Abstract
N-linked glycans not only orchestrate the folding and intracellular transport of viral glycoproteins but also modulate their function. We have characterized the three glycans attached to fusion (F) proteins of the morbilliviruses canine distemper virus and measles virus. The individual Morbillivirus glycans have similar functional properties: the glycan at position 68 is essential for protein transport, and those at positions 36 and 75 modulate fusion (numbering according to the Newcastle disease virus [NDV] F protein sequence). Based on the crystal structure of the NDV F protein, we then predicted the locations of the Morbillivirus glycans: the glycan at position 36 is located in the F protein head, and those at positions 68 and 75 are located near the neck-stalk interface. NDV position 36 is not occupied by a glycan; the only glycan in that F protein head also has a fusion control function and grows from residue 366, located only 6 Å from residue 36. We then exchanged the glycan at position 36 with the glycan at position 366 and showed functional complementation. Thus, structural information about the F proteins of Paramyxoviridae coupled with functional analysis disclosed a location in the protein head into which fusion-modulating glycans independently evolved.
N-linked glycans not only orchestrate folding and intracellular transport of viral glycoproteins but also modulate their function. Maturation of the influenza virus hemagglutinin (HA) protein has been extensively studied, making it a paradigm for understanding the cellular secretory pathway (10, 15). In addition, N-linked glycans have direct effects on protein function: certain glycans located near the receptor binding site on the top of the HA protein regulate receptor affinity (30). Others located in the stem region maintain the protein in a metastable conformation required for fusion activity (31, 36, 40).
The effects of different N-linked glycans on fusion modulation have been studied extensively in another virus family, the Paramyxoviridae. In these viruses, the receptor attachment and cell fusion functions are distributed on two glycoproteins, the fusion (F) protein and the attachment protein. The F protein is a trimeric type I integral membrane glycoprotein that is synthesized initially as an inactive precursor (F0). After being glycosylated, it is transported to the trans-Golgi network, where it is cleaved by furin, or other cellular proteases, into the disulfide-linked subunits F1 and F2 (20). In different subfamilies of the Paramyxoviridae, the attachment protein is named hemagglutinin (H) or hemagglutinin-neuraminidase (HN) to reflect its function. The trimeric F and tetrameric H or HN protein complexes mediate cell-cell and virus-cell fusion in concert (19).
Despite the exact conservation of the number and spacing of cysteines among the F proteins of Paramyxoviridae (26), the number, location and functional importance of the attached glycans are variable (2, 24, 35, 41) and little evidence for the functional conservation of certain glycans is available. However, until recently, no tertiary structure of any paramyxovirus F protein was available, and the spatial locations of different chains could not be predicted. This situation changed with the publication of the Newcastle disease virus (NDV) F protein structure (8).
For functional studies of N-linked glycans, the F protein of the Morbillivirus genus of Paramyxoviridae is of particular interest because of its limited complexity: it harbors only three or four N-linked glycans. The roles of the different glycans of the F protein of the human morbillivirus measles virus (MV) have been examined. It has been demonstrated that all three potential glycosylation sites located in the F2 subunit are used (1, 16). Agreement about the importance of different glycans for function was incomplete, possibly because different mutations were introduced and different expression systems were used. On the other hand, by incubation of MV-infected cells with glucosidase or mannosidase inhibitors, it was shown that trimming of glucose residues and the presence but not the trimming of terminal mannose residues are required for syncytium formation and infectivity (4, 23). It is also known that mechanisms other than N glycosylation influence the fusion activity of the MV glycoprotein complex: in particular, the membrane-associated matrix protein controls fusion (6), and lateral interactions between F and H protein complexes modulate it (34).
In this study, we defined the principal functions of the individual glycans in the F proteins of MV and another morbillivirus, canine distemper virus (CDV), as protein folding or fusion control. We predicted the locations of these glycans based on the assumption that the structure of the morbillivirus F protein is similar to that of the NDV F protein (8), and we noted that the only morbillivirus fusion-controlling glycan in the protein head is predicted to be located close (6 Å) to an NDV fusion-controlling glycan. We then tested whether these two glycans are functionally interchangeable.
MATERIALS AND METHODS
Cells.
All experiments were performed with Vero cells (ATCC CCL-81). The cells were cultivated in Dulbecco′s modified Eagle′s medium (DMEM) with 5% fetal calf serum (both from Invitrogen).
Construction of expression plasmids containing different glycosylation mutants.
Plasmid pCG-FOS (F protein) (39) constituted the basis for all the CDV F protein mutants. The consensus sequences for N-linked glycosylation [N-X-(S/T)] at the four predicted sites that are part of the mature F protein were altered by changing the asparagine into a glutamine residue by site-directed mutagenesis, yielding pCG-FOSg1 (referred to below as g1, and carrying the mutation N141Q), pCG-FOSg2 (g2; N173Q), pCG-FOSg3 (g3; N179Q), pCG-FOSg1/3 (g1/3; N141Q and N179Q), and pCG-FOSg4 (g4; N517Q). A second set of mutants was generated to introduce NDV F protein glycosylation sites into the CDV F glycosylation mutants. The position of the NDV site, which was identified as located in close proximity to the respective CDV site in the structural NDV model, was identified in the superimposed CDV sequence, and the residues were mutated to generate a consensus sequence for N-linked glycosylation. The following compensation mutants were generated: pCG-FOSn1 (referred to below as n1, and carrying mutations N141Q and D474N), pCG-FOSn2 (n2; N173Q plus E189N and L191S), and pCG-FOSn3 (n3; N179Q plus A301N and Q303S).
A consensus sequence for endoplasmic reticulum retention of transmembrane proteins, KSKTH (18, 33), was added to the carboxy terminus of the CDV F protein cytoplasmic tail by PCR (Expand High Fidelity PCR system; Roche Biochemicals), yielding pCG-FOSRet (R). Mutant pCG-FOSTryps (T), in which the furin cleavage motif RRQR(R/F) was changed into the trypsin cleavage motif RNHN(R/F), was also generated by site-directed mutagenesis. This mutation was also introduced into the respective furin motif of pCG-FMV (FMV) (5), the expression plasmid containing the MV F protein open reading frame, yielding pCG-FMVTryps (TMV). This plasmid corresponds to pCG-Fcl (22). In addition, the three predicted N-glycosylation sites that are part of the mature MV F protein were abolished separately, yielding pCG-FMVg1 (referred to below as g1MV, and carrying the mutation N29Q), pCG-FMVg2 (g2MV; N61Q), and pCG-FMVg3 (g3MV; N67Q). The sequences of all primers are available upon request. The correct sequences of all constructs were confirmed (ABI PRISM 377 DNA sequencer; Perkin-Elmer Applied Biosystems).
