Abstract
Sequences of retroviral origin occupy approximately 8% of the human genome. Most of these “retroviral” genes have lost their coding capacities since their entry into our ancestral genome millions of years ago, but some reading frames have remained open, suggesting positive selection. The complete sequencing of the human genome allowed a systematic search for retroviral envelope genes containing an open reading frame and resulted in the identification of 16 genes that we have characterized. We further showed, by quantitative reverse transcriptase PCR using specifically devised primers which discriminate between coding and noncoding elements, that all 16 genes are expressed in at least some healthy human tissues, albeit at highly different levels. All envelope genes disclose significant expression in the testis, three of them have a very high level of expression in the placenta, and a fourth is expressed in the thyroid. Besides their primary role as key molecules for viral entry, the envelope genes of retroviruses can induce cell-cell fusion, elicit immunosuppressive effects, and even protect against infection, and as such, endogenous retroviral envelope proteins have been tentatively identified in several reports as being involved in both normal and pathological processes. The present study provides a comprehensive survey of candidate genes and tools for a precise evaluation of their involvement in these processes.
Completion of the sequencing of the human genome has led to the conclusion that a significant fraction (approximately 8%) of our genome is of retroviral origin, with thousands of proviral sequences disclosing similarities with the integrated form of infectious retroviruses (23). These elements—also called human endogenous retroviruses (HERV)—are most probably the traces of “ancient” infections of the germ line by active retroviruses, which have thereafter been transmitted in a Mendelian manner. According to sequence homologies, these elements can be grouped into distinct families, with copy numbers ranging from a few to several hundred per haploid genome. Families have been tentatively named according to the tRNA normally used to prime reverse transcription in a retroviral replicative cycle (reviewed in references 25 and 43). In agreement with the proposed evolutionary scheme for the presence of these proviral elements, strong similarities between HERV and the present-day infectious retroviruses can be observed at the sequence level and in several instances at the functional level. Actually, phylogenetic analyses based on either the highly conserved reverse transcriptase (RT) domain of the pol gene or the transmembrane (TM) moiety of the envelope gene reveal interspersion of HERVs and infectious elements, suggesting a common history and shared ancestors (4, 40). HERV-encoded retrovirus-like particles have been detected in some tissues by electronic microscopy, revealing structural similarities to exogenous retroviruses (10). It has also recently been demonstrated that the HERV-K(HML-2) family can encode a regulatory protein (called Rec or cORF) which is functionally homologous to the Rev protein encoded by the human immunodeficiency retrovirus (26, 44) and that the coding envelope protein of the HERV-W family can still interact with the cellular receptor of the present-day D-type retroviruses (8). Finally, it has been shown for endogenous retroviruses of other species that their replicative cycle is closely related to that of exogenous retroviruses, with evidence for retrovirus-like recombination in the course of the reverse transcription step (20). As a consequence of the close relationship between HERVs and exogenous infectious retroviruses, it can be proposed that HERVs may still possess some of the functions of infectious retroviruses and as such have pathogenic effects, provided that they are transcriptionally active. Conversely, it is plausible that HERV proteins may have been subverted by the host for its benefit. Along this line, it has been proposed that the HERV envelope proteins could play a role in several processes, including (i) protection against infection by closely related exogenous retroviruses via receptor interference (5, 9, 37), (ii) protection of the fetus from the mother's immune system via a domain (the immunosuppressive domain) located in the TM subunit of most retroviral envelope proteins and known to inhibit immune effector functions (12, 28), and (iii) placenta formation via envelope protein-mediated fusogenic effects that could be involved in the generation of the syncytiotrophoblast (8, 31). An assessment of these putative roles of HERV is rendered extremely difficult as a result of the multicopy nature of these elements, which precludes classical genetic approaches. However, it has to be taken into account that a large fraction of endogenous retroviral genes are no longer coding genes, due to the accumulation of mutations, frameshifts, and deletions. Accordingly, we have made an exhaustive survey of the human genome for complete proviral elements, and specifically those containing a complete and coding env gene. Interestingly, this survey has led to the identification of a limited number of genes that we have characterized. To get insight into their potential functions, we devised a quantitative RT-PCR assay using primers allowing amplification of the coding copies specifically and provide the transcriptome of these human genes of retroviral origin in a large panel of healthy tissues. This analysis results in the unraveling of a series of new transcriptionally active human genes whose functions can now be appraised by classical molecular genetic approaches.
MATERIALS AND METHODS
Computer sequence analyses.
Homology searches were performed using the BLASTN program at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST), screening the finished (nr [nonredundant]) and unfinished (htgs [high-throughput genomic sequence]) databases with consensus envelope genes for each family. Hydropathy of the envelope proteins was calculated by the Kyte-Doolittle method implemented by the DNA Strider program (29). Alignments were performed with the CLUSTALW multialignment program at Infobiogen (http://www.infobiogen.fr). Chromosomal localizations of the coding env genes were performed at the Ensembl web site (http://www.ensembl.org/Homo_sapiens).
In vitro transcription-translation assay.
