Abstract
The productive cycle of human papillomaviruses (HPVs) can be divided into discrete phases. Cell proliferation and episomal maintenance in the lower epithelial layers are followed by genome amplification and the expression of capsid proteins. These events, which occur in all productive infections, can be distinguished by using antibodies to viral gene products or to surrogate markers of their expression. Here we have compared precancerous lesions caused by HPV type 16 (HPV16) with lesions caused by HPV types that are not generally associated with human cancer. These include HPV2 and HPV11, which are related to HPV16 (supergroup A), as well as HPV1 and HPV65, which are evolutionarily divergent (supergroups E and B). HPV16-induced low-grade squamous intraepithelial lesions (CIN1) are productive infections which resemble those caused by other HPV types. During progression to cancer, however, the activation of late events is delayed, and the thickness of the proliferative compartment is progressively increased. In many HPV16-induced high-grade squamous intraepithelial lesions (CIN3), late events are restricted to small areas close to the epithelial surface. Such heterogeneity in the organization of the productive cycle was seen only in lesions caused by HPV16 and was not apparent when lesions caused by other HPV types were compared. By contrast, the order in which events in the productive cycle were initiated was invariant and did not depend on the infecting HPV type or the severity of disease. The distribution of viral gene products in the infected cervix depends on the extent to which the virus can complete its productive cycle, which in turn reflects the severity of cervical neoplasia. It appears from our work that the presence of such proteins in cells at the epithelial surface allows the severity of the underlying disease to be predicted and that markers of viral gene expression may improve cervical screening.
Papillomaviruses cause a variety of epithelial lesions, which range in severity from benign warts to invasive cervical cancer. More than 200 different types of human papillomavirus (HPV) have so far been identified on the basis of sequence analysis (22, 54). Each HPV type shows a tropism for a certain epithelial site and is associated with a particular type of skin lesion (54, 73). Palmar and plantar warts are caused by viruses such as HPV type 1 (HPV1) and HPV2, while genital warts are caused by viruses such as HPV11.
Although the reason for the site specificity of papillomaviruses is not understood, it is clear that the different papillomavirus types must replicate and produce infectious virions if they are to be successfully maintained in the population. The effectiveness with which they do this reflects their infection site and transmission route (73). HPV1, which is transmitted by indirect contact, produces lesions that are highly productive (5, 28). Genital warts caused by viruses such as HPV11 produce fewer infectious particles (73). Despite this heterogeneity, the productive cycles of all papillomaviruses are organized in a similar way (73). The viral genome is maintained as a low-copy-number episome in cells of the basal and parabasal layers and is amplified as the infected cell migrates towards the epithelial surface. The amplified genomes are subsequently packaged into infectious virions, which are lost from the epithelial surface during desquamation (42). Although the timing of life cycle events can vary, their order must be preserved if infectious virions are to be produced.
Viruses such as HPV6 and HPV11 are classified as low-risk papillomavirus types. In addition to causing external genital warts, these viruses can infect cervical tissue, producing benign epithelial lesions or condyloma. Of the 30 or so HPV types that can infect cervical epithelium, a subset are classified as high risk because the lesions they produce can progress to high-grade cervical intraepithelial neoplasia (CIN) and cancer (62). HPV16 is the most prevalent of the high-risk types and is found in more than 50% of all cervical cancers. Most cervical cancers (>99.7%) contain HPV DNA, and it is widely accepted that papillomavirus infection is a necessary factor in the development of the disease (90). Cancer of the cervix is the second most common female cancer worldwide and the primary female cancer in developing countries.
In countries with cervical screening programs, the incidence of cervical cancer is low. Cervical cancer is the ninth most common female cancer in the United Kingdom and the eighth most common in the United States (32). The reduction in cervical cancer incidence seen in many developed countries during the past few decades is largely attributed to the introduction of cervical screening programs, which allow the identification of precancerous cervical lesions before they become life-threatening. Since its introduction in the 1960s, the United Kingdom cervical screening program has saved an estimated 20,000 lives. Despite this success, the current strategy for cervical screening is far from perfect, failing to detect cervical abnormalities in approximately 20% (10 to 40%) of women (3).