Western blot analysis and surface biotinylation.
Vero cells were seeded into 12-well plates, transfected with the different constructs by using Lipofectamine 2000 (Invitrogen), and incubated at 37°C for 36 h. For Western blot analysis, cells were washed twice with phosphate-buffered saline (PBS) before addition of 0.2 ml of radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris-HCl [pH 8.0]) with protease inhibitors (Complete; Roche Biochemicals) to each well. After incubation for 20 min at 4°C, the lysates were cleared by centrifugation at 5,000 × g for 15 min at 4°C and the supernatant was mixed with an equal amount of 2× Laemmli sample buffer (Bio-Rad) containing 100 mM dithiothreitol (DTT). Samples were incubated for 10 min at 95°C and subsequently fractionated on SDS-10% polyacrylamide gels (Bio-Rad) and blotted onto polyvinylidene difluoride (PVDF) membranes (Millipore). After being blocked with 1% blocking reagent (Roche Biochemicals) overnight, the membranes were incubated with a rabbit antiserum against the cytoplasmic tail peptide (anti-Fcyt) that corresponds to the 14 carboxy-terminal residues of the CDV and MV F proteins (7). After incubation with a peroxidase-conjugated goat anti-rabbit immunoglobulin G antiserum, the membranes were subjected to ECL detection (Amersham Pharmacia Biotech).
For surface biotinylation, cells were shifted to 4°C and washed once with cold PBS before 125 mg of EZ-Link Sulfo-NHS-LC-biotin (Pierce) dissolved in 0.3 ml of cold PBS was added to each well. After incubation for 20 min on a rocker platform at 4°C, cells were washed with 0.5 M glycine in PBS, followed by addition of 1 ml of 0.5 M glycine-PBS and incubation for 20 min at 4°C to quench the excess biotin. Samples were then processed as described above. Instead of adding the 2× sample buffer directly, the supernatant was mixed with 50 μl of protein A-agarose beads (Bio-Rad), and the antibody was added in the appropriate concentration. After incubation at 4°C overnight, the beads were washed three times in RIPA buffer before 30 μl of 2× Laemmli sample buffer (Bio-Rad) containing 100 mM DTT was added. The samples then underwent Western blot analysis as described above. The membranes were incubated with peroxidase-conjugated streptavidin (Amersham Pharmacia Biotech) and subjected to ECL detection.
Fusion assay.
The fusion assay was performed as described previously (38). Briefly, Vero cells were transfected with the different F expression plasmids together with pCG-HOS or pCG-HMV and pTM1-luc by using a molar ratio of 1:1:0.7. For each well transfected, a second well of Vero cells was infected with modified vaccinia virus Ankara expressing the T7 polymerase (MVA-T7) (27) at a multiplicity of infection (MOI) of 1 at the time of transfection. Twelve hours after transfection or infection, the cells were washed twice with PBS (Gibco BRL), and 50 μl of 0.25% trypsin-EDTA (Gibco BRL) was added to detach the cells. After incubation at 37°C for 5 min, each well of cells was resuspended in 1 ml of DMEM supplemented with 5% fetal calf serum, transferred into 2 wells of a 24-well plate, and incubated for 36 h at 37°C. Following the visual grading of the fusion activity, the luciferase activity was determined by using the Luciferase Assay System (Promega) and a 96-well plate reading luminometer (Microlumat LB96P; EG & G Berthold).
Radioimmunoprecipitation and endoglycosidase digestion.
Two wells of a six-well plate seeded with Vero cells were transfected with each construct and incubated at 37°C for 36 h. The cells were washed twice with PBS, and 2 ml of DMEM without glutamine, methionine, or cysteine was added. After incubation at 37°C for 1 h, the medium was replaced with 1 ml of DMEM without glutamine, methionine, or cysteine, and 100 μCi of Redivue Pro-mix [35S] in vitro cell labeling mix (methionine-cysteine; Amersham Pharmacia Biotech) was added to each well. Following incubation for 4 h at 37°C, cells were washed three times with cold PBS and lysed with 500 μl of RIPA buffer with protease inhibitors (Complete; Roche Biochemicals) for 20 min at 4°C. Lysates from wells transfected with the same construct were pooled and centrifuged at 5,000 × g for 15 min at 4°C, the supernatant was mixed with 150 μl of protein A-agarose beads (Bio-Rad), and antibodies were added in the appropriate concentration. Following incubation at 4°C overnight, the beads were washed four times in RIPA buffer, and 50 μl of denaturation buffer (final concentrations, 0.5% SDS and 1% β-mercaptoethanol; New England Biolabs) was added. After incubation at 95°C for 10 min, the beads were spun down and the supernatant was split into three equal aliquots. The first was kept as an untreated control, the second was mixed with endoglycosidase H (endo H) deglycosylation buffer (final concentration, 50 mM sodium citrate [pH 5.5]) and 2 U of endo H, and the third aliquot was mixed with endo F glycosylation buffer (final concentrations, 50 mM sodium phosphate [pH 7.5] and 1% NP-40) and 2 U of endo F and then adjusted to 1% NP-40. All samples were incubated for 1 h at 37°C, followed by addition of 2× Laemmli sample buffer (Bio-Rad) containing 100 mM DTT and subjection to SDS-polyacrylamide gel electrophoresis (PAGE) analysis using a gel with a polyacrylamide concentration appropriate for the protein of interest. The gels were dried for 1 to 1.5 h at 70°C and exposed for 3 to 16 days by using Biomax films (Kodak).
Protein modeling and software.
A structural model of the CDV F protein was built by using SYBYL's Biopolymer Module (version 6.8; Tripos, Inc., St. Louis, Mo.). The amino acid sequence of CDV F was aligned with the sequence of NDV F by using the BestFit algorithm of the Genetics Computer Group (GCG; University of Wisconsin, Madison) package; the alignment was straightforward because of the exactly conserved number and spacing of cysteines. Nonidentical residues in the crystal structure of the NDV F protein were replaced by simple mutation to residues corresponding to those of the CDV F protein. Steric minimization of all replaced side chains in the preliminary CDV F protein model was performed before energy minimization to the final structure. Final refinement was performed using AMBER 40 with an AMBER force field engine for 300 iterations.
RESULTS
The second CDV F protein glycan is essential for correct processing.
