Abstract
Hepatitis C virus (HCV) is the leading cause of chronic liver disease worldwide. HCV is also the major cause of mixed cryoglobulinemia, a B-lymphocyte proliferative disorder. Direct experimentation with native viral proteins is not feasible. Truncated versions of recombinant E2 envelope proteins, used as surrogates for viral particles, were shown to bind specifically to human CD81. However, truncated E2 may not fully mimic the surface of HCV virions because the virus encodes two envelope glycoproteins that associate with each other as E1E2 heterodimers. Here we show that E1E2 complexes efficiently bind to CD81 whereas truncated E2 is a weak binder, suggesting that truncated E2 is probably not the best tool with which to study cellular interactions. To gain better insight into virus-cell interactions, we developed a method by which to isolate E1E2 complexes that are properly folded. We demonstrate that purified E1E2 heterodimers bind to cells in a CD81-dependent manner. Furthermore, engagement of B cells by purified E1E2 heterodimers results in their aggregation and in protein tyrosine phosphorylation, a hallmark of B-cell activation. These studies provide a possible clue to the etiology of HCV-associated B-cell lymphoproliferative diseases. They also delineate a method by which to isolate biologically functional E1E2 complexes for the study of virus-host cell interaction in other cell types.
Hepatitis C virus (HCV) infection is a major health problem affecting an estimated 160 million people worldwide (36). It is a major cause of chronic hepatitis, liver cirrhosis, hepatocellular carcinoma (53), and mixed cryoglobulinemia, a B-lymphocyte proliferative disorder (reviewed in references 6 and 49). HCV is a small enveloped virus that belongs to the Hepacivirus genus in the Flaviviridae family (33). Its genome encodes a single ∼3,000-amino-acid polyprotein that is co- and posttranslationally processed by viral and cellular proteases to yield the mature structural and nonstructural proteins (33, 37). The structural proteins—the core protein and envelope glycoproteins E1 and E2—are believed to be the major constituents of HCV particles. The E1 and E2 envelope proteins are N glycosylated in their large N-terminal ectodomains and are anchored into membranes by their hydrophobic C-terminal transmembrane domains (TMDs) (39). These domains have been shown to be endoplasmic reticulum (ER) retention signals (10, 12, 20, 23). E1 and E2 associate to form two types of complexes: properly folded E1E2 heterodimers stabilized by noncovalent interactions and misfolded disulfide-linked aggregates (for a review, see reference 39).
The E1E2 noncovalent heterodimer, comprising the viral envelope (reviewed in reference 39), is involved in viral entry (3, 30); however, the mechanism of HCV cell entry is not clear. Several putative cell surface receptors of HCV or recombinant E2 proteins have been identified (1, 25, 34, 45, 46, 50, 51). Among these receptors, human CD81 has been repeatedly shown to interact with recombinant soluble E2, the E1E2 complex, HCV-like particles, and HCV particles from infectious plasma (3, 8, 17, 22, 27, 30, 31, 35, 42, 45, 48, 55, 59).
CD81 is a member of the tetraspanin family, which contains four TMDs, short intracellular domains, and two extracellular loops designated the small extracellular loop and the large extracellular loop (LEL). CD81 is widely expressed and is associated with other membrane proteins that vary in different cell lineages (32). The CD81 binding site for E2 has been localized within the LEL (45), and specific LEL amino acid residues essential for this interaction have been identified (17, 29). By contrast, the E2 regions involved in CD81 interaction are not well defined. The E2 glycoprotein is one of the most variable HCV proteins and is characterized by hypervariable region 1 (HVR1) and HVR2. Previous reports have shown that HVR2 and/or adjacent residues are involved in the interaction with CD81 (22, 48, 60). In contrast, E2 lacking or containing HVR1 binds equally well to CD81 (24, 43). A more recent study demonstrated that a region comprising residues 613 to 618 is essential for binding to CD81 and that a complex interplay between HVR1 and HVR2 may modulate E2-CD81 interactions (48).
The exact mechanisms whereby HCV establishes and maintains its persistence, which, in turn, leads to liver damage and immune modulation, are not well understood. Nevertheless, it has been shown that CD81 engagement by a truncated form of E2 (E2661) provides a costimulatory signal for human T cells (57) and an inhibitory effect on natural killer functions (14, 56).
Because of the lack of a suitable cell culture system for propagation of HCV in vitro and the unavailability of virions in sufficient quantities, truncated, secreted versions of E2 have been used as surrogates for native virus particles. Indeed, the identification of CD81 as a putative cellular receptor for HCV is based on its binding to E2661 (45). Intriguingly, intracellular forms of truncated E2, enriched for the presence of monomeric, nonaggregated E2, were found to bind CD81 with greater affinity than the secreted forms (21, 28), suggesting that structural differences exist between the intracellular and secreted forms of the E2 glycoprotein. Indeed, several monoclonal antibodies (MAbs) that recognize conformation-dependent epitopes within E2 have provided insight into the conformational state of the HCV envelope glycoproteins (5, 9, 12, 15, 16, 26, 27, 38, 43). Studies with these MAbs have demonstrated structural differences between truncated forms of E2 and full-length E1E2 complexes. Most recently, we identified a human MAb (CBH-2) that recognizes E2 only when complexed with E1. This suggests that the presence of E1 may influence the folding of E2 and that structural differences exist between E1E2 heterodimers and E2 expressed in the absence of E1 (13).
CD81-LEL binds weakly to truncated E2.