The coding env genes for in vitro transcription-translation assays were amplified with the Expand long-template PCR system (Roche, Indianapolis, Ind.). PCRs were carried out for 35 cycles (1 min at 94°C, 30 s at 58°C, 2 min at 68°C) using 1 ng of bacterial artificial chromosome (BAC) DNA. Primers were as follows: envHT7.3 (GCTAATACGACTCACTATAGGAACAGACCACCATGCACCACAGTATCAACCTTAC)and flR2B1 (TTCTGTTTCAGCTACAACTCTGT) for envH1; envHT7.3 and flRB2 (AGCAATAGTTTGTTAAATTC) for envH2; envHT7.2 (GCTAATACGACTCACTATAGGAACAGACCACCATGAGGGCACCCTCCAATACTTC) and flRB3 (ACCCCATGTTCTAGTCTTCC) for envH3; envKT7 (GCTAATACGACTCACTATAGGAACAGACCACCATGGAGATGCAAAGAAAAGCA) and LTRenvsK (GTGAACAAAGGTCTTTGCATCATAG) for envK1, envK3, envK5, and envK6; envKT7 and 3′fl51C12 (GAATTAGGCTTTCGGGACTTGAA) for envK2; envKT7 and KLTR3′ (C/TTTAAC/G/AG/AAGCATGCTGC/AC) for envK4; envTT7.2 (GCTAATACGACTCACTATAGGAACAGACCACCATGTTGGATTCATCACTCCCA) and envTflanq2 (CTGAAGGGAGTTCCTCCTAGG) for envT; envWT7 (GCTAATACGACTCACTATAGGAACAGACCACCATGGCCCTCCCTTATCAT) and envW64flR2 (ACAGCCAAGCAGGTACAG) for envW; envFRDT7.2 (GCTAATACGACTCACTATAGGAACAGACCACCATGCTCCTGCTGGTTCTCATTC) and envFRDfl3′ (CTGCAGCAGACTCCATCCTTG) for envFRD; enverv3T7 (GCTAATACGACTCACTATAGGAACAGACCACCATGACTAAAACCCTGTTGTATCA) and enverv3AS (GTTAATACTTAGTTAGGGCC) for envR; HS89F2T7 (GCTAATACGACTCACTATAGGAACAGACCACCATGGATCCACTACACACGATTGA) and HS89R3fl (TGTTTTGGGACACCACGAAT) for envR(b); envF(c)2T7 (GCTAATACGACTCACTATAGGAACAGACCACCATGAATTCTCCATGTGAC) and envF(c)2flR3 (GACACTTAATAGTTGCGACA) for envF(c)2; and envF(c)1T7 (GCTAATACGACTCACTATAGGAACAGACCACCATGGCCAGACCTTCCCCACTATGC) and envF(c)1fl3′ (GCCTTGGCAACTAAACCATTC) for envF(c)1. T7 promoter-containing PCR products were ethanol precipitated, and 200 ng of the amplification products was used in the TNT coupled reticulocyte lysate system (Promega Corp., Madison, Wis.) according to the manufacturer's instructions, with [3H]methionine (ICN Biomedicals Inc., Irvine, Calif.) for protein labeling. After electrophoresis of the translation products, sodium dodecyl sulfate-polyacrylamide gels were impregnated with Amplify (Amersham Biosciences, Piscataway, N.J.), rinsed with water, dried, and autoradiographed.
Tissue samples, RNA calibration, and reverse transcription.
Human BAC clones containing the identified coding envelope genes were purchased from BACPAC Resources (Oakland, Calif.). RNAs from various human tissues were purchased from Clontech (Palo Alto, Calif.), Stratagene (La Jolla, Calif.), and Ambion (Austin, Tex.). Quality of the RNA was assessed on an RNA LabChip (Agilent 2100 Bioanalyzer), and RNA concentration was quantified spectrophotometrically. Five micrograms of each RNA sample was subjected to DNase treatment (DNA-free; Ambion) to eliminate DNA contaminants. One microgram of RNA from each sample was reverse transcribed in a 20-μl reaction using 50 U of Moloney murine leukemia virus RT and 20 U of RNuclease inhibitor (Applied Biosystems, Foster City, Calif.) per reaction, 1 mmol of dA/T/G/C (Amersham-Pharmacia Biotech, Uppsala, Sweden) per liter, 5 mmol of MgCl2 per liter, 10 mmol of Tris-HCl (pH 8.3) per liter, 10 mmol of KCl per liter, and 2.5 μmol of random hexamers (Applied Biosystems) per liter. The cDNAs were then diluted 1/25 in nuclease-free H2O (Promega Corp.).
Real-time quantitative PCR.
Oligonucleotides were designed with the computer program Oligo (MedProbe, Oslo, Norway). The special requirements were a melting temperature of 60°C and an amplicon length between 80 and 350 bp. Oligonucleotides were purchased from MWG (Ebersberg, Germany).
Real-time quantitative PCR was achieved by using a cDNA equivalent of 20 ng of total RNA. The reaction was performed in 25 μl using SYBR green PCR core reagents (Applied Biosystems) according to the manufacturer's instructions. PCR was developed with the ABI PRISM 7000 sequence detection system (Applied Biosystems). Amplification was performed using a 2-min step at 50°C and then a 10-min denaturation step at 95°C, followed by 40 cycles of 15 s of denaturation at 95°C, 1 min of primer annealing, and a polymerization step at 60°C. To normalize for differences in the amount of total RNA added to the reaction, amplification of 18S total rRNA was performed as an endogenous control (variation was less than a factor of 3.5). The primers and probe from 18S RNA were purchased from Applied Biosystems. The relative expression in each sample was calculated with respect to a standard calibration curve (the dilution series of genomic DNA). Each sample was analyzed at least twice.
The control plasmid (p11env) containing the 11 envelope amplicons obtained with the primers listed in Table 1 was constructed first by cloning each amplicon into the pGEMT vector and then by successive three-fragment ligations using fragments from these 11 plasmids and from construct intermediates. A control series of p11env plasmid dilutions ranging from 1 to 0.008 ng was amplified with each primer set to measure relative yield (variation of less than a factor of 2). A control tube containing 1 ng of the p11env plasmid was included in each real-time PCR assay as a reference.
TABLE 1.