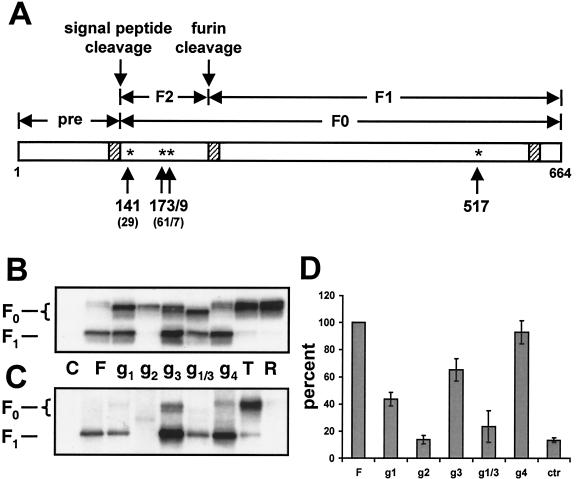
Four N-glycosylation motifs exist in the open reading frame of the CDV mature F protein; three of these are located in the F2 subunit at positions 141, 173, and 179, and one is located at position 517 in the F1 subunit (Fig. 1A). The three motifs in the F2 subunit are conserved between CDV and MV (positions 29, 61, and 67 in the MV open reading frame). The extended signal peptide of the CDV F protein accounts for the additional N-terminal residues (38). These positions correspond to 36, 68, and 75 in the NDV numbering.
FIG. 1.
Scheme of the CDV F protein (A), and analysis of the processing (B), cell surface expression (C), and fusion activity (D) of different F protein mutants. (A) Linear drawing of the CDV F protein. Hatched boxes, hydrophobic regions; stars, potential glycosylation sites. Numbers below the stars correspond to the positions of the residues in the CDV F open reading frame. The corresponding residues in the MV F open reading frame are given in parentheses. In constructs g1 through g4 (panels B, C, and D), one potential N-glycosylation site was inactivated by mutating the respective asparagine residue to glutamine. In mutant g1/3, the asparagines at positions 141 and 179 were mutated. (B) Characterization of the different F proteins by Western blot analysis. Proteins were extracted from Vero cells transfected with the different F constructs, separated by reducing SDS-PAGE, and blotted onto PVDF membranes. The F proteins were revealed with an anti-cytoplasmic tail serum (anti-Fcyt). The F0 and F1 subunits are indicated on the left. (C) Surface biotinylation. Duplicate wells of the cells used for Western blot analysis were shifted to 4°C, biotin labeled, lysed, and immunoprecipitated overnight with the anti-Fcyt antiserum. Samples were separated by reducing SDS-PAGE (10% acrylamide), blotted onto PVDF membranes, and probed with peroxidase-coupled streptavidin. (D) Quantitative fusion assays. Vero cell monolayers were either infected with MVA-T7 (MOI, 1) or transfected with the different F constructs, a plasmid coding for the H protein, and a plasmid containing the luciferase gene under the control of the T7 promoter. At 12 h after transfection, the two cell populations were mixed and seeded into fresh plates. After 36 h at 37°C, fusion was quantified by measuring luciferase activity. For each experiment, the value measured for the parental F protein was set to 100%. Means and standard deviations for four independent experiments in duplicate are shown.
To determine which of the potential CDV glycosylation motifs are used, the asparagines to which glycans are added were mutated to glutamines, mutations known to prevent glycosylation. To verify whether glycosylation was prevented, the migration patterns of the four mutants were assessed (Fig. 1B). The uncleaved F0 precursor of the three single-amino-acid mutants (Fig. 1B, mutants g1, g2, and g3) migrated faster than the unaltered parental protein (Fig. 1B, mutant F), indicating that all three sites are used. An additional shift was observed with the mutant in which only the second glycosylation site remained intact, confirming glycosylation at the first and third positions in the standard protein (Fig. 1B, g1/3). In contrast, the mutant in which the consensus glycosylation sequence at the fourth position located in the F1 subunit was mutated displayed a migration pattern identical to that of the parental protein, suggesting that this site is not used (Fig. 1B, g4). Compared to the parental protein, less-efficient cleavage of the F0 precursor into the F1 and F2 subunits was observed for mutants g1 and g1/3. Similar quantities of the F protein were monitored for the mutants and the parental protein. In contrast, the protein lacking glycosylation of g2 was detected in small amounts and precursor cleavage did not occur, suggesting incorrect folding and reduced stability, probably due to retention and degradation of the protein by quality control mechanisms. Lack of glycosylation at g3 had no negative influence on the amount of F1 protein detected; in fact, higher levels of protein were detected than in the control (see below).
Since cell surface expression is necessary for F protein function, we then verified whether glycan-defective CDV F proteins reach the cell surface (Fig. 1C). CDV F protein mutants retained in the endoplasmic reticulum (R) or expressed at the cell surface in the precursor form (T) were produced by addition of the KSKTH endoplasmic reticulum retention sequence to the cytoplasmic tail (33) or according to procedures established for the MV F protein mutations of the furin-dependent cleavage sequence (RRQRR) to a trypsin-dependent sequence (RNHNR) (22), respectively. These proteins were included as controls. As expected, the cleaved F1 subunit of the standard F protein and the uncleaved F0 precursor of the control T protein were detected on the cell surface, whereas the control R mutant was retained intracellularly (Fig. 1C). F protein mutants g1, g3, g1/3, and g4 all reached the cell surface in the processed form (Fig. 1C). In contrast, mutant g2 was not transported to the cell surface (Fig. 1C), indicating that the absence of the second glycan leads to protein retention in a compartment preceding the trans-Golgi network, where cleavage by furin-like enzymes occurs.
In summary, all three N-glycosylation motifs in the F2 subunit are used, whereas the motif in the F1 subunit is not. The mutant protein lacking the second glycosylation motif is not processed into its mature form consisting of the disulfide-linked F1 and F2 subunits, appears to be less stable, and is not transported to the cell surface. The other single mutants g1, g3, and g4, as well as the combination mutant g1/3, are processed and transported normally. The quantities of these mutated F proteins reaching the cell surface are comparable to or exceed that of the unmodified F protein control.
The first and third glycans of the CDV F protein are necessary for optimal fusion activity.