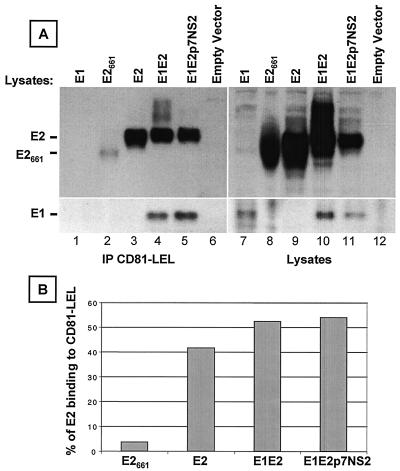
Most studies of E2-cell interactions have used truncated, secreted versions of the recombinant E2 glycoprotein, possibly because of the relative ease of production. However, it is likely that such truncated versions do not fully mimic the corresponding glycoprotein structures on HCV virions. To explore whether folding of the HCV envelope glycoprotein could influence their function, we tested various forms of these recombinant glycoproteins for the ability to bind the putative HCV receptor, CD81. Human embryonic kidney 293T cells cultured in Dulbecco modified Eagle medium-10% fetal calf serum (FCS) were transiently transfected (with the GenePORTER 2 transfection reagent [Gene Therapy Systems, Inc., San Diego, Calif.]) with 20 μg of plasmids encoding the following forms of HCV glycoproteins from genotype 1a, strain H: full-length E1 (E1; amino acids 171 to 383), truncated E2 (E2661; amino acids 364 to 661), full-length E2 (E2; amino acids 364 to 746), E1 and E2 (E1E2; amino acids 171 to 746), and E1E2p7NS2 (amino acids 171 to 1026). Intracellular glycoproteins were analyzed in a glutathione S-transferase pulldown assay as described previously (13). Expression of E2 (Fig. 1A, lane 3) or coexpression of E1 and E2 (Fig. 1A, lanes 4 and 5) resulted in efficient binding of E2 by the CD81-LEL fusion protein, i.e., 41 and 54%, respectively (Fig. 1B). In contrast, binding of truncated E2 (E2661) was inefficient at 3.6% (Fig. 1A, lane 2, and B). This result suggests that intracellular E2661, which was previously shown to be superior to its secreted form (21, 28), is probably not the best tool with which to study HCV binding to CD81. As expected, coexpression of E1 and E2 resulted in efficient precipitation of E1 by CD81-LEL (Fig. 1A, lanes 4 and 5, bottom), whereas E1 expressed alone did not bind CD81 (lane 1). It should be noted that similar amounts of E1 were present in the E1 and E1E2p7NS2 lysates (Fig. 1A, lanes 7 and 11, bottom). Because the E1 and E2 proteins form complexes (16) and because coexpression of E1 with E2 was most efficient in binding to CD81 (Fig. 1B), the E1E2 glycoprotein heterodimer is a more appropriate tool with which to study HCV attachment.
FIG. 1.
Interaction of soluble CD81 with different forms of HCV glycoproteins. (A) Human embryonic kidney 293T cells transfected with a plasmid encoding E1 (lane 1), truncated E2661 (lane 2), E2 (lane 3), E1E2 (lane 4), or E1E2p7NS2 (lane 5) or with an empty vector (lane 6) were lysed at 72 h posttransfection. Cleared lysates (5 × 106 cell equivalents) were precipitated with a recombinant fusion protein containing the LEL of human CD81 fused to glutathione S-transferase (CD81-LEL) preadsorbed onto glutathione-Sepharose 4B beads. Precipitations (lanes 1 to 6) and lysates (lanes 7 to 12; 3 × 105 cell equivalents) were separated by SDS-10% PAGE under reducing conditions, and immunoblots were analyzed with anti-E2 MAb 3/11 (top). The blots were then stripped and reprobed with the anti-E1 MAb, A4 (bottom) as previously described (13). (B) To compare the binding of various forms of E2 to CD81, the intensities of monomeric E2 were measured with ImageQuant 3.3 (Molecular Dynamics). The percentage of E2 binding to CD81 was calculated as follows: [(E2 precipitated by CD81-LEL)/(total E2 in cell lysate)] × 100.
It has previously been shown that folding and glycosylation of E1 are helped by the presence of E2 downstream of E1 on the polyprotein (18, 38, 44). Insertion of alanine substitutions within the TMDs of E1 and E2 disrupted E1E2 heterodimer formation and decreased the amount of properly folded E1 (40). These results indicate that E2 possesses a chaperone-like function to facilitate proper folding of E1 (for a review, see reference 39). In contrast to E1, E2 expressed in the absence of E1 was shown to fold properly (38). However, coexpression of E1 either in cis or in trans was required for stable association of E2 with the ER membrane, suggesting that interaction between the hydrophobic TMDs of these proteins is required for efficient ER membrane insertion and complex formation (11). Recently, we have shown that a conformation-dependent human MAb, CBH-2, recognizes E2 only when it is coexpressed with E1 (13). Together, these data suggest that the presence of E1 may also influence the folding of E2 and therefore modulate receptor binding by E2. This is confirmed by the observation that pseudoparticles containing unmodified HCV envelope glycoproteins require both E1 and E2 for infectivity (3, 30).
Purification and characterization of HCV envelope E1E2 heterodimers.