Sequences of the primers used to detect the expression of the coding envelopes by quantitative RT-PCR analysis
| Gene | Primers sequence (5′-3′) |
|---|---|
| envH1 | F: TTCACTCCATCCTTGGCTAT |
| R: CGTCGAGTATCTACGAGCAAT | |
| envH2 | F: ACTACACACATCACTGAAACAAA |
| R: GGATGGAGTGAAATACAGGAC | |
| envH3 | F: CCCTCCTCCACATTTATTTG |
| R: GTTGGGCTTTGGAGATGG | |
| envK | F: CACAACTAAAGAAGCTGACG |
| R: CATAGGCCCAGTTGGTATAG | |
| envT | F: CCAGGATTTGATGTTGGG |
| R: GGGGTGAGGTTAAGGAGATGG | |
| envW | F: CCCCATCGTATAGGAGTCTT |
| R: CCCCATCAGACATACCAGTT | |
| envFRD | F: GCCTGCAAATAGTCTTCTTT |
| R: ATAGGGGCTATTCCCATTAG | |
| envR | F: CCATGGGAAGCAAGGGAACT |
| R: CTTTCCCCAGCGAGCAATAC | |
| envR(b) | F: GGACAGTGCCGACATACTAT |
| R: TAGAGTGCAGCATCCTAACC | |
| envF(c)2 | F: ATGGAGGACTATATGAGCACAA |
| R: ATTAAAGTTAACCACGAGAAGC | |
| envF(c)1 | F: GGGCCACTAAGTTACTAGGTC |
| R: AGTTAGGAGGGAGTTACTGGG |
RESULTS
Screening of human databases for coding envelope genes.
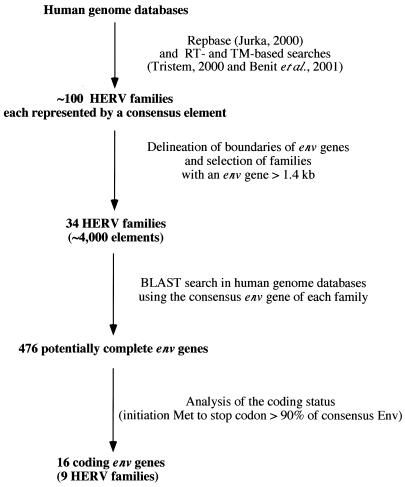
The rationale for the screening procedure to identify the coding envelope genes in the human genome is illustrated in Fig. 1. Basically, we used the Repbase database (final control search with update 8.2.1, Oct. 2002) (22), in which each family is represented by a “consensus” element, built from an alignment of all the proviruses belonging to the same family (i.e., copies which cluster together in phylogenetic trees). We implemented it by using RT- and TM-based searches as described in references 4 and 40, which revealed two additional families, the F(c)1 and F(c)2 families. Overall, approximately 100 HERV families were identified. For each family, the envelope gene was tentatively delineated, with the pol gene (which was easily positioned due to its high degree of conservation among retroviral elements) at its 5′ end and with the retroviral long terminal repeat (LTR) at its 3′ end. When the distance between the pol gene and LTR was higher than 1.4 kb (the average length of an envelope gene being 1.9 kb), the envelope gene was considered potentially complete and the HERV family was selected. The resulting list of the 34 corresponding HERV families is given in Table 2. For each family, we then performed a BLAST query on human genome databases using the envelope gene as a probe. Overall, it yielded 476 potentially complete envelope genes (see Table 2 for their distribution among the HERV families). The coding status of each identified env gene was finally assessed, and only those with an open reading frame (ORF) beginning at the first Met codon of the consensus envelope gene and uninterrupted over >90% of this gene were retained. Among the 476 envelope genes that were individually analyzed, only 16 genes were found to potentially encode envelope proteins. These 16 genes are listed and described in Table 3 and Fig. 2. Ten of these genes had previously been identified (2, 7, 13, 14, 24, 30, 38, 41), and six new genes emerged from this screen, including genes from the FRD, T, R(b), F(c)1, and F(c)2 families and one supplementary gene from the HERV-K(HML-2) family. The chromosomal localization was determined or confirmed by using the Ensembl web site and the corresponding accession numbers. It is noteworthy that one of these genes, envK2, can be found in duplicate in some individuals, since the corresponding provirus is organized as a tandem repeat (35). The two env sequences are 100% identical at the nucleotide level and are both referred to here as the envK2 gene. As can be observed in Fig. 2, which describes the 16 putative envelope proteins in comparison with the Moloney leukemia virus envelope protein, the overall length of the proteins is variable, with a minimum of 514 amino acids for EnvR(b) and maximum of 699 amino acids for EnvK. This size variability is also found among exogenous retroviral envelope proteins since, for example, human T-cell leukemia virus type 1 and human immunodeficiency virus type 1 envelope proteins are 488 and 861 amino acids long, respectively.
FIG. 1.
Flow chart for the identification of the 16 coding envelope genes of the human genome (see Results).
TABLE 2.