To compare the fusion functions of our collection of mutants, they were coexpressed with the CDV H protein and their activities were determined in a quantitative fusion assay (29). For each experiment, the fusion activity of the unaltered F protein (FCDV) measured 3 days after transfection was set to 100% (Fig. 1D, column F). The fusion activities of the mutants were expressed as percentages of the fusion activity of the unaltered F protein. The g4 protein, in which the inactive glycosylation site in the F1 subunit was mutated, displayed a fusion activity similar to that of the unaltered protein (Fig. 1D), indicating that the amino acid change from asparagine to glutamine at this position does not affect function. In contrast, the fusion activities of all proteins with altered glycosylation were affected negatively to different degrees. The g3 mutant, lacking the third glycan, fused with about 70% the efficiency of the wild type, whereas the fusion activity of the g1 mutant, lacking the first glycan, was reduced to approximately 45% (Fig. 1D). The higher level of cell surface expression of g3 relative to the control may make it appear to be more fusogenic than it actually is. The fusion activity observed for the protein lacking the first and third glycans (g1/3) was further reduced to levels close to background (Fig. 1D; compare the g1/3 and control columns). As expected, the protein without the second glycan (Fig. 1D, g2) was fusion inactive, suggesting that coexpression with H did not have a beneficial effect on its processing or surface expression. This fusion assay demonstrates that loss of glycosylation can lead to impaired protein function despite correct intracellular processing and efficient surface expression, as observed for the three constructs g1, g3, and g1/3.
Analysis of the glycans present on the mutant F proteins.
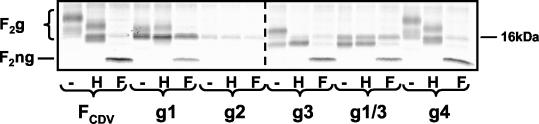
To further investigate the intracellular transport of F protein, we characterized the types of glycans present on the collection of mutants. Cells were labeled with 35S, and following immunoprecipitation, proteins were analyzed with endo H and endo F. Endo H-resistant glycans are acquired in the Golgi compartment, and proteins containing these oligosaccharides are generally transported rapidly to the cell surface (32). Control digestion with endo F completely eliminates all N-linked oligosaccharide chains.
Cell extracts were split into three aliquots, of which one was left untreated (Fig. 2, lanes −), another was digested with endo H (Fig. 2, lanes H), and the third was digested with endo F (Fig. 2, lanes F). In the parental FCDV protein and in all the mutants except g2, digestion with endo F caused a 3- to 9-kDa shift to the size of the nonglycosylated F2 subunit (Fig. 2, all bands labeled F2ng). For the g2 mutant, only a background band was detected, as expected, because this protein is not proteolytically cleaved (Fig. 1B). Two broad bands were detected for the unaltered F protein (Fig. 2, FCDV lane F), which may represent two differently glycosylated populations. Endo H digestion led to a slight shift of both bands to a lower molecular weight (Fig. 2, FCDV lanes − and H). A similar pattern was observed for the g4 mutant, which has all standard glycans (Fig. 2, g4 lanes − and H). In the g1 protein, without the first glycan, the bands migrated faster, and the shift after endo H digestion was minor (Fig. 2, g1 lanes). Thus, in this mutant, processing of glycans 2 and 3 was nearly complete. However, proteolytic cleavage was less efficient than that of the parental protein.
FIG. 2.
Endoglycosidase digestion analysis of the CDV F protein glycosylation mutants. Vero cells were transfected, incubated for 36 h, and labeled for 4 h with 35S. Cell extracts were immunoprecipitated with an anti-F2 rabbit anti-peptide serum and either digested with endo H (lanes H) or endo F (lanes F) or kept as untreated controls (lanes −), followed by SDS-PAGE separation (on 8 to 16% acrylamide gradient gels). Different gels are separated by a dashed line. The positions of the glycosylated (F2g) and nonglycosylated (F2ng) forms of F2 are indicated on the left. The origin of the weak 16-kDa band is unknown.
Absence of the third glycan (g3) also caused faster migration. Treatment with endo H caused the upper band to disappear and resulted in an increase in the intensity of the lower band (Fig. 2, g3 lanes − and H). Thus, in this mutant, one oligosaccharide was fully processed but the other was not. In view of the results with the g1 protein, the first oligosaccharide must be incompletely processed. In the g1/3 double mutant, a major band similar in size to the lower band detected in g3 was detected. The position and strength of this band did not change after digestion with endo H (Fig. 2, g1/3 lanes − and H). Thus, oligosaccharide 2 is completely processed. In summary, the oligosaccharides in position 1 are more heterogeneous than those in position 2 or 3.
The properties of the MV and CDV F protein glycans are similar.
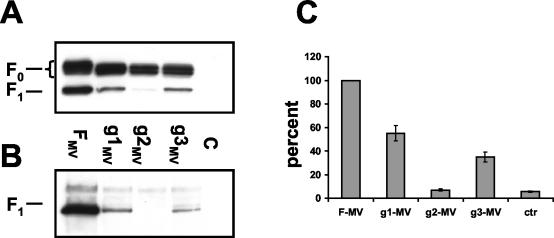
To determine if these findings are generalizable to another morbillivirus, we produced matched MV F protein glycosylation mutants (see scheme in Fig. 1A) and characterized their intracellular transport and function in the same system. As observed for the CDV F mutants, the F0 precursor and the F1 subunit were detected for the parental F protein and the g1MV and g3MV mutants by Western blot analysis, whereas only the F0 precursor was detected for the g2MV mutant (Fig. 3A). It is noteworthy that cleavage or activation of all MV F protein mutants was less efficient than that of the CDV F protein glycosylation mutants. Moreover, in all three mutants, a smaller F0 precursor was monitored, confirming that oligosaccharides are indeed attached to each of the three sites.
FIG. 3.
Analysis of the processing (A), cell surface expression (B), and fusion activities (C) of different MV F protein mutants. (A) Western blot analysis. Proteins were extracted from Vero cells transfected with the different F constructs, separated by reducing SDS-PAGE, and blotted onto PVDF membranes. The membranes were incubated with the anti-Fcyt antiserum. The positions of F0 and F1 are indicated on the left. (B) Surface biotinylation. Duplicate wells of the cells used for Western blot analysis was shifted to 4°C, biotin labeled, lysed, and immunoprecipitated overnight with the anti-Fcyt antiserum. Samples were separated by reducing SDS-PAGE, blotted onto PVDF membranes, and probed with peroxidase-coupled streptavidin. (C) Quantitative fusion assays were performed as described in the legend for Fig. 1, with the MV H protein replacing the CDV H protein. For each experiment, the value measured for the parental MV F protein was set to 100%. Means and standard deviations for four independent experiments in duplicate are shown.