Several reports have implicated HVR1 as having a role in eliciting neutralizing antibodies and as a region essential for E2 interaction with SR-BI (51) and with ASGP-R (50). In contrast, HVR1 was shown to negatively regulate binding to CD81 (48, 51). We used this property to design a construct (pShuttle/E1FLAGE2) expressing E1 and E2 proteins (genotype 1a) in which HVR1 was exchanged with a FLAG epitope (DYKDDDDK). COS-7 cells were transiently transfected with 10 μg of pShuttle/E1FLAGE2 or pShuttle/E1E2 (expressing the parental E1E2 proteins; amino acids 171 to 746) by using the GenePORTER 2 transfection reagent. After 48 h, cells were lysed in 50 mM Tris-150 mM NaCl, pH 7.4 (TBS)-1% Triton X-100-20 mM iodoacetamide-protease inhibitors.
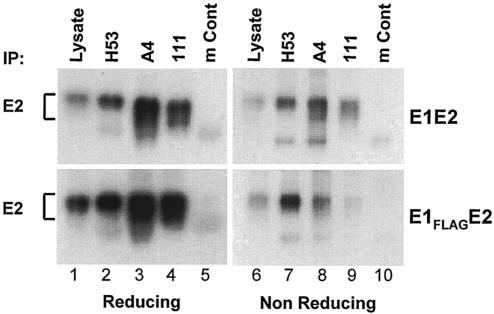
To determine whether the interaction between E1 and E2 is preserved in the context of the FLAG epitope, we immunoprecipitated E1FLAGE2 proteins with different MAbs and compared the reactivity of E1FLAGE2 to that of the parental E1E2 proteins (Fig. 2). For this purpose, MAbs were preadsorbed onto protein A-Sepharose CL-4B beads (Sigma, St. Louis, Mo.) (human MAb 111 [Z. Y. Keck, S. Rajyaguru, J. Rowe, S. Perkins, and S. K. H. Foung, 9th Int. Meet. HCV Relat. Virus, abstr. P-209, 2002], kindly provided by S. K. H. Foung) or onto rabbit anti-mouse-protein A beads (mouse MAbs H53 [12, 16] and A4 [19] and a mouse control) overnight at 4°C and washed twice in phosphate-buffered saline (PBS) containing 0.2% Triton X-100 (Sigma). Agarose-immobilized MAbs were incubated from 2 h to overnight at 4°C with clarified cell lysates. Immunoprecipitations and aliquots of lysate were analyzed by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) under reducing and nonreducing conditions and immunoblotted with anti-E2 MAb 3/11 (Fig. 2) or anti-E1 MAb A4 (data not shown). Anti-E1 MAbs A4 and 111 coimmunoprecipitated E2 proteins comparably from cell lysates expressing E1FLAGE2 or E1E2 (Fig. 2, lanes 3 and 4 and lanes 8 and 9), suggesting that the FLAG epitope did not alter E1E2 complex formation. Conversely, coimmunoprecipitation of E1 by anti-E2 MAb H53 was also comparable in E1E2 and E1FLAGE2 lysates (data not shown). It is worth noting that E2 in the context of E1FLAGE2 or native E1E2 was recognized equally well by conformation-sensitive anti-E2 MAbs H53 (Fig. 2, lanes 2 and 7) and CBH-2 (data not shown) under reducing or nonreducing conditions. Because these MAbs recognize properly folded E1E2 heterodimers (12, 13, 16), E1FLAGE2 complexes are likely to represent a native form.
FIG. 2.
FLAG epitope does not alter folding and E1E2 complex formation. COS-7 cells transfected with a plasmid encoding E1FLAGE2 or E1E2 were lysed 48 h posttransfection. Clarified lysates (1.25 × 106 cell equivalents) were immunoprecipitated (IP) with mouse conformation-dependent anti-E2 MAb H53, mouse anti-E1 MAb A4, or human anti-E1 MAb 111. Irrelevant mouse immunoglobulins (mouse control [mCont]) were used as a negative control. Bound proteins and lysates (105 cell equivalents) were run under reducing and nonreducing conditions and analyzed by Western blotting with anti-E2 MAb 3/11.
E1FLAGE2 complexes were then purified with an anti-FLAG affinity column. For this purpose, COS-7 cells were transiently transfected with pShuttle/E1FLAGE2, washed three times with ice-cold PBS, and lysed in TBS-1% Triton X-100-10 mM CaCl2-20 mM iodoacetamide-protease inhibitors. Clarified cell lysates were preadsorbed onto a protein A agarose column (Sigma) and then applied to a calcium-dependent anti-FLAG M1 affinity column (Sigma). The column was successively washed with 20 column volumes of lysis buffer and 20 column volumes of TBS-0.5% Triton X-100-10 mM CaCl2. E1FLAGE2 heterodimers were eluted with TBS-0.5% Triton X-100 containing 2 mM EDTA. Purified HCV heterodimers were then dialyzed against PBS overnight at 4°C and concentrated 30 times with a filter device (Centricon YM-3; Millipore, Bedford, Mass.). Purified proteins were loaded into three adjacent lanes, separated by 10% PAGE, and analyzed by silver staining (GelCode SilverSNAP; Pierce, Rockford, Ill.) or by Western blotting with MAb 3/11 or A4.