Recent work has suggested that biomarkers could improve the accuracy and cost-effectiveness of cervical screening. Among the markers examined are Ki-67 (48), PCNA, minichromosome maintenance protein (MCM), p16 (51, 52, 81), and cyclins (48), which are not usually expressed above the basal and parabasal layers in normal cervical epithelium. An important cause of their increased expression in CIN is HPV infection and the expression of viral oncogenes such as E6 and E7. During productive papillomavirus infection, such surrogate markers of HPV infection (49) are expressed prior to the assembly of virus particles (73). In cervical cancer, such markers are present throughout the epithelium. Cervical cancer develops from areas of productive HPV infection (HPVI) through a series of well-defined stages that are classified as CIN1 to CIN3, or low- or high-grade squamous intraepithelial lesions (LSIL or HSIL). LSIL is equivalent to HPVI or CIN1, whereas CIN2 and CIN3 are equivalent to HSIL. Although markers of oncogene activity are not expressed in the upper layers of productive papillomas, the viral E4 protein is abundant (15, 27, 73). The expression of E4 follows that of viral gene products involved in cell proliferation (such as E6 and E7) and is itself followed by the expression of virion structural proteins such as L1 (27, 73). As markers of HPV infection, the E4 and L1 proteins are separate and distinct from those that allow the visualization of cells expressing viral oncogenes. Their abundance (that of E4 in particular [8, 15, 25-27, 73]) in productive lesions caused by many different papillomavirus types suggests that they may be used as specific markers of HPV-associated LSIL.
In this study we have compared the distribution of E4, L1, and markers of viral oncogene activity (MCM, PCNA, and cyclin A) in productive papillomavirus infections (caused by HPV1, -2, -65, -11, and -16) with the distribution of these markers during cervical cancer progression. In regions of LSIL caused by HPV16, the distribution of these markers is similar to that seen in productive lesions caused by other papillomavirus types and is consistent with such lesions being the site of virus synthesis. HSIL are abortive papillomavirus infections in which the life cycle of the virus either is not supported or is supported only poorly. E4 expression is restricted to isolated pockets near the epithelial surface, and viral oncogene expression is widespread. The expression of these markers follows a highly predictable pattern during cancer progression. The simultaneous detection of these three markers during cervical screening may allow a more accurate assessment of cervical disease than can be achieved with markers of viral oncogene activity alone.
MATERIALS AND METHODS
Clinical material and raft tissue.
Biopsy specimens were obtained from 25 patients with cervical neoplasia caused by HPV16. These lesions contained 36 discrete epithelial foci that were subsequently classified as CIN1 to CIN3 (and LSIL or HSIL) by at least two independent observers. Archival biopsy material was collected from four different centers without reference to patient details. These comprised the Queens Medical Centre (Nottingham, United Kingdom), the University of Tubingen (Tubingen, Germany), Addenbrooke's Hospital (Cambridge, United Kingdom), and the University of Otago (Dunedin, New Zealand). In most cases (n = 17), the presence of HPV DNA was first established using the general HPV primers GP5+ and GP6+ (83). PCR products were then separated by agarose gel electrophoresis (1.5%), and autoradiography was performed using a cocktail of 14 digoxigenin (DIG)-labeled high-risk HPV types (types 16, 18, 31, 33, 35, 45, 51, 52, 56, 58, 59, 62, 66, and 68) and 2 low-risk HPV types (types 6 and 11). The probes were then used separately to identify the specific infecting HPV type. In some instances, typing was carried out using a smaller selection of probes, consisting of HPV types 6, 11, 16, 18, 31, 33, 35, 42, 43, 44, 45, 52, 56, and 58 (n = 5). A small number of cases (n = 3) were screened for the presence of active HPV16 infection by immunostaining using antibodies to the HPV16 E4 protein. Differentiated raft tissue containing HPV16 was produced by methods described previously (33, 34). Apart from those caused by HPV11, cervical lesions caused by papillomavirus types other than HPV16 were not selected for this study, because high-affinity antibodies to their E4 proteins were not available. Normal (i.e., HPV-negative) cervical tissue was obtained from patients undergoing hysterectomy for diseases unrelated to the cervix. The source of lesions caused by HPV1, -2, -11, and -65 has been described previously (73). The images shown in the figures are typical of those seen following the analysis of at least 6 individual biopsy specimen (22 specimens for HPV1, 6 for HPV65, 25 for HPV2, and 6 for HPV11). Raft tissue was prepared from NIKS cells or NIKS cells harboring the HPV16 (W12) genome by using established protocols (33, 34).
