies.
Acknowledgments
The authors wish to thank Barbara Lamb and Diane Miller for their excellent technical support and Drs. Robin Leach and Stephen Wood, University of Texas at San Antonio for mapping the D4 fragment by radiation hybrid analysis. This study was funded by National Institutes of Health Grants CA71606 (to D.G.B.), CA26803 (to S.H.), Training Grant CA09672-06 (to S.J.H.), and the Frederick A. Coller Society (to S.J.H.).
ABBREVIATIONS
- 2D
two-dimensional
- BAC
bacterial artificial chromosome
- CTSB
cathepsin B
- FISH
fluorescent in situ hybridization
- FDFT1
farnesyl-diphosphate farnesyltransferase
- LPL
lipoprotein lipase
- NAT-1
arylamine N-acetyltransferase
- EST
expressed sequence tag
References
- 1. Reid B J. In: Textbook of Gastroenterology. Yamada Y, Alpers D H, Owyang C, Powel D W, Silverstein F E, editors. Philadelphia: Lippincott; 1991. pp. 1159–1178. [Google Scholar]
- 2.Blot W J, Devesa S S, Kneller R W, Fraumeni J F. J Am Med Assoc. 1991;265:1287–1289. [PubMed] [Google Scholar]
- 3.Spechler S J, Goyal R K. N Engl J Med. 1986;315:362–371. doi: 10.1056/NEJM198608073150605. [DOI] [PubMed] [Google Scholar]
- 4.Winters D, Spurling T J, Chobainian S J, Curtis D J, Esposito R L, Hacker J F, Johnson D A, Cruess D F, Cotelingham J D, Gurney M S, et al. Gastroenterology. 1987;92:188–194. [PubMed] [Google Scholar]
- 5.Wang X, Chan S J, Eddy R L, Byers M G, Fukushima Y, Henry W M, Haley L L, Steiner D F, Shows J B. Cytogenet Cell Genet. 1998;59:710–711. [Google Scholar]
- 6.Sloane B F, Moin K, Krepela E, Rozhin J. Cancer Metastasis Rev. 1990;9:333–352. doi: 10.1007/BF00049523. [DOI] [PubMed] [Google Scholar]
- 7.Keppler D, Sloane B F. Enzyme Protein. 1996;49:94–105. doi: 10.1159/000468619. [DOI] [PubMed] [Google Scholar]
- 8.Sloane B F. Semin Cancer Biol. 1990;1:137–152. [PubMed] [Google Scholar]
- 9.Sukoh N, Abe S, Ogura S, Isobe H, Takekawa H, Inoue K, Kawakami Y. Cancer. 1994;74:46–51. doi: 10.1002/1097-0142(19940701)74:1<46::aid-cncr2820740109>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 10.Campo E, Munoz J, Miquel R, Palacin A, Cardesa A, Sloane B F, Emmert-Buck M R. Am J Pathol. 1994;145:301–309. [PMC free article] [PubMed] [Google Scholar]
- 11.Foekens J A, Kos J, Peters H A, Krasovec M, Look M P, Cimerman N, Meijer-van Gelder M E, Henzen-Logmans S C, van Putten W L, Klijn J G. J Clin Oncol. 1198;16:1013–1021. doi: 10.1200/JCO.1998.16.3.1013. [DOI] [PubMed] [Google Scholar]
- 12.Blin N, Stafford D W. Nucleic Acids Res. 1976;3:2303. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuick R, Asakawa J-I, Neel J V, Satoh C, Hanash S M. Genomics. 1995;25:345–353. doi: 10.1016/0888-7543(95)80032-h. [DOI] [PubMed] [Google Scholar]
- 14.Almeida A, Zhu X X, Vogt N, Tyagi R, Muleris M, Dutrillaux A-M, Dutrillaux B, Ross D, Malfoy B, Hanash S. Oncogene. 1998;16:2997–3002. doi: 10.1038/sj.onc.1201828. [DOI] [PubMed] [Google Scholar]
- 15.Wilke C M, Hall B K, Hoge A, Paradee W, Smith D I, Glover T W. Hum Mol Genet. 1996;5:187–195. doi: 10.1093/hmg/5.2.187. [DOI] [PubMed] [Google Scholar]
- 16.Wilke C M, Guo S-W, Hall B K, Boldog F, Gemmill R M, Chandrasekharappa S C, Barcroft C L, Drabkin H A, Glover T W. Genomics. 1994;22:319–326. doi: 10.1006/geno.1994.1390. [DOI] [PubMed] [Google Scholar]
- 17.Lemieux N, Dutrillaux B, Viegas-Pequignot E. Cytogenet Cell Genet. 1992;59:311–312. doi: 10.1159/000133277. [DOI] [PubMed] [Google Scholar]
- 18.Chan S J, San Segundo B, McCormick M B, Steiner D F. Proc Nat Acad Sci USA. 1986;83:7721–7725. doi: 10.1073/pnas.83.20.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Summers C, Karst F, Charles A D. Gene. 1993;136:185–192. doi: 10.1016/0378-1119(93)90462-c. [DOI] [PubMed] [Google Scholar]
- 20.Ohsako S, Deguchi T. J Biol Chem. 1990;265:4630–4634. [PubMed] [Google Scholar]
- 21.Hughes S J, Nambu Y, Soldes O S, Hamstra D, Rehemtulla A, Iannettoni M D, Orringer M B, Beer D B. Cancer Res. 1997;57:5571–5578. [PubMed] [Google Scholar]
- 22.Wion K L, Kirchgessner T G, Lusis A J, Schotz M C, Lawn R M. Science. 1987;235:1638–1641. doi: 10.1126/science.3823907. [DOI] [PubMed] [Google Scholar]
- 23.Gantz I, Konda Y, Yang Y K, Miller D E, Dierick H A, Yamada T. Cytogenet Cell Genet. 1996;74:286–290. doi: 10.1159/000134436. [DOI] [PubMed] [Google Scholar]
- 24.Hanson L A, Nuzum E O, Jones B C, Malkinson A M, Beer D G. Exp Lung Res. 1993;17:371–387. doi: 10.3109/01902149109064425. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka S, Mori M, Akiyoshi T, Tanaka Y, Mafune K, Wands J R, Sugimachi K. Cancer Res. 1997;57:28–31. [PubMed] [Google Scholar]
- 26.Akiyama N, Sasaki H, Ishizuka T, Kishi T, Sakamoto H, Onda M, Hirai H, Yazaki Y, Sugimura T, Terada M. Cancer Res. 1997;57:3548–3553. [PubMed] [Google Scholar]
- 27.Zenser T V, Lakshmi V M, Rustan T D, Doll M A, Deitz A C, Davis B B, Hein D W. Cancer Res. 1996;56:3941–3947. [PubMed] [Google Scholar]
- 28.Matas N, Thygesen P, Stacey M, Risch A, Sim E. Cytogenet Cell Genet. 1997;77:290–295. doi: 10.1159/000134601. [DOI] [PubMed] [Google Scholar]
- 29.Guan G, Jiang G, Koch R L, Shechter I. J Biol Chem. 1995;270:21958–21965. doi: 10.1074/jbc.270.37.21958. [DOI] [PubMed] [Google Scholar]
- 30.Sinha A A, Wilson M J, Gleason D F, Reddy P K, Sameni M, Sloane B F. Prostate. 1995;26:171–178. doi: 10.1002/pros.2990260402. [DOI] [PubMed] [Google Scholar]
- 31.Inoue T, Ishida T, Sugio K, Sugimach K. Cancer Res. 1994;54:6133–6136. [PubMed] [Google Scholar]