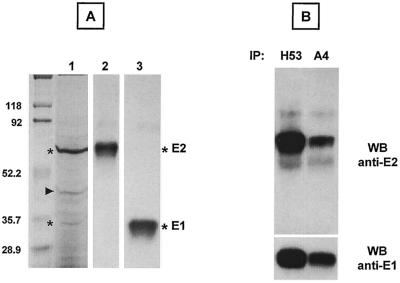
Silver staining showed bands corresponding to the sizes of E1 and E2 (Fig. 3A, lane 1, asterisks). The identities of these proteins were confirmed by simultaneous Western blotting with anti-E2 (Fig. 3A, lane 2) and anti-E1 (Fig. 3A, lane 3) MAbs. The Western blot analysis (Fig. 3A, lanes 2 and 3) suggests that the additional band with an apparent molecular mass of ∼45 kDa (Fig. 3A, lane 1, indicated by an arrow) is not related to glycoproteins E1 and E2. Purified E1FLAGE2 was further analyzed by immunoprecipitation with MAbs to E1 and E2. Conformation-sensitive anti-E2 MAb H53 immunoprecipitated E2 and coimmunoprecipitated E1 (Fig. 3B, top and bottom, respectively). Similarly, anti-E1 MAb A4 immunoprecipitated E1 and coimmunoprecipitated E2 (Fig. 3B, bottom and top, respectively). This analysis indicates that E1 and E2 remained associated after purification. The purified E1FLAGE2 complex was also recognized by CBH-2, a human MAb that recognizes only native E1E2 heterodimers (13) (data not shown). As expected, analysis under nonreducing conditions also revealed the presence of disulfide-linked E1 and E2 aggregates in the FLAG immunoaffinity-purified proteins (data not shown). Taken together, the results demonstrate that the FLAG epitope approach permits easy and efficient isolation of HCV E1E2 complexes.
FIG. 3.
Characterization of purified E1FLAGE2 complexes. (A) Purified glycoproteins were separated by SDS-PAGE under reducing conditions. The same gel was analyzed by silver staining (lane 1) or by Western blotting (WB) with anti-E2 MAb 3/11 (lane 2) or anti-E1 MAb A4 (lane 3). Sizes (in kilodaltons) of protein molecular mass markers are indicated on the left. The E1 and E2 proteins are indicated by asterisks, and the copurified protein is indicated by an arrowhead. (B) Purified E1FLAGE2 proteins were immunoprecipitated (IP) with mouse conformation-sensitive anti-E2 MAb H53 or mouse linear anti-E1 MAb A4, followed by reducing SDS-PAGE and Western blotting with MAb 3/11 (top) or A4 (bottom).
Recently, two groups have successfully generated pseudoparticles that are assembled by displaying unmodified and functional HCV glycoproteins onto retrovirus and lentivirus core particles (3, 30). These pseudoparticles are infectious for some human hepatoma cell lines, and infectivity depends on coexpression of both glycoproteins E1 and E2 (3, 30). Future experiments will determine the ability of purified E1FLAGE2 complexes to neutralize pseudotype virus infection.
Isolated E1FLAGE2 heterodimers bind to cells in a CD81-dependent manner.
MAbs to CD81 have been shown to induce cell-cell aggregation and an antiproliferative effect in B-cell lines (54). Similarly, ligation of CD81 by glycoprotein E2661 was shown to induce B-cell aggregation and inhibit cell proliferation (22). Studies done with other lymphoid cells showed that CD81 engagement by truncated E2 provides a costimulatory signal for human T cells, whereas it inhibits natural killer functions (14, 56, 57).
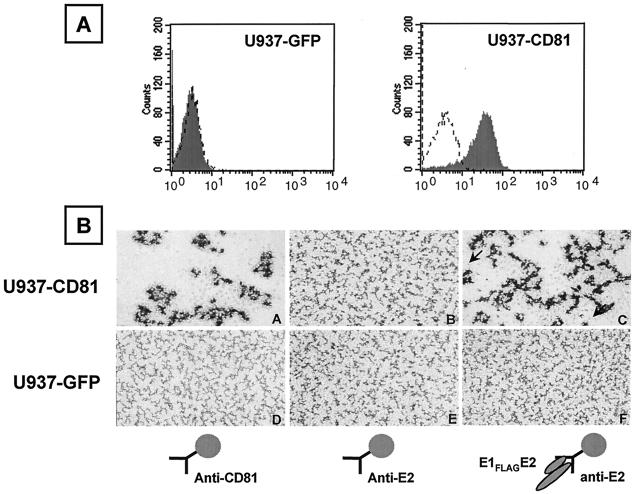
To determine whether purified E1FLAGE2 specifically interacts with cell surface-expressed CD81, we used the U937 cell line, which does not express CD81 (Fig. 4A), and engineered U937 cells expressing CD81. U937 cells were transfected with the mouse ecotropic viral receptor as detailed previously (2), and a single-cell clone was isolated for subsequent infection by retroviral vectors. Human CD81 was amplified with the FOR primer GGTATGAATTCGCCGCCATGGGAGTGGAGGG and the REV primer GGTACTCGAGCTCAGTACACGGAGCTGTTC (italicized are the EcoRI and XhoI sites, and underlined are the start and stop codons). The amplified CD81 DNA was digested with EcoRI and XhoI and inserted into the respective sites in the multicloning region of the retroviral vector pBMN-IRES-GFP. This retroviral vector encodes an IRES downstream of the multicloning region, followed by the green fluorescent protein (GFP), and was kindly provided by G. Nolan, Stanford University. Production of high-titer ecotropic retrovirus in θNX-Eco cells and infection with a retrovirus encoding GFP alone or CD81 and GFP was performed as detailed at http://www.stanford.edu/group/nolan/retroviral_systems/phx.html. Isolated U937 subclones that constitutively express CD81 (U937-CD81) or GFP (U937-GFP) were tested by flow cytometry of cells washed twice with PBS-2% BSA and incubated with an anti-CD81 MAb (5A6) (41) or an immunoglobulin G1 (IgG1) isotype control. After two washes with PBS-2% BSA, cells were incubated with phycoerythrin-conjugated anti-mouse IgG1 (Becton Dickinson, San Jose, Calif.). Flow cytometry data were acquired with a Becton Dickinson FACScalibur. U937-GFP cells, like the parental uninfected cells, do not express CD81 (Fig. 4A, left side), whereas U937-CD81 cells express CD81 on their surface (Fig. 4A, right side). These cell lines were used to determine if the binding of E1FLAGE2 is CD81 dependent.