Generation and labeling of antibodies.
The synthetic Fab TVG 405, which is specific for the HPV16 E4 protein, was expressed from the bacterial expression vector pUC119.His.myc (27) before being labeled with DIG or Alexa 488 by using commercially available kits (Roche or Molecular Probes). To maximize expression, bacteria were transferred directly from a frozen stock (stored at −70°C) into 2× tryptone-yeast extract (TYE) medium containing 1% glucose (and 100 μg of ampicillin/ml) in order to maintain repression of the lac promoter. After overnight incubation at 37°C, the culture was diluted 100-fold and grown to an optical density at 600 nm of 0.8 in 750 ml of 2× TYE medium containing ampicillin but no glucose. Expression of TVG 405 was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubation at 30°C for 3 h. After induction, periplasmic proteins were extracted by resuspension of the bacterial pellet in 40 ml of ice cold 20% sucrose-1 mM EDTA-30 mM Tris-HCl (pH 7.9) for 20 min, followed by recentrifugation and reextraction of the bacterial pellet in 40 ml of ice-cold 5 mM MgSO4. Fab was purified from the periplasmic preparations by using nickel-nitrilotriacetic acid chromatography (Qiagen) before being immediately dialyzed against phosphate-buffered saline (PBS) containing 0.2 mM EDTA. This optimized procedure typically produced 2 to 3 mg of Fab per liter of bacteria. Polyclonal antibodies to the HPV11 E4 protein were prepared by immunization of rabbits with a GST-E1^E4 fusion protein expressed from plasmid pGEX.11E1^E4 by using established protocols. Antibodies to the E4 proteins of other HPV types (HPV1, -2, -63, and -65) have been described previously (25, 26, 73), as have antibodies to L1 (Camvir1) (57). The anti-E7 polyclonal antibody (hen 16E7 [24]) was a gift from A. Venuti (Laboratory of Virology, Regina Elena Institute for Cancer Research, Rome, Italy).
Immunofluorescence staining.