HERV families containing full-length envelope genes
| Familya | Repbase identifier | Class | No. of elements
|
||
|---|---|---|---|---|---|
| Totalb | With full-length env genes | With coding env genes | |||
| HERV-H | HERVH | I | 1,000 | 43 | 3 |
| HERV-IP | HERVIP10FH | I | 60 | 38 | 0 |
| HERV-9 | HERV9 | I | 1,000 | 37 | 0 |
| HERV-K(HML-2) | HERVK | II | 50 | 35 | 6 |
| HERV-K(HML-8) | HERVK11 | II | 100 | 31 | 0 |
| HERV-K(HML-5) | HERVK22 | II | 15 | 25 | 0 |
| HERV-E | HERVE | I | 150 | 22 | 0 |
| HERV-T | HERVS71 | I | 100 | 19 | 1 |
| HERV-S | HERV18 | III | 60 | 19 | 0 |
| HERV-Z69907 | MER66 | I | 100 | 18 | 0 |
| HERV-ADP | HERVP71A | I | 50 | 17 | 0 |
| HERV-K(HML-6) | HERVK3 | II | 100 | 17 | 0 |
| HERV-P | HUERS-P3 | I | 100 | 13 | 0 |
| HERV-W | HERV17 | I | 100 | 13 | 1 |
| HERV-F | HERVFH19 | I | 50 | 11 | 0 |
| HERV-U3 | HERV-L66 | III | 30 | 10 | 0 |
| HERV-K(HML-7) | HERVK11D | II | 20 | 10 | 0 |
| MER57(A) | I | 150 | 10 | 0 | |
| RRHERV-I | HERV15 | I | 30 | 10 | 0 |
| HERV-U2 | PRIMA41 | III | 100 | 10 | 0 |
| HERV-I/FTD | HERVI | I | 50 | 8 | 0 |
| PRIMA4 | I | 100 | 8 | 0 | |
| HERV-FRD | MER50 | I | 100 | 8 | 1 |
| HERV-K(HML-10) (C4) | HERVK14 | II | 20 | 7 | 0 |
| HERV-K(HML-3) | HERVK9 | II | 150 | 6 | 0 |
| HERV-R/erv3 | HERV3 | I | 100 | 6 | 1 |
| HERV-R(b) | PABL_B | I | 10 | 6 | 1 |
| MER70 | III | 100 | 6 | 0 | |
| HERV-XA34 | HERVFH21 | I | 30 | 5 | 0 |
| HERV-F(c)2 | I | 15 | 5 | 1 | |
| HERV-U4 | MER83 | I | 10 | 4 | 0 |
| HERV30 | I | 40 | 1 | 0 | |
| HERV-K(HML-4) | HERVK13 | II | 20 | 1 | 0 |
| HERV-F(c)1 | I | 2 | 1 | 1 | |
| Total | ∼4,000 | 476 | 16 | ||
TABLE 3.
The 16 coding envelope genes of the human genome
| Gene | Chromosome location | Accession no. | Position in sequence entrya | Bibliographic name | Reference |
|---|---|---|---|---|---|
| envH1 | 2q24.3 | AJ289709 | 6313-8067 (+) | envH/p62 | 14 |
| H19 | 24 | ||||
| envH2 | 3q26 | AJ289710 | 5393-7084 (+) | envH/p60 | 14 |
| envH3 | 2q24.1 | AJ289711 | 5204-6871 (+) | envH/p59 | 14 |
| envK1 | 12q14.1 | AC074261 | 93508-95604 (+) | This study | |
| envK2 | 7p22.1 | AC072054 | 30365-32464 (−) | HML-2.HOM | 30 |
| 38869-40968 (−)b | K(C7) | 38 | |||
| HERV-K108 | 2 | ||||
| envK3 | 19q12 | Y17833 | 5581-7680 (+) | HERV-K (C19) | 38 |
| envK4 | 6q14.1 | AF164615 | 6412-8508 (+) | HERV-K109 | 2 |
| envK5 | 19p13.11 | AY037928 | 6451-8550 (+) | HERV-K113 | 41 |
| envK6 | 8p23.1 | AY037929 | 6442-8541 (+) | HERV-K115 | 41 |
| envT | 19p13.11 | AC078899 | 154738-156618 (+) | This study | |
| envW | 7q21.2 | AC000064 | 35879-37495 (+) | Syncytin gene | 7 |
| envFRD | 6p24.1 | AL136139 | 21355-22972 (−) | This study | |
| envR | 7q11.21 | AC073210 | 54963-56978 (−) | erv3 | 13 |
| envR(b) | 3p24.3 | AC093488 | 78681-80225 (+) | This study | |
| envF(c)2 | 7q36.2 | AC016222 | 85216-86963 (+) | This study | |
| envF(c)1 | Xq21.33 | AL354685 | 46744-48717 (−) | This study |
+ and −, orientation in the sequence entry.
The HML-2.HOM/K(C7)/HERV-K108 locus is organized as a tandem repeat in some individuals, with 100% nucleotide identity for the two envelope genes.
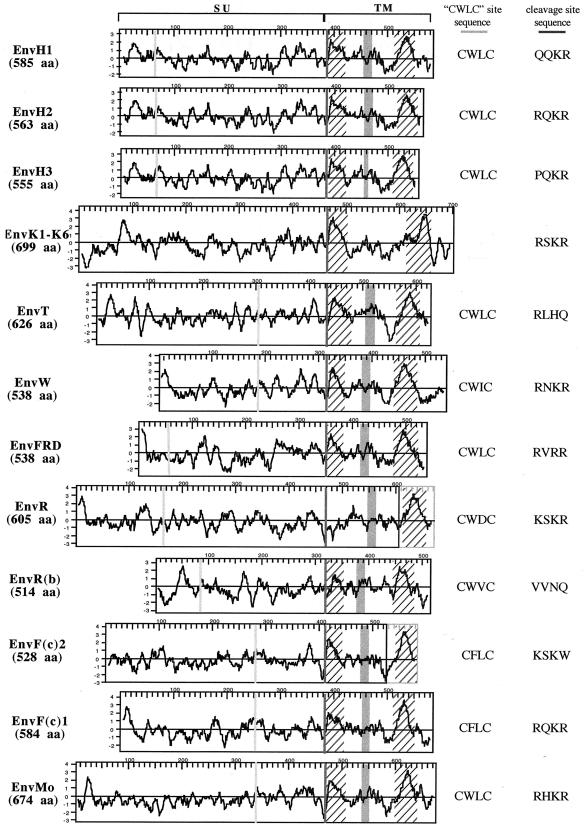
FIG. 2.
Structural organization and characteristic features of the 16 identified HERV envelope proteins. Hydrophobicity profiles and characteristic domains of the 16 HERV envelope proteins (the six HERV-K HML2 envelope proteins that are almost identical are represented by a single protein) and of the Moloney murine leukemia virus envelope protein (EnvMo) are shown. The black frames delineate the ORFs, and the gray frames at the end of the R and F(c)2 envelope proteins represent the short reading frames present downstream of the stop codon. Hatched areas correspond to hydrophobic regions associated with the fusion peptide and the transmembrane region, respectively; the CWLC motifs (consensus, C-X-X-C) and the proteolytic cleavage sites (consensus, R/K-X-R/K-R) are represented by light and dark gray vertical bars, respectively, and their sequences are indicated on the right; the putative immunosuppressive domain is represented with a gray box.