FIG. 4.
E1FLAGE2 binding is dependent on cell surface-expressed CD81 and induces cell-cell aggregation. (A) Flow cytometry analysis of binding of anti-CD81 MAb 5A6 (shaded curve) or a mouse IgG1 isotype control (dashed curved) to U937 cells expressing GFP (left side) or CD81-GFP (right side). (B) U937-CD81 (top) or U937-GFP (bottom) cells (1.5 × 105 cells/well) were incubated at 37°C with magnetic beads (10:1 ratio) precoated with H53-captured E1FLAGE2 (C and F), anti-CD81 MAb 5A6 (A and D), or anti-E2 MAb H53 (B and E). Photographs were taken after incubation for 8 h (magnification, ×100). The arrow in the right bottom of part C points to a cell. The arrow in the left top of part C points to one of the magnetic beads (4.5-μm diameter) used for immobilization of the antibodies and proteins.
We have previously shown that anti-CD81 MAbs induce cell aggregation (54). To test whether E1FLAGE2 could have a similar effect, we used an additional step to isolate and separate the noncovalently linked heterodimers from the disulfide-linked E1E2 aggregates. This was done by immunoaffinity on conformation-sensitive MAb H53. Magnetic beads precoated with goat anti-mouse IgG (Dynabeads; Dynal, Lake Success, N.Y.) were washed in accordance with the manufacturer's instructions and incubated with 10 μg of purified anti-CD81 MAb 5A6 or anti-E2 MAb H53 for 1 h at room temperature. Following incubation, the beads were washed twice in PBS supplemented with 0.1% bovine serum albumin (PBS-BSA) by exposure to a hand-held magnet. H53 beads were then incubated or not overnight at 4°C with purified E1FLAGE2 and washed twice in PBS-BSA supplemented with 0.1% Triton X-100. Magnetic beads immobilized with 5A6, with H53, or with E1FLAGE2 bound to H53 were resuspended in PBS-BSA supplemented with 0.1% Triton X-100. Magnetic beads were added to U937 cell lines suspended in 96-well flat-bottom plates (1.5 × 105 cells/well, 10:1 bead-to-cell ratio), and cell aggregation was observed after 8 h of incubation.
Purified E1FLAGE2 heterodimers, isolated this way, induced aggregation of U937-CD81 cells but not of U937-GFP cells (Fig. 4 B, right side). As expected, immobilized anti-CD81 MAb induced U937-CD81, but not U937-GFP, cell aggregation (Fig. 4B, left side). Neither cell reacted to beads immobilized with H53 alone (Fig. 4B, middle). These experiments indicate that binding of E1FLAGE2 heterodimers is CD81 dependent and leads to cell aggregation. Unlike U937 cells, which are among the few human cell lines that do not express CD81, human B-cell lines express this molecule. Immobilized E1FLAGE2 heterodimers induced the aggregation of all of the B-cell lines tested (data not shown). Taken together, these data demonstrate that E1FLAGE2 complexes are biologically functional and induce a biological effect upon binding to cells.
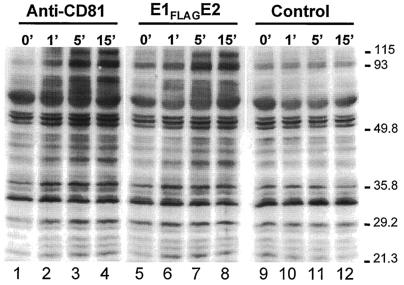
Because engagement of B cells by the anti-CD81 MAb was previously shown to induce an increase in protein tyrosine phosphorylation (52), we next questioned whether E1FLAGE2 can induce a similar effect. For this purpose, OCI-LY8 cells were washed and resuspended in RPMI medium-1% FCS (RPMI-FCS). Magnetic beads coated with 5A6, H53, or H53-E1FLAGE2 were prepared as described above, washed, and resuspended in RPMI-FCS. Samples of 4 × 106 OCI-LY8 cells were preincubated with beads for 15 min at 4°C. Rosetted cells were then washed and resuspended in 0.4 ml of RPMI-FCS. Samples of 0.1 ml were incubated for the indicated times at 37°C. The cells were lysed in 0.1 ml of lysis buffer (PBS [pH 7.4], 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, phosphatase inhibitor cocktail [Sigma], protease inhibitor cocktail) for 30 min on ice. Clarified cell lysates were then loaded onto 10% acrylamide gel, and tyrosine phosphoproteins were analyzed by Western blotting for the presence of tyrosine-phosphorylated proteins with an antiphosphotyrosine antibody (4G10; Upstate Biotechnology, Lake Placid, N.Y.).
Incubation of OCI-LY8 cells with E1FLAGE2 or with anti-CD81 MAb 5A6 induced a similar increase in the level of protein tyrosine phosphorylation similar to that in control cells (Fig. 5). The increase in tyrosine phosphorylation of several protein bands was seen after as little as 1 min of incubation (Fig. 5, lanes 2 and 6). Maximal tyrosine phosphorylation was seen after 15 min of exposure of the cells to the beads-E1FLAGE2 or beads-5A6 (lanes 4 and 8).