Surface biotinylation analysis revealed that the F1 subunit of the g2MV mutant is not transported to the cell surface (Fig. 3B), a finding very similar to those for the CDV g2 mutant (Fig. 1C). The other two single mutants, g1MV and g3MV, are processed and transported normally, even though the amounts of protein detected are lower than that of the parental F protein (Fig. 3B).
The fusion activities of the MV F mutant proteins were quantified after coexpression with the MV H protein by using the fusion assay described above. As expected, the g2MV mutant, which is not transported to the cell surface, did not show any fusion activity (Fig. 3C). The activities of both the g1MV and g3MV mutants were lower than that of the parental protein (Fig. 3C), corresponding to the findings in the CDV system, but the relative degrees of reduction were reversed (Fig. 1D). This suggests that the roles of the individual glycans in modulating fusion may differ slightly between the MV and CDV proteins.
Because in one of the previous studies of MV F protein glycosylation, asparagines were mutated to serine rather than to glutamine (16), we tested if the type of mutation introduced influences the outcome of the experiment. We generated CDV and MV F protein mutants in which the asparagine at position 173 or 61, respectively, was changed to serine (g2S and g2MVS). The resulting proteins had the same characteristics as the g2 and g2MV mutants (data not shown), indicating that the nature of the amino acid introduced does not significantly affect processing and function.
Similar locations of glycans on the F proteins of Paramyxoviridae.
The structure of the F protein of the avulavirus NDV, sharing about 28% identity and exactly conserved cysteine spacing with CDV F, has been determined recently (8). In this protein four oligosaccharides are added (24). The glycan at position 85 is essential for folding, and lack of the oligosaccharide at position 191 has a negative effect on protein transport, whereas the other two do not alter folding or transport but modulate fusion (positions 336 and 447) (24). Based on the primary protein structure, the distribution of the glycans appears to be different: all three CDV F protein oligosaccharides (positions 36, 68 and 75 in the NDV numbering [Fig. 4A]) are attached to the amino-terminal F2 subunit, whereas three of the four NDV glycans are on the F1 subunit (Fig. 4A).
FIG. 4.
Comparison of the primary and tertiary structures of the NDV and CDV F proteins. (A) Linear comparison of the two sequences, which are 28% identical and 46% homologous (26). The positions of the CDV glycans are indicated at the top using the CDV numbering and the g1, g2, g3 code used for Fig. 1. The positions of the NDV glycans are shown below, using the NDV numbering and an n1, n2, n3 code devised in analogy with the CDV code. SP, signal peptide; FP, fusion peptide; TM, transmembrane region. F2 and F1, regions coding for the small and large F protein subunits, respectively. (B) Paramyxovirus F protein structure. (Left) Backbone of the NDV F protein as determined by X-ray crystallography (8). (Center left) Schematic diagram of the NDV F structure, encompassing a head, neck and stalk. The F2 subunit (85 amino acids) does not form a separate domain but runs like a thread between subdomains of the larger F1 subunit. A short loop near the F2 protein N terminus and a larger loop near the C terminus have been observed. Glycans are numbered with the n code used above. Cysteines that constitute the disulfide bond between the F2 and F1 subunit are shown as heavy dots joined by a line. (Center right) Schematic diagram of the superimposed CDV F protein drawn with the same symbols. Glycans are numbered with the g code. (Right) Backbone of the CDV F protein modeled after the NDV structure. Green, head region; red, neck region; blue, stalk region. Numbers next to black dots indicate the position of the respective residue on the backbone.
However, when the CDV F backbone is superimposed on the known NDV F protein structure, a striking similarity becomes apparent (Fig. 4B). The NDV oligosaccharide that is essential for efficient folding (n2) is situated in the lower half of the protein “neck,” as is the CDV oligosaccharide with a similar function (g2) (Fig. 4B; compare the two center schemes). The α-carbon atom of the asparagine to which the NDV n2 glycan is attached is predicted to be located 11.4 Å from the α-carbon atom of the homologous residue to which the g2 glycan is attached. Distances between residues in the three-dimensional model were determined by using a three-dimensional structure viewer (CN3D; National Center for Biotechnology Information, Bethesda, Md.). More strikingly, the NDV F1 n1 glycan at the top of the “head” and the corresponding CDV F2 g1 glycan (Fig. 4B, head region) are predicted to be anchored to the backbone at asparagines located 6.2 Å apart. A third oligosaccharide (n3 in NDV and g3 in CDV) is anchored near the neck-stalk interface in both molecules, but in this case the two asparagines are predicted to be located 14.4 Å apart (Fig. 4B). Finally, the fourth NDV glycan is situated in a fragment of the protein that was not crystallized, but it must reside in the stalk in view of its relative proximity to the transmembrane segment. Thus, an unsuspected conservation of location and function emerges from the comparative analysis of the oligosaccharides on the NDV and CDV F proteins. We term glycans with conserved location homotopic.
Two homotopic glycans with fusion control function are exchangeable.
The observations described above suggested the hypothesis that certain glycans, especially the closest pair on the top of the molecule, evolved independently into the same location to execute the same function. To test this hypothesis, we generated three mutants based on the single CDV glycosylation mutants g1 through 3. In each mutant, an N-glycosylation consensus sequence was introduced at the position of the homotopic NDV glycan. For g1, the homotopic oligosaccharide is attached to N366 (one-letter amino acid code) on the NDV sequence, which translates as position D474 on the CDV sequence. Since an S is located 2 residues downstream, the D474N mutation was sufficient to generate the consensus glycosylation sequence N-X-S for n1. The NDV homotopic glycan for g2 is attached to N85 on the NDV sequence, corresponding to position E189 on the CDV F protein. In this case, the E189N and L191S mutations were introduced to obtain a consensus glycosylation sequence. In the case of the compensation mutant for g3, direct transfer of the corresponding NDV glycan is complicated by P300, a residue incompatible with glycosylation of the upstream residue. Therefore, we moved the consensus sequence by 2 residues (A301N and Q303S), thereby increasing the predicted distance between the asparagines slightly, by 0.5 Å.
We then investigated whether mutagenesis introduced functional glycosylation sites. As shown in Fig. 5B, the apparent molecular weight of the F0 bands of mutants n1 and n2 reverted to that of the parental F protein, suggesting that the corresponding glycosylation sites are indeed used. However, the site introduced in n3 did not induce a decrease in F0 protein mobility and thus may not be used (Fig. 5B). This conclusion is corroborated by the fact that the F1 subunit of mutant n1 has a slower mobility than the parental subunit (Fig. 5B and C).