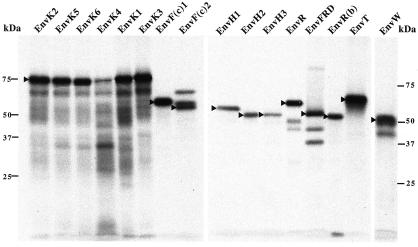
To ascertain the existence of the ORFs inferred from the nucleotide sequences, an in vitro transcription-translation assay was performed with human BAC clones containing the identified coding envelope genes. The 16 BACs were obtained from BACPAC Resources, except for the H1-, H2-, and H3-containing BACs that had been cloned previously (14). The in vitro transcription-translation assay was performed directly on the env PCR products (generated by using a forward T7-containing primer at the env 5′ end and a reverse primer downstream of the stop codon). In all cases, translation products of the size expected from the sequences in the database were obtained (Fig. 3). For some envelope proteins, additional bands of lower molecular weight were observed, compatible with initiation at internal sites, and interestingly, in one case [F(c)2], additional bands at higher molecular weights were detected, most probably associated with the presence of a frameshift signal at the gene's 3′ end (3).
FIG. 3.
SDS-PAGE analysis of the radiolabeled proteins produced upon direct in vitro transcription-translation assays of the amplification products of the coding retroviral envelope genes obtained from the corresponding BAC clones. Arrowheads point to the translation products of the expected size for each envelope gene. Bands of higher molecular weight are observed for EnvF(c)2, most probably associated with the presence of a frameshift signal at the env gene 3′ end (3).
The predicted hydrophobic profiles of the 16 proteins represented in Fig. 2 allow the identification of characteristic domains of these envelope proteins, namely, the fusion peptide, located just downstream of the proteolytic cleavage site between the Surface (SU) and TM moieties of the envelope proteins, and the transmembrane domain of the TM subunit, which permits the anchorage of the envelope protein in the membrane. Noteworthy, this analysis revealed that one envelope protein (EnvR) seems to be devoid of a fusion peptide and that two envelope proteins [EnvR and EnvF(c)2] disclose a premature stop codon just upstream of their transmembrane hydrophobic domain. Two other domains are delineated in Fig. 2, which are characteristic features of the envelope proteins belonging to the C-type and D-type retroviruses: the CWLC domain, involved in the interaction between the SU and TM moieties (34), and the CKS17-like immunosuppressive domain (12). It is noteworthy that, like B-type retrovirus, lentivirus, and spumavirus envelope proteins, the six EnvK proteins lack these two domains.
Transcriptome of the coding envelope genes. (i) Strategy.
To get insight into the expression profile of these genes in a quantitative manner, we devised a real-time RT-PCR strategy which uses specific primers designed in such a way that only envelope genes with an ORF should be amplified among all the envelope genes of a given family. To do so, for each HERV family containing a member with a coding element, env nucleotide sequences were aligned with the CLUSTALW program and primers were designed within domains of maximal divergence between the coding copy and the other copies. Primers for Sybr green amplification were devised with their 3′ ends forced at nucleotide positions with, again, maximum divergence between the coding sequence and the others. For the HERV-K(HML-2) family, which contains six coding env genes, this strategy could not be applied due to the too-high sequence conservation between copies. In this case, specific primers were devised that matched all six coding genes, tentatively excluding most other HERV-K(HML-2) envelope genes. The complete list of devised primers is given in Table 1.
To determine whether the resulting primers fulfilled the requirements for both efficiency and specificity, a first series of assays was performed by PCR amplification of human genomic DNA. As expected, in all cases a single band was observed, thus excluding nonspecific amplifications or amplifications of elements of unusual size (data not shown). Then, PCR products for each couple of primers were cloned into a pGEM-T vector, and at least six clones per amplicon were sequenced for each envelope gene [26 clones for the HERV-K(HML-2) family; see below]. In all cases, the six clones for a given envelope gene were identical and unambiguously corresponded to the coding sequence, being different from all the other aligned sequences within each HERV family. For the HERV-K(HML-2) family, 26 clones were sequenced, of which 21 had an identical sequence corresponding to the sequence of the six coding envelope genes, and 5 corresponded to two other HERV-K(HML-2) envelope genes. Despite this parasitic amplification of noncoding envelope genes, the HERV-K(HML-2) coding envelope primer set was retained since it allowed the amplification of all the coding envelope genes.
A second series of assays was then performed to determine the yield of each pair of primers to normalize env expression levels. To do so, we constructed a single plasmid (see Materials and Methods) which contained the complete set of 11 amplicons [the six envK(HML-2) genes being amplified by the same primer set]. This plasmid, used as a “control” matrix for each couple of primers, actually allowed a refined normalization for possible differences in the yield of the various primers and was used in each real-time amplification below.
(ii) Survey of human tissues.