FIG. 5.
Engagement of B cells by E1FLAGE2 complexes induces protein tyrosine phosphorylation. Human OCI-LY8 B cells were exposed to magnetic beads precoated with H53-captured E1FLAGE2 (middle), anti-CD81 MAb 5A6 (left side), or anti-E2 MAb H53 (control, right side) for the indicated times (minutes) at 37°C. The cells were lysed, and proteins containing phosphotyrosines were revealed by Western blotting with a mouse antiphosphotyrosine horseradish peroxidase-conjugated antibody 4G10. The values on the right are molecular sizes in kilodaltons.
Taken together, our results show that engagement of B cells by E1FLAGE2 heterodimers induces cell aggregation and an increase in protein tyrosine phosphorylation, a hallmark of B-cell activation, suggesting that the interaction of B cells with HCV may lead to their activation.
HCV infection is associated with B-cell lymphoproliferative disorders (reviewed in reference 58) and is the major cause for mixed cryoglobulinemia (MC), a disorder characterized by serum immunoglobulin complexes that precipitate in the cold. It is therefore of note that in B cells, CD81 is a component of a multimeric protein complex, which includes the signaling molecule CD19, complement receptor 2 (CD21), and the interferon-inducible Leu-13 (CD225) protein (4). Coengagement of this CD19-CD21-CD81 complex together with the B-cell antigen receptor reduces the threshold of B-cell activation (7). Binding of HCV to CD81 and at the same time to a specific B-cell receptor could similarly reduce their activation threshold. Indeed, we have previously shown that the B-cell receptor expressed by a B-cell lymphoma of an HCV-infected patient had specificity for the E2 glycoprotein (47). Such dual binding by HCV to B-cell signaling complexes would lower their activation threshold, which in turn may promote cell proliferation, resulting in B-lymphocyte proliferative disorders (58). Other lymphoid cells could also be activated by the virus; thus, binding of CD81 on human T cells by anti-CD81 MAbs or by the truncated E2 glycoprotein provided a costimulatory signal to the engagement of CD3 (57). This binding led to a sustained increase in interleukin-2 production and enhanced gamma interferon production (57). Although HCV affects the immune system, the liver is the organ most damaged by the virus. Future studies may benefit from the use of functional E1E2 heterodimers for the study of virus-hepatocyte interactions.
Acknowledgments
This work was supported by National Institutes of Health grants CA 34233 (S.L.), by grant 2110046 from the California Cancer Research Program (S.L.), by grant QLRT-2000-01120 from the European Union (J.D.), and by a grant from the Association pour la Recherche sur le Cancer (J.D.). L.C. was supported by a Dean's Fellowship from Stanford University.
We thank Elizabeth R. Quinn for developing the aggregation assay, J. A. McKeating for the anti-E2 MAb 3/11, and S. K. H. Foung for the anti-E1 MAb 111.
REFERENCES
- 1.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q.-X. Zhang. 1999. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, B. W., D. Boettiger, E. Spooncer, and J. D. Norton. 1992. Efficient retroviral-mediated gene transfer into human B lymphoblastoid cells expressing mouse ecotropic viral receptor. Nucleic Acids Res. 20:5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartosch, B., J. Dubuisson, and F.-L. Cosset. 2003. Highly infectious hepatitis C pseudoviruses containing functional E1E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradbury, L. E., G. S. Kansas, S. Levy, R. L. Evans, and T. F. Tedder. 1992. The CD19/CD21 signal transducing complex of human B lymphocytes includes the target of antiproliferative antibody-1 and Leu-13 molecules. J. Immunol. 149:2841-2850. [PubMed] [Google Scholar]
- 5.Burioni, R., P. Plaisant, A. Manzin, D. Rosa, V. Delli Carri, F. Bugli, L. Solforosi, S. Abrignani, P. E. Varaldo, G. Fadda, and M. Clementi. 1998. Dissection of human humoral immune response against hepatitis C virus E2 glycoprotein by repertoire cloning and generation of recombinant Fab fragments. Hepatology 28:810-814. [DOI] [PubMed] [Google Scholar]
- 6.Cacoub, P., P. Hausfater, L. Musset, and J. C. Piette. 2000. Mixed cryoglobulinemia in hepatitis C patients. Ann. Med. Interne 151:20-29. [PubMed] [Google Scholar]
- 7.Carter, R. H., and D. T. Fearon. 1992. CD19: lowering the threshold for antigen receptor stimulation of B lymphocytes. Science 256:105-107. [DOI] [PubMed] [Google Scholar]
- 8.Chan-Fook, C., W. R. Jiang, B. E. Clarke, N. Zitzmann, C. Maidens, J. A. McKeating, and I. M. Jones. 2000. Hepatitis C virus glycoprotein E2 binding to CD81: the role of E1E2 cleavage and protein glycosylation in bioactivity. Virology 273:60-66. [DOI] [PubMed] [Google Scholar]
- 9.Clayton, R. F., A. Owsianka, J. Aitken, S. Graham, D. Bhella, and A. H. Patel. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76:7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocquerel, L., S. Duvet, J. C. Meunier, A. Pillez, R. Cacan, C. Wychowski, and J. Dubuisson. 1999. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocquerel, L., J. C. Meunier, A. Op de Beeck, D. Bonte, C. Wychowski, and J. Dubuisson. 2001. Coexpression of hepatitis C virus envelope proteins E1 and E2 in cis improves the stability of membrane insertion of E2. J. Gen. Virol. 82:1629-1635. [DOI] [PubMed] [Google Scholar]