Tissue sections were dewaxed in xylene (four times for 5 min each time), hydrated in industrial methylated spirits (three times for 5 min each time), and rinsed in deionized water. Slides were then microwaved at high power (800 W) for 15 min in 500 ml of antigen retrieval buffer (10 mM citric acid, pH 6.0), followed by cooling in the buffer for 20 min. After being washed in PBS, sections were blocked in 10% goat serum in 1% bovine serum albumin (BSA)-PBS for 2 h at room temperature. Primary antibodies (anti-Mcm7 [Neomarkers], 1:25; anti-cyclin A [Novacastra], 1:10; anti-PCNA [Neomarkers], 1:200) were diluted in PBS-1% BSA before being applied to the sections and incubated overnight at 4°C in a humidified chamber. Stringent washes in 0.05% Tween 20-PBS were followed by a 2-h incubation with a biotin-labeled secondary antibody (DAKO; 1:250) mixed with Alexa 488-conjugated TVG 405 and a nuclear counterstain diluted in 1% BSA-PBS. Following washing in PBS, the biotin signal was amplified with streptavidin-alkaline phosphatase (AP; Dako) and developed with a fast red substrate (Sigma) according to the manufacturer's instructions. The reaction was halted after 10 min by rinsing in water. Samples were mounted in Citifluor mounting medium (Agar Scientific), and images were captured using a SenSys monochrome camera and IPLab imaging software (Roper Scientific). Immunofluorescence staining was also attempted with TVGY701 and TVGY703 (anti-E7 [94]), C1P5 and 618 (anti-E6 [4, 47]), E1-N1 and E1-C1 (anti-E1 [55]), and TVG 261 and TVG 271 (anti-E2 [40]) by using the approach described above. HPV16 L1 protein and the E4 proteins of HPV types other than HPV16 were detected by using secondary antibodies (Amersham) conjugated to either fluorescein isothiocyanate, Alexa 488, or Texas red. For S-phase analysis, the method outlined by Mills et al. was followed, with the inclusion of a blocking step (with 1:20 goat serum for 1 h at room temperature) prior to incubation with the anti-DIG antibody (60). An anti-DIG monoclonal antibody (MAb) (Roche) (used at 1:25 for 1 h at room temperature in PBS) was used to detect the DIG signal, followed by detection with an anti-mouse Alexa 594 antibody (35).
Immunoenzymatic staining.
Tissue sections were dewaxed as described above and heated in a pressure cooker for 2 min in 2 liters of citrate buffer (10 mM citric acid, pH 6.0) before being washed in Tris-buffered saline (TBS). Blocking and immunostaining were carried out as previously described (35, 91) using MAbs to Mcm2 and Mcm5 or polyclonal antibodies to E7 (hen 16E7; 1:100 [24]). DIG-labeled TVG 405 diluted 1:50 was added, and detection was carried out using an anti-DIG secondary antibody conjugated to either AP or horseradish peroxidase (dilution, 1:50) followed by development using either fast red or diamino benzidine (DAB) according to the manufacturer's instructions (Sigma). Hen antibodies were detected using biotinylated anti-chicken immunoglobulin G (IgG) (1:1,000; Vector Laboratories) followed by streptavidin-AP (1:100; Dako) and developed using fast red. Immunofluorescence images were captured after fast red staining following double staining with TVG 405-Alexa 488 as described above. Sections were dehydrated through graded alcohol and mounted in DPX mounting medium (Gurr, BDH). Appropriate controls were performed by omission of antibody layers.
RESULTS
Visualization of events during productive papillomavirus infection using antibodies to MCM, E7, E4, and L1.
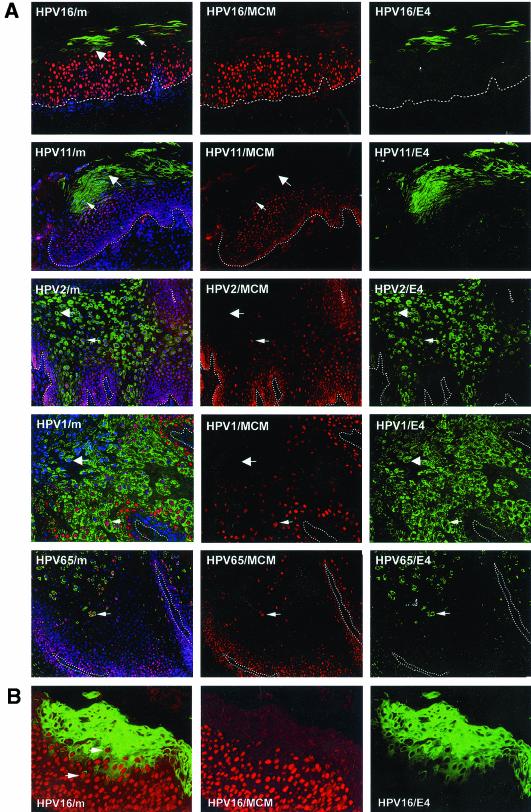