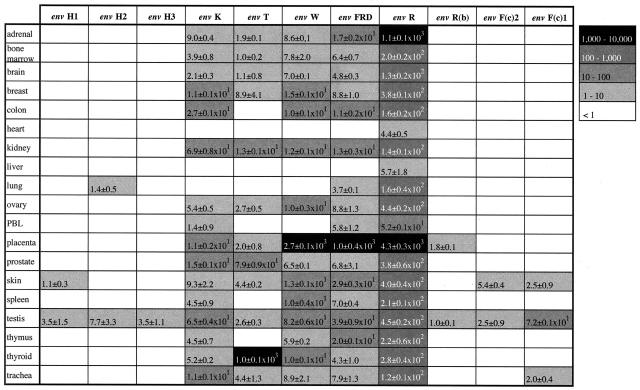
A systematic screening of the expression level of the 16 coding env genes present in the human genome was achieved with the primers listed above, on a series of 19 healthy human tissues (Fig. 4). RNA were subjected to DNase treatment and reverse transcribed using murine leukemia virus RT, and a first real-time quantitative PCR was performed using primers for 18S rRNA to normalize for differences in the amount of total RNA added to the reaction mixture (less than a 3.5-fold variation among samples). Real-time PCR was then performed for each pair of specific primers. Control PCRs performed on RNA without the RT step never resulted in any amplification, as expected. The expression levels represented in Fig. 4 using a logarithmic gray scale point to several interesting features. Firstly, there is an enormous variation in the level of expression (up to 4 log) among the different tissues for a given gene as well as among the different env genes. Secondly, and at a more refined level, it appears that all genes are transcribed at a significant level in the testis, still with variations (over a 100-fold) depending on the gene tested. Thirdly, the placenta is the organ where maximal expression can be observed for envR, envW, and envFRD, but the other env genes are expressed very poorly or not at all in this organ. Fourthly, thyroid exhibits a specific and high-level expression of the envT gene not observed for the other env genes in this organ. Fifthly, there is no expression of coding env genes in heart and liver, except for envR (the lowest level of expression for this gene). Finally, two groups of coding envelope genes can be inferred from these transcriptional data: those which display severe tissue specificity together with an overall low expression level, namely, the three envH genes, envR(b), envF(c)1, and envF(c)2, and those which are transcribed in the majority of the tissues, namely, envK, envT, envW, envFRD, and envR. The latter gene is singular, as its level of expression is extremely high in all tissues tested, with the highest value among all coding env genes (with the exception of the thyroid for envT).
FIG. 4.
Transcript levels of the coding retroviral envelope genes of the human genome in a panel of 19 healthy human tissues, as determined by real-time quantitative PCR. Each value is expressed as the mean ± standard deviation relative to the value for a reference control plasmid assayed in each real-time PCR experiment (see Materials and Methods), after correction for total RNA content in each tissue extract (using the 18S transcript as an internal control). The code for the logarithmic gray scale is indicated on the right. PBL, peripheral blood lymphocytes.
DISCUSSION
Limited number of retroviral coding envelope genes.
The extensive survey of the human genome performed here reveals that among the >10,000 retroviral elements clustered into approximately 100 families, only 16 possess a coding retroviral envelope gene (Table 2 and Fig. 2). The most important contributor to the coding envelope genes is the HERV-K(HML-2) family, since it comprises six coding envelope genes out of 35 full-length envelope gene copies. The status of the HERV-K(HML-2) family is unique among the coding env-containing families in several respects. Although elements of this family first entered the primate genome more than 30 million years ago (36), new proviral copies have been generated in the recent past. Actually, the date of entry into the human genome of four of the six HERV-K(HML-2) proviruses with a coding env gene (K1 [data not shown] and K2, K3, and K4 [2]) has been estimated to be less than 5 million years ago. In addition, the proviruses K5 and K6 are polymorphic in humans with allele frequencies of 0.04 and 0.19 (41). Furthermore, the six HERV-K(HML-2) proviruses with coding env have ORFs in other genes [all six have ORFs in their gag genes, four have ORFs in their pro genes, and four have ORFs in their pol genes, resulting in three completely coding HERV-K(HML-2) proviruses], and among the 29 other env-containing proviruses of this family, 10 additional retroviral coding genes can be found.
In comparison with the HERV-K(HML-2) family, the other env-containing families can be considered “old families,” with no human-specific integrations. The date of entry into the primate genome of the coding env gene-containing proviruses varies from 10 million years (for the HERV-Hp62 provirus [15]) to more than 45 million years [for the coding env-containing HERV-R(b) provirus (data not shown)]. Furthermore, no entirely coding gene besides the env genes has been found so far among those families.
Transcriptional status of the coding envelope genes.
We have devised an efficient method to detect the expression of specific genes belonging to large multigenic families with high homology between their members. It allowed us to quantitatively and specifically monitor the expression level of these endogenous retroviral genes, a task which would be impossible using the Northern blot or classical RT-PCR method, which detect the overall expression of the complete set of elements among each family. The first important outcome in the observed transcriptional pattern is that all genes are transcribed, at least in the testis. Despite the low (although unambiguous) transcriptional level of some of the env genes in this organ [e.g., envR(b), envF(c)2, and the three envH genes], this indicates that all promoters are active. The fact that testis is an organ in which all coding envelope genes are transcribed is not a totally unexpected result, since germ line expression is a common feature of transposable elements in other species, including mice (17, 39) and Drosophila (reviewed in references 11 and 18). In these species, expression in the germ line is associated with a high transpositional activity which may result in stably inherited mutations and most probably plays a role in the generation of genetic diversity in the course of evolution. For the placenta, expression of the coding env genes is much more heterogeneous than in the testis, with an extremely high expression level detected in this organ for three envelope genes, i.e., for the newly identified envFRD gene and for envR and envW (for the latter two, high-level expression had previously been observed at the protein level by using antibodies [8, 42]). Conversely, for five coding envelope genes [namely, the envH genes, envF(c)2, and envF(c)1], expression is undetectable. Several studies on HERV transcription have been performed by Northern blot analyses (reviewed in reference 25), which revealed a preferential expression of all HERV families in the placenta (except for the HERV-K family). It was suggested that a release of retroelements expression might take place in this organ without deleterious consequences for the individual due to the “provisional” status of this organ. Our results suggest that such a “generalized” expression is likely not to stem from all HERV members among each family but rather results from the activation of a limited number of proviral copies (possibly noncoding), for instance via position effects (for an example, see reference 17). Finally, the high-level expression of the newly identified envT gene detected in the thyroid is intriguing because it is one of the rare coding env genes highly expressed in a “permanent” healthy tissue and it is highly expressed only in this organ. This expression might be relevant to the hormone-producing status of the thyroid gland (and this interpretation might similarly hold for envR in the adrenal gland) and to specific sequences in the HERV LTR or in the vicinity of the proviral insertion site.