- 12.Cocquerel, L., J. C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocquerel, L., E. R. Quinn, M. Flint, K. G. Hadlock, S. K. Foung, and S. Levy. 2003. Recognition of native hepatitis C virus E1E2 heterodimers by a human monoclonal antibody. J. Virol. 77:1604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crotta, S., A. Stilla, A. Wack, A. D'Andrea, S. Nuti, U. D'Oro, M. Mosca, F. Filliponi, R. M. Brunetto, F. Bonino, S. Abrignani, and N. M. Valiante. 2002. Inhibition of natural killer cells through engagement of CD81 by the major hepatitis C virus envelope protein. J. Exp. Med. 195:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.da Silva Cardoso, M., K. Siemoneit, D. Sturm, C. Krone, D. Moradpour, and B. Kubanek. 1998. Isolation and characterization of human monoclonal antibodies against hepatitis C virus envelope glycoproteins. J. Med. Virol. 55:28-34. [PubMed] [Google Scholar]
- 16.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drummer, H. E., K. A. Wilson, and P. Poumbourios. 2002. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 76:11143-11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubuisson, J., S. Duvet, J. C. Meunier, A. Op De Beeck, R. Cacan, C. Wychowski, and L. Cocquerel. 2000. Glycosylation of the hepatitis C virus envelope protein E1 is dependent on the presence of a downstream sequence on the viral polyprotein. J. Biol. Chem. 275:30605-30609. [DOI] [PubMed] [Google Scholar]
- 19.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duvet, S., L. Cocquerel, A. Pillez, R. Cacan, A. Verbert, D. Moradpour, C. Wychowski, and J. Dubuisson. 1998. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J. Biol. Chem. 273:32088-32095. [DOI] [PubMed] [Google Scholar]
- 21.Flint, M., J. Dubuisson, C. Maidens, R. Harrop, G. R. Guile, P. Borrow, and J. A. McKeating. 2000. Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J. Virol. 74:702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flint, M., and J. A. McKeating. 1999. The C-terminal region of the hepatitis C virus E1 glycoprotein confers localization within the endoplasmic reticulum. J. Gen. Virol. 80:1943-1947. [DOI] [PubMed] [Google Scholar]
- 24.Forns, X., T. Allander, P. Rohwer-Nutter, and J. Bukh. 2000. Characterization of modified hepatitis C virus E2 proteins expressed on the cell surface. Virology 274:75-85. [DOI] [PubMed] [Google Scholar]
- 25.Gardner, J. P., R. J. Durso, R. R. Arrigale, G. P. Donovan, P. J. Maddon, T. Dragic, and W. C. Olson. 2003. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 100:4498-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habersetzer, F., A. Fournillier, J. Dubuisson, D. Rosa, S. Abrignani, C. Wychowski, I. Nakano, C. Trépo, C. Desgranges, and G. Inchauspé. 1998. Characterization of human monoclonal antibodies specific of the hepatitis C virus glycoprotein E2 with in vitro binding neutralization properties. Virology 249:32-41. [DOI] [PubMed] [Google Scholar]
- 27.Hadlock, K. G., R. E. Lanford, S. Perkins, J. Rowe, Q. Yang, S. Levy, P. Pileri, S. Abrignani, and S. K. H. Foung. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 74:10407-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heile, J. M., Y.-L. Fong, D. Rosa, K. Berger, G. Saletti, S. Campagnoli, G. Bensi, S. Capo, S. Coates, K. Crawford, C. Dong, M. Wininger, G. Baker, L. Cousens, D. Chien, P. Ng, P. Archangel, G. Grandi, M. Houghton, and S. Abrignani. 2000. Evaluation of hepatitis C virus glycoprotein E2 for vaccine design: an endoplasmic reticulum-retained recombinant protein is superior to secreted recombinant protein and DNA-based vaccine candidates. J. Virol. 74:6885-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higginbottom, A., E. R. Quinn, C. C. Kuo, M. Flint, L. H. Wilson, E. Bianchi, A. Nicosia, P. N. Monk, J. A. McKeating, and S. Levy. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 7271-7276. 100: [DOI] [PMC free article] [PubMed]
- 31.Lambot, M., S. Fretier, A. Op De Beeck, B. Quatannens, S. Lestavel, V. Clavey, and J. Dubuisson. 2002. Reconstitution of hepatitis C virus envelope glycoproteins into liposomes as a surrogate model to study virus attachment. J. Biol. Chem. 277:20625-20630. [DOI] [PubMed] [Google Scholar]
- 32.Levy, S., S. C. Todd, and H. T. Maecker. 1998. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89-109. [DOI] [PubMed] [Google Scholar]
- 33.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1042. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 34.Lozach, P. Y., H. Lortat-Jacob, A. De Lacroix De Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houles, F. Fieschi, O. Schwartz, J. L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 278:20358-20366. [DOI] [PubMed] [Google Scholar]
- 35.Lucas, M., E. Tsitoura, M. Montoya, B. Laliotou, E. Aslanoglou, V. Kouvatsis, C. Entwisle, J. Miller, P. Klenerman, A. Hadziyannis, S. Hadziyannis, P. Borrow, and P. Mavromara. 2003. Characterization of secreted and intracellular forms of a truncated hepatitis C virus E2 protein expressed by a recombinant herpes simplex virus. J. Gen. Virol. 84:545-554. [DOI] [PubMed] [Google Scholar]