FIG. 5.
Structure and function of mutant proteins with homotopic glycan substitutions. (A) Linear representation of the three CDV mutant proteins. Positions of added glycans are indicated with glycan symbols above the box representing the reading frame. Newly introduced NDV glycans are indicated by dotted glycan symbols. Numbers above glycan symbols indicate the asparagine position in the CDV sequence; n1 through 3 refer to the CDV glycan that is complemented by the respective mutant (as in Fig. 4). SP, signal peptide; FP, fusion peptide; TM, transmembrane region; F2 and F1, regions coding for the small and large F protein subunits, respectively. (B) Characterization of the different F proteins by Western blot analysis. Proteins were extracted from Vero cells transfected with the different F expression plasmids, separated by reducing SDS-PAGE (10% acrylamide), and blotted onto PVDF membranes. The membranes were incubated with the rabbit anti-peptide serum anti-Fcyt. The positions of F0 and F1 are indicated on the left. (C) Surface biotinylation. Duplicate wells of the cells used for Western blot analysis were shifted to 4°C, biotin labeled, lysed, and immunoprecipitated overnight with the rabbit anti-peptide antibody anti-Fcyt. Samples were separated by reducing SDS-PAGE, blotted onto PVDF membranes, and probed with peroxidase-coupled streptavidin. (D) Quantitative fusion assays were performed as described in the legend for Fig. 1. For each experiment, the value measured for the parental F protein was set to 100%. Means and standard deviations for four independent experiments in duplicate are shown.
We then investigated if the glycan introduced in n2 elicited functional complementation. Elimination of g2 on CDV F results in intracellular retention (Fig. 1C), and the n2 mutant did not correct this phenotype, as determined by Western blotting of biotinylated cell surface proteins (Fig. 5C) and by cell fusion assays (Fig. 5D). In contrast, the n1 mutant did compensate for g1 function: the NDV homotopic glycan introduced into n1 led to an increase of fusion activity from 30% to almost 80% (Fig. 5D), indicating that the glycans located on the F protein head are able to perform each other's functions at least partially. Even though no glycan was added at the artificial consensus sequence in n3, the mutation led to an increase in total protein levels detected, but that did not translate into improved function. We then compared the stabilities of the g1-g3 and n1-n3 F protein mutants in a pulse-chase experiment and found that all proteins that reach the cell surface have similar transport rates (data not shown).
In summary, the attempt to reconstitute the function of CDV F proteins by transferring the homotopic NDV glycan was successful only for the fusion-modulating glycan located on the top of the molecule. The function of the neck glycan g2, which is necessary for folding, was not compensated for by the introduction of its homotopic partner. The third mutation did not produce a functional glycan attachment site. In view of the uncertainties of structural predictions, more extensive mutagenesis may be necessary to reconstitute the functions of the g2 and g3 glycans.
DISCUSSION
Correct N glycosylation is a prerequisite for folding and influences the function of many viral envelope proteins. In this study, we present a systematic analysis of the relevance of N-glycans for the maturation and function of a viral fusion protein. We found that in the F proteins of the morbilliviruses MV and CDV, only three glycans are added. The low number of glycans compared to other model glycoproteins such as influenza virus HA or human immunodeficiency virus Env facilitated the functional characterization of each individual chain: we showed that one glycan is essential for proper protein folding, whereas the absence of the other two does not interfere with this process. However, even when proteins lacking the first and third glycans are efficiently folded, proteolytically cleaved, and transported to the cell surface, they are less efficient at supporting fusion function than the parental proteins. The effects of eliminating the first and third glycans are cumulative.
The process of N glycosylation is closely interconnected with protein folding, since the glycan addition occurs while the nascent polypeptide chain folds into its native three-dimensional structure. Correct folding involves binding of chaperones to monoglycosylated trimming intermediates (14, 37). For MV the interaction of the F protein with GRP78 and calnexin has been demonstrated (3), and it is highly likely that the same chaperones interact with the CDV F proteins. In addition to catalysis of protein assembly and folding, chaperones also prevent further transport of misfolded proteins into the trans-Golgi network, thus performing a quality control function. Therefore, proteins transported to the cell surface usually correspond to the correctly folded and processed native conformation (12). It has been shown that complete inhibition of N glycosylation leads to loss of stability and function of most viral glycoproteins (11). Sometimes, one individual N-linked glycan can have a profound effect on intracellular transport (28), but more often the removal of oligosaccharides has a cumulative effect (13, 17, 21). One notable exception is the respiratory syncytial virus F protein, which is efficiently transported to the cell surface even when all three N-glycosylation sites are eliminated (9, 41).
A considerable amount of information about the function of the individual glycans added to the F proteins of different paramyxoviruses has been gained. As illustrated in Fig. 6A, the functions of the F protein glycans of NDV (Avulavirus genus), MV and CDV (Morbillivirus genus), simian virus 5 (SV5; Rubulavirus genus), and Sendai virus (SeV; Respirovirus genus) have been characterized (2, 24, 35). Glycans that are essential for folding and transport (T), essential for fusion control (F), or nonessential (N) have been defined (Fig. 6C). We interpreted this information after integrating it with the structural data recently presented for the NDV F protein (8). In the absence of an experimentally determined structure for the CDV F protein, we made the assumption that this structure is similar to that of the NDV F protein. In retrospect, that assumption is fully compatible with our results.
FIG. 6.
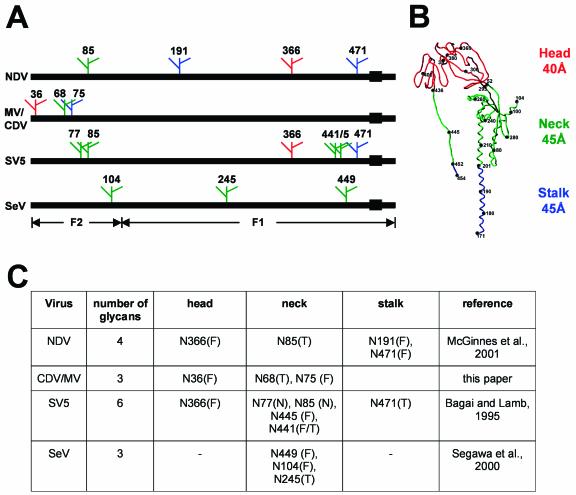
Locations and functions of glycans in the F proteins of Paramyxoviridae. (A) Primary structures of the F proteins of NDV, MV and CDV, SV5, and SeV and locations of individual glycans. Numbers refer to the NDV residues homologous to those of the related virus to which a glycan is added. Green, head glycans; red, neck glycans; blue, stalk glycans. Solid boxes, transmembrane regions. (B) Paramyxovirus F protein tertiary structure. Shown is the backbone of the CDV F protein, modeled after the NDV structure determined by X-ray crystallography (8). Head, neck, and stalk regions are indicated on the right and in the backbone with the same color code as in panel A. Numbers next to black dots indicate the position of the respective residue on the backbone. (C) Tabular comparison of locations and functions of the individual oligosaccharides. Glycans essential for folding and transport are labeled with a T, glycans that modulate fusion activity are labeled with an F, and dispensable glycans are labeled with an N.