Biological relevance of env expression in human tissues.
Detection of an active transcription of the coding envelope genes in healthy tissues together with the probable positive selective pressure responsible for the conservation of ORFs throughout millions of years raises the question of a putative role of these genes in normal cell physiology. The envelope proteins of exogenous counterparts of HERVs possess several functions besides their primary role as key molecules for viral entry: most of them elicit immunosuppressive effects, and some of them have the capacity to create cell-cell fusion. Consequently, the high-level expression of three endogenous coding env genes observed in the placenta (namely, envR, envW, and the newly identified envFRD) could be implicated in two major physiological processes of this organ: fusion of the cytotrophoblast cells to form the syncytiotrophoblast layer and local immunosuppression at the materno-fetal barrier. Along the first line, it has been shown that the envW gene product is a highly fusogenic protein (8). Furthermore, inhibition of envW expression in primary cultures of human villous cytotrophoblasts leads to a decrease in trophoblast fusion and differentiation, suggesting a role in syncytiotrophoblast layer formation (19). Interestingly, the protein encoded by envFRD (also highly expressed in the placenta) might have a similar role, as it can also generate cell-cell fusion (S. Blaise, personal communication). In the case of envR, in contrast, we had previously ruled out its possible implication in fundamental processes of placenta formation by the discovery of a premature stop mutation present in 16% of Caucasians in the heterozygous state and in 1% in the homozygous state (16). Another property of retroviral envelope proteins is to inhibit infection by retroviruses sharing the same receptor—i.e., receptor interference (reviewed in references 5 and 37). It is therefore plausible, though again difficult to demonstrate, that some of the identified endogenous envelope proteins, as demonstrated in the mouse for the Fv4 locus (21), protect cells against infections by exogenous retroviruses. Interestingly, it has been demonstrated that EnvW interacts with the type D mammalian retrovirus receptor (8), and this envelope protein could therefore play a protective role at the placental barrier.
HERV-encoded proteins have also been tentatively involved in several human pathologies, including cancer, immune disorders, and neurological diseases (reviewed in references 25 and 32). By analogy with animal models, it is particularly tempting to implicate endogenous retroviral envelope proteins in tumorigenesis as a result of a transforming (1, 33) or immunosuppressive (6, 27, 28) effect. Numerous studies report on expressed endogenous retroviral sequences in pathological tissues, but in most cases the techniques used (Northern blot or RT-PCR using degenerate primers) did not allow the specific detection of coding sequences. Accordingly, the significance of these expressions remains elusive, and an association with diseases is only speculative. With the help of the primers devised in this study, a systematic and quantitative evaluation of the transcription levels of coding envelope genes in pathological versus healthy tissues should now be possible.
In conclusion, the present survey of coding env genes of the human genome provides a comprehensive list of candidate genes for the possible involvement of HERVs in human physiology and physiopathology. Furthermore, we show that all of the identified genes can be expressed at least in some tissues, and we provide tools to specifically evaluate their transcriptional status in physiological and pathological conditions. The identified genes should allow further studies of their function via classical genetic approaches (identification of susceptibility loci and searches for polymorphisms among the human population as previously performed for the HERV-R locus [16]), while the limited number of unraveled coding sequences reduces a hitherto insoluble multigenic analysis to a simpler one.
Acknowledgments
This work was supported by the CNRS and by grants from the Ligue Nationale contre le Cancer (Equipe Labellisée).
REFERENCES
- 1.Alberti, A., C. Murgia, S. L. Liu, M. Mura, C. Cousens, M. Sharp, A. D. Miller, and M. Palmarini. 2002. Envelope-induced cell transformation by ovine betaretroviruses. J. Virol. 76:5387-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. [DOI] [PubMed] [Google Scholar]
- 3.Benit, L., A. Calteau, and T. Heidmann. 2003. Characterization of the low copy HERV-Fc family: evidence for recent integrations in primates of elements with coding envelope genes. Virology 312:159-168. [DOI] [PubMed]
- 4.Benit, L., P. Dessen, and T. Heidmann. 2001. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J. Virol. 75:11709-11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best, S., P. R. Le Tissier, and J. P. Stoye. 1997. Endogenous retroviruses and the evolution of resistance to retroviral infection. Trends Microbiol. 5:313-318. [DOI] [PubMed] [Google Scholar]
- 6.Blaise, S., M. Mangeney, and T. Heidmann. 2001. The envelope of Mason-Pfizer monkey virus has immunosuppressive properties. J. Gen. Virol. 82:1597-1600. [DOI] [PubMed] [Google Scholar]
- 7.Blond, J.-L., F. Besème, L. Duret, O. Bouton, F. Bedin, H. Perron, B. Mandrand, and F. Mallet. 1999. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J. Virol. 73:1175-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blond, J. L., D. Lavillette, V. Cheynet, O. Bouton, G. Oriol, S. Chapel-Fernandes, B. Mandrand, F. Mallet, and F.-L. Cosset. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeke, J. D., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p. 343-436. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 10.Boller, K., H. König, M. Sauter, N. Mueller-Lantzsch, R. Löwer, J. Löwer, and R. Kurth. 1993. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology 196:349-353. [DOI] [PubMed] [Google Scholar]