- 36.Major, M. E., B. Rehermann, and S. M. Feinstone. 2001. Hepatitis C viruses, p. 1127-1162. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 37.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michalak, J. P., C. Wychowski, A. Choukhi, J. C. Meunier, S. Ung, C. M. Rice, and J. Dubuisson. 1997. Characterization of truncated forms of hepatitis C virus glycoproteins. J. Gen. Virol. 78:2299-2306. [DOI] [PubMed] [Google Scholar]
- 39.Op De Beeck, A., L. Cocquerel, and J. Dubuisson. 2001. Biogenesis of hepatitis C virus envelope glycoproteins. J. Gen. Virol. 82:2589-2595. [DOI] [PubMed] [Google Scholar]
- 40.Op De Beeck, A., R. Montserret, S. Duvet, L. Cocquerel, R. Cacan, B. Barberot, M. Le Maire, F. Penin, and J. Dubuisson. 2000. The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization. J. Biol. Chem. 275:31428-31437. [DOI] [PubMed] [Google Scholar]
- 41.Oren, R., S. Takahashi, C. Doss, R. Levy, and S. Levy. 1990. TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol. Cell. Biol. 10:4007-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Owsianka, A., R. F. Clayton, L. D. Loomis-Price, J. A. McKeating, and A. H. Patel. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877-1883. [DOI] [PubMed] [Google Scholar]
- 43.Patel, A. H., J. Wood, F. Penin, J. Dubuisson, and J. A. McKeating. 2000. Construction and characterization of chimeric hepatitis C virus E2 glycoproteins: analysis of regions critical for glycoprotein aggregation and CD81 binding. J. Gen. Virol. 81:2873-2883. [DOI] [PubMed] [Google Scholar]
- 44.Patel, J., A. H. Patel, and J. McLauchlan. 2001. The transmembrane domain of the hepatitis C virus E2 glycoprotein is required for correct folding of the E1 glycoprotein and native complex formation. Virology 279:58-68. [DOI] [PubMed] [Google Scholar]
- 45.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 46.Pohlmann, S., J. Zhang, F. Baribaud, Z. Chen, G. J. Leslie, G. Lin, A. Granelli-Piperno, R. W. Doms, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 77:4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinn, E. R., C. H. Chan, K. G. Hadlock, S. K. Foung, M. Flint, and S. Levy. 2001. The B-cell receptor of a hepatitis C virus (HCV)-associated non-Hodgkin lymphoma binds the viral E2 envelope protein, implicating HCV in lymphomagenesis. Blood 98:3745-3749. [DOI] [PubMed] [Google Scholar]
- 48.Roccasecca, R., H. Ansuini, A. Vitelli, A. Meola, E. Scarselli, S. Acali, M. Pezzanera, B. B. Ercole, J. McKeating, A. Yagnik, A. Lahm, A. Tramontano, R. Cortese, and A. Nicosia. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 77:1856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasso, E. H. 2000. The rheumatoid factor response in the etiology of mixed cryoglobulins associated with hepatitis C virus infection. Ann. Med. Interne (Paris) 151:30-40. [PubMed] [Google Scholar]
- 50.Saunier, B., M. Triyatni, L. Ulianich, P. Maruvada, P. Yen, and L. D. Kohn. 2003. Role of the asialoglycoprotein receptor in binding and entry of hepatitis C virus structural proteins in cultured human hepatocytes. J. Virol. 77:546-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schick, M. R., V. Q. Nguyen, and S. Levy. 1993. Anti-TAPA-1 antibodies induce protein tyrosine phosphorylation that is prevented by increasing intracellular thiol levels. J. Immunol. 151:1918-1925. [PubMed] [Google Scholar]
- 53.Strader, D. B., and L. B. Seeff. 2001. Hepatitis C: a brief clinical overview. Inst. Lab. Anim. Resour. 42:107-116. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi, S., C. Doss, S. Levy, and R. Levy. 1990. TAPA-1, the target of an antiproliferative antibody, is associated on the cell surface with the Leu-13 antigen. J. Immunol. 145:2207-2213. [PubMed] [Google Scholar]
- 55.Triyatni, M., J. Vergalla, A. R. Davis, K. G. Hadlock, S. K. Foung, and T. J. Liang. 2002. Structural features of envelope proteins on hepatitis C virus-like particles as determined by anti-envelope monoclonal antibodies and CD81 binding. Virology 298:124-132. [DOI] [PubMed] [Google Scholar]
- 56.Tseng, C. T., and G. R. Klimpel. 2002. Binding of the hepatitis C virus envelope protein E2 to CD81 inhibits natural killer cell functions. J. Exp. Med. 195:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wack, A., E. Soldaini, C. Tseng, S. Nuti, G. Klimpel, and S. Abrignani. 2001. Binding of the hepatitis C virus envelope protein E2 to CD81 provides a co-stimulatory signal for human T cells. Eur. J. Immunol. 31:166-175. [DOI] [PubMed] [Google Scholar]
- 58.Weng, W. K., and S. Levy. Hepatitis C virus (HCV) and lymphomagenesis. Leuk. Lymphoma, in press. [DOI] [PubMed]
- 59.Wunschmann, S., J. D. Medh, D. Klinzmann, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 74:10055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yagnik, A. T., A. Lahm, A. Meola, R. M. Roccasecca, B. B. Ercole, A. Nicosia, and A. Tramontano. 2000. A model for the hepatitis C virus envelope glycoprotein E2. Proteins 40:355-366. [DOI] [PubMed] [Google Scholar]