The table shown in Fig. 6C allows the following generalizations. First, in three of four F proteins, most glycans are located in the neck region. Second, one F protein (that of SV5) has more glycans than the other F proteins, but two glycans can be eliminated without detrimental effects on function, probably reflecting functional redundancy. Third, the glycans supporting efficient folding and transport are located in the neck and stalk regions. Analogously, in the influenza virus HA molecule, the glycans protecting the fusion-active metastable form are located in the stalk region (31). Fourth, in each F protein at least one glycan is essential for folding and transport; other glycans modulate function to varying degrees. Fifth, a glycan with a fusion control function is located in the head region in the F proteins of three genera. In fact, the noncolinear top glycans of morbilliviruses (position 36 by NDV numbering) and NDV (position 366) reside very close to each other; the α-carbon atoms of their asparagines are separated by only 6 Å.
This remarkable spatial concurrence suggested the hypothesis that these two glycans evolved independently into the same location to execute the same function. Indeed, this hypothesis was confirmed experimentally: introduction of a glycan in the morbillivirus F1 subunit at the position homologous to NDV residue 366 compensated for the loss of the homotopic F2 CDV glycan (position 36) and restored fusion efficiency almost completely. We refer to glycans occupying the same location as homotopic. We note that homotopic substitutions may be straightforward only for fusion-regulating glycans. These exert their function after completion of translation and folding, and thus, only their location on the surface of the native protein is relevant. We also note that SeV (Fig. 6C) lacks a head glycan, indicating that not all paramyxoviruses use the same mechanism to modulate fusion.
It was not possible to compensate for the elimination of morbillivirus glycan 68 by introducing a glycan at position 85; these two glycans, which are necessary for efficient folding and transport of the respective F proteins, may function in different contexts. The three morbillivirus F protein glycans are clustered near the N terminus of the F2 subunit, whereas the single NDV F2 glycan is located somewhat more distantly. It was previously suggested that a cluster of glycans within about 50 amino acids of a protein's N terminus may determine the initial choice of chaperonins and therefore the folding pathway (25). It is possible that the exact structure of this glycan cluster influences the choice of chaperonins and that moving the position of a glycan 17 residues downstream, as was done here for the g2 and n2 mutants, disrupts the chaperonin selection function.
It is worth noting that two of the three morbillivirus F protein glycans are located near a cysteine involved in a long-distance disulfide bond (residues 76 to 199). All other eight cysteines are vicinal (positions 338 to 425) and may form four short loops in two compact domains, as in NDV. The organization of N-glycans near the N terminus of the influenza virus HA protein is similar to that of the morbillivirus F protein, with three vicinal glycans at position 8, 22, and 38 that surround a cysteine at position 14 involved in a large disulfide loop (residues 14 to 466). These glycans selectively slow down the oxidation of the long-range cysteine bond, retarding the folding of the N-terminal part of this protein while its C-terminal portion is being translated (10). By analogy, folding of the morbillivirus F protein N-terminal region may occur after folding of the C-terminal portion.
In summary, our studies disclosed a location in the head of the trimeric F protein of certain paramyxoviruses into which fusion-modulating glycans have independently evolved. In the morbillivirus F protein, the F2 subunit contributes the fusion-regulating glycan. In the avula- and rubulaviruses, the F1 subunit contributes the homotopic glycan.
Acknowledgments
We thank Daniel McCormick for help with structural modelling and Sompong Vongpunsawad for excellent technical support. We are grateful to Daniel Hebert, Georg Herrler, and Eric Poeschla for comments on the manuscript.
This work was supported by grants of the Mayo and Siebens Foundations to R.C. and by an Emmy Noether award of the German Research Foundation (DFG) to V.V.M.
REFERENCES
- 1.Alkhatib, G., S. H. Shen, D. Briedis, C. Richardson, B. Massie, R. Weinberg, D. Smith, J. Taylor, E. Paoletti, and J. Roder. 1994. Functional analysis of N-linked glycosylation mutants of the measles virus fusion protein synthesized by recombinant vaccinia virus vectors. J. Virol. 68:1522-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagai, S., and R. A. Lamb. 1995. Individual roles of N-linked oligosaccharide chains in intracellular transport of the paramyxovirus SV5 fusion protein. Virology 209:250-256. [DOI] [PubMed] [Google Scholar]
- 3.Bolt, G. 2001. The measles virus (MV) glycoproteins interact with cellular chaperones in the endoplasmic reticulum and MV infection upregulates chaperone expression. Arch. Virol. 146:2055-2068. [DOI] [PubMed] [Google Scholar]
- 4.Bolt, G., I. R. Pedersen, and M. Blixenkrone-Moller. 1999. Processing of N-linked oligosaccharides on the measles virus glycoproteins: importance for antigenicity and for production of infectious virus particles. Virus Res. 61:43-51. [DOI] [PubMed] [Google Scholar]
- 5.Cathomen, T., C. J. Buchholz, P. Spielhofer, and R. Cattaneo. 1995. Preferential initiation at the second AUG of the measles virus F mRNA: a role for the long untranslated region. Virology 214:628-632. [DOI] [PubMed] [Google Scholar]
- 6.Cathomen, T., B. Mrkic, D. Spehner, R. Drillien, R. Naef, J. Pavlovic, A. Aguzzi, M. A. Billeter, and R. Cattaneo. 1998. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J. 17:3899-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure (Cambridge) 9:255-266. [DOI] [PubMed] [Google Scholar]
- 9.Collins, P. L., and G. Mottet. 1991. Post-translational processing and oligomerization of the fusion glycoprotein of human respiratory syncytial virus. J. Gen. Virol. 72:3095-3101. [DOI] [PubMed] [Google Scholar]
- 10.Daniels, R., B. Kurowski, A. E. Johnson, and D. N. Hebert. 2003. N-linked glyans direct the cotranslational folding pathway of influenza hemagglutinin. Mol. Cell 11:79-90. [DOI] [PubMed] [Google Scholar]