- 11.Bucheton, A., C. Vaury, M.-C. Chaboissier, P. Abad, A. Pélisson, and M. Simonelig. 1993. I elements and the Drosophila genome, p. 173-188. In J. F. McDonald (ed.), Transposable elements and evolution. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 12.Cianciolo, G. J., T. Copeland, S. Orozlan, and R. Snyderman. 1985. Inhibition of lymphocyte proliferation by a synthetic peptide homologous to retroviral envelope protein. Science 230:453-455. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, M., M. Powers, C. O'Connell, and N. Kato. 1985. The nucleotide sequence of the env gene from the human provirus erv3 and isolation and characterization of an erv3-specific cDNA. Virology 147:449-458. [DOI] [PubMed] [Google Scholar]
- 14.de Parseval, N., J. F. Casella, L. Gressin, and T. Heidmann. 2001. Characterization of the three HERV-H proviruses with an open envelope reading frame encompassing the immunosuppressive domain and evolutionary history in primates. Virology 279:558-569. [DOI] [PubMed] [Google Scholar]
- 15.de Parseval, N., H. Alkabbani, and T. Heidmann. 1999. The long terminal repeats of the HERV-H human endogenous retrovirus contain binding sites for transcriptional regulation by the myb protein. J. Gen. Virol. 80:841-845. [DOI] [PubMed] [Google Scholar]
- 16.de Parseval, N., and T. Heidmann. 1998. Physiological knock-out of the envelope gene of the single copy ERV-3 human endogenous retrovirus in a fraction of the Caucasian population. J. Virol. 72:3442-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dupressoir, A., and T. Heidmann. 1996. Germ line-specific expression of intracisternal A-particle retrotransposons in transgenic mice. Mol. Cell. Biol. 16:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engels, W. R. 1989. P elements in Drosophila melanogaster, p. 437-484. In D. Berg and M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 19.Frendo, J. L., D. Olivier, V. Cheynet, J. L. Blond, O. Bouton, M. Vidaud, M. Rabreau, D. Evain-Brion, and F. Mallet. 2003. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell. Biol. 23:3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidmann, O., and T. Heidmann. 1991. Retrotransposition of a mouse IAP sequence tagged with an indicator gene. Cell 64:159-170. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda, H., and H. Sugimura. 1989. Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference properties. J. Virol. 63:5405-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurka, J. 2000. Repbase update, a database and an electronic journal of repetitive elements. Trends Genet. 16:418-420. [DOI] [PubMed] [Google Scholar]
- 23.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 24.Lindeskog, M., D. Mager, and J. Blomberg. 1999. Isolation of a human endogenous retroviral HERV-H element with an open env reading frame. Virology 258:441-450. [DOI] [PubMed] [Google Scholar]
- 25.Löwer, R., J. Löwer, and R. Kurth. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. USA 93:5177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magin, C., R. Lower, and J. Lower. 1999. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J. Virol. 73:9496-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mangeney, M., N. de Parseval, G. Thomas, and T. Heidmann. 2001. The full-length envelope of an HERV-H human endogenous retrovirus has immunosuppressive properties. J. Gen. Virol. 82:2515-2518. [DOI] [PubMed] [Google Scholar]
- 28.Mangeney, M., and T. Heidmann. 1998. Tumor cells expressing a retroviral envelope escape immune rejection in vivo. Proc. Natl. Acad. Sci. USA 95:14920-14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marck, C. 1988. “DNA Strider”: a “C” program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 16:1829-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer, J., M. Sauter, A. Racz, D. Scherer, N. Mueller-Lantzsch, and E. Meese. 1999. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat. Genet. 21:257-258. [DOI] [PubMed] [Google Scholar]
- 31.Mi, S., X. Lee, X. Li, G. M. Veldman, H. Finnerty, L. Racie, E. LaVallie, X. Y. Tang, P. Edouard, S. Howes, J. C. J. Keith, and J. M. McCoy. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785-788. [DOI] [PubMed] [Google Scholar]
- 32.Nakagawa, K., and L. C. Harrison. 1996. The potential roles of endogenous retroviruses in autoimmunity. Immun. Rev. 152:193-236. [DOI] [PubMed] [Google Scholar]
- 33.Palmarini, M., and H. Fan. 2001. Retrovirus-induced ovine pulmonary adenocarcinoma, an animal model for lung cancer. J. Nat. Cancer Inst. 93:1603-1614. [DOI] [PubMed] [Google Scholar]
- 34.Pinter, A., R. Kopelman, Z. Li, S. C. Kayman, and D. A. Sanders. 1997. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J. Virol. 71:8073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reus, K., J. Mayer, M. Sauter, D. Scherer, N. Muller-Lantzsch, and E. Meese. 2001. Genomic organization of the human endogenous retrovirus HERV-K(HML-2.HOM) (ERVK6) on chromosome 7. Genomics 72:314-320. [DOI] [PubMed] [Google Scholar]
- 36.Reus, K., J. Mayer, M. Sauter, H. Zischler, N. Muller-Lantzsch, and E. Meese. 2001. HERV-K(OLD): ancestor sequences of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 75:8917-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg, N., and P. Jolicoeur. 1997. Retroviral pathogenesis, p. 475-586. In J. M. Coffin, C. L. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 38.Tönjes, R. R., F. Czauderna, and R. Kurth. 1999. Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K. J. Virol. 73:9187-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trelogan, S. A., and S. L. Martin. 1995. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc. Natl. Acad. Sci. USA 92:1520-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tristem, M. 2000. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J. Virol. 74:3715-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner, G., M. Barbulescu, M. Su, M. I. Jensen-Seaman, K. K. Kidd, and J. Lenz. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531-1535. [DOI] [PubMed] [Google Scholar]
- 42.Venables, S., M. Brookes, W. Fan, E. Larsson, R. N. Maini, and M. T. Boyd. 1995. Abundance of an endogenous retroviral envelope protein in placental trophoblasts suggests a biological function. Virology 211:589-592. [DOI] [PubMed] [Google Scholar]
- 43.Wilkinson, D. A., D. L. Mager, and J.-A. C. Leong. 1994. Endogenous human retroviruses, p. 465-535. In J. A. Levy (ed.), The Retroviridae, vol. 3. Plenum Press, New York, N.Y.
- 44.Yang, J., H. P. Bogerd, S. Peng, H. Wiegand, R. Truant, and B. R. Cullen. 1999. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc. Natl. Acad. Sci. USA 96:13404-13408. [DOI] [PMC free article] [PubMed] [Google Scholar]