- 11.Doms, R. W., R. A. Lamb, J. K. Rose, and A. Helenius. 1993. Folding and assembly of viral membrane proteins. Virology 193:545-562. [DOI] [PubMed] [Google Scholar]
- 12.Ellgaard, L., M. Molinari, and A. Helenius. 1999. Setting the standards: quality control in the secretory pathway. Science 286:1882-1888. [DOI] [PubMed] [Google Scholar]
- 13.Gallagher, P. J., J. M. Henneberry, J. F. Sambrook, and M. J. Gething. 1992. Glycosylation requirements for intracellular transport and function of the hemagglutinin of influenza virus. J. Virol. 66:7136-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond, C., I. Braakman, and A. Helenius. 1994. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. USA 91:913-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hebert, D. N., J. X. Zhang, W. Chen, B. Foellmer, and A. Helenius. 1997. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J. Cell Biol. 139:613-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, A., T. Cathomen, R. Cattaneo, and E. Norrby. 1995. Influence of N-linked oligosaccharide chains on the processing, cell surface expression and function of the measles virus fusion protein. J. Gen. Virol. 76:705-710. [DOI] [PubMed] [Google Scholar]
- 17.Hu, A., R. Cattaneo, S. Schwartz, and E. Norrby. 1994. Role of N-linked oligosaccharide chains in the processing and antigenicity of measles virus haemagglutinin protein. J. Gen. Virol. 75:1043-1052. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, M. R., T. Nilsson, and P. A. Peterson. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 9:3153-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 20.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 21.Machamer, C. E., R. Z. Florkiewicz, and J. K. Rose. 1985. A single N-linked oligosaccharide at either of the two normal sites is sufficient for transport of vesicular stomatitis virus G protein to the cell surface. Mol. Cell. Biol. 5:3074-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maisner, A., B. Mrkic, G. Herrler, M. Moll, M. A. Billeter, R. Cattaneo, and H. D. Klenk. 2000. Recombinant measles virus requiring an exogenous protease for activation of infectivity. J. Gen. Virol. 81:441-449. [DOI] [PubMed] [Google Scholar]
- 23.Malvoisin, E., and F. Wild. 1994. The role of N-glycosylation in cell fusion induced by a vaccinia recombinant virus expressing both measles virus glycoproteins. Virology 200:11-20. [DOI] [PubMed] [Google Scholar]
- 24.McGinnes, L., T. Sergel, J. Reitter, and T. Morrison. 2001. Carbohydrate modifications of the NDV fusion protein heptad repeat domains influence maturation and fusion activity. Virology 283:332-342. [DOI] [PubMed] [Google Scholar]
- 25.Molinari, M., and A. Helenius. 2000. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science 288:331-333. [DOI] [PubMed] [Google Scholar]
- 26.Morrison, T., and A. Porter. 1991. Structure, function, and intracellular processing of the glycoproteins of Paramyxoviridae, p. 347-382. In D. W. Kingsbury (ed.), The Paramyxoviruses. Plenum Press, New York, N.Y.
- 27.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. Product review. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 28.Ng, D. T., S. W. Hiebert, and R. A. Lamb. 1990. Different roles of individual N-linked oligosaccharide chains in folding, assembly, and transport of the simian virus 5 hemagglutinin-neuraminidase. Mol. Cell. Biol. 10:1989-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nussbaum, O., C. C. Broder, B. Moss, L. B. Stern, S. Rozenblatt, and E. A. Berger. 1995. Functional and structural interactions between measles virus hemagglutinin and CD46. J. Virol. 69:3341-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohuchi, M., R. Ohuchi, A. Feldmann, and H. D. Klenk. 1997. Regulation of receptor binding affinity of influenza virus hemagglutinin by its carbohydrate moiety. J. Virol. 71:8377-8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohuchi, R., M. Ohuchi, W. Garten, and H. D. Kenk. 1997. Oligosaccharides in the stem region maintain influenza virus hemagglutinin in the metastable form required for fusion activity. J. Virol. 71:3719-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeffer, S. R., and J. E. Rothman. 1987. Biosynthetic protein transport and sorting by the endoplasmic reticulum and Golgi. Annu. Rev. Biochem. 56:829-852. [DOI] [PubMed] [Google Scholar]
- 33.Plemper, R. K., A. L. Hammond, and R. Cattaneo. 2001. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J. Biol. Chem. 276:44239-44246. [DOI] [PubMed] [Google Scholar]
- 34.Plemper, R. K., A. L. Hammond, D. Gerlier, A. K. Fielding, and R. Cattaneo. 2002. Strength of envelope protein interaction modulates cytopathicity of measles virus. J. Virol. 76:5051-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segawa, H., T. Yamashita, M. Kawakita, and H. Taira. 2000. Functional analysis of the individual oligosaccharide chains of Sendai virus fusion protein. J. Biochem. (Tokyo) 128:65-72. [DOI] [PubMed] [Google Scholar]
- 36.Steinhauer, D. A., J. Martin, Y. P. Lin, S. A. Wharton, M. B. Oldstone, J. J. Skehel, and D. C. Wiley. 1996. Studies using double mutants of the conformational transitions in influenza hemagglutinin required for its membrane fusion activity. Proc. Natl. Acad. Sci. USA 93:12873-12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trombetta, E. S., and A. Helenius. 1998. Lectins as chaperones in glycoprotein folding. Curr. Opin. Struct. Biol. 8:587-592. [DOI] [PubMed] [Google Scholar]
- 38.von Messling, V., and R. Cattaneo. 2002. Amino-terminal precursor sequence modulates canine distemper virus fusion protein function. J. Virol. 76:4172-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Messling, V., G. Zimmer, G. Herrler, L. Haas, and R. Cattaneo. 2001. The hemagglutinin of canine distemper virus determines tropism and cytopathogenicity. J. Virol. 75:6418-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner, R., D. Heuer, T. Wolff, A. Herwig, and H. D. Klenk. 2002. N-glycans attached to the stem domain of haemagglutinin efficiently regulate influenza A virus replication. J. Gen. Virol. 83:601-609. [DOI] [PubMed] [Google Scholar]
- 41.Zimmer, G., I. Trotz, and G. Herrler. 2001. N-glycans of F protein differentially affect fusion activity of human respiratory syncytial virus. J. Virol. 75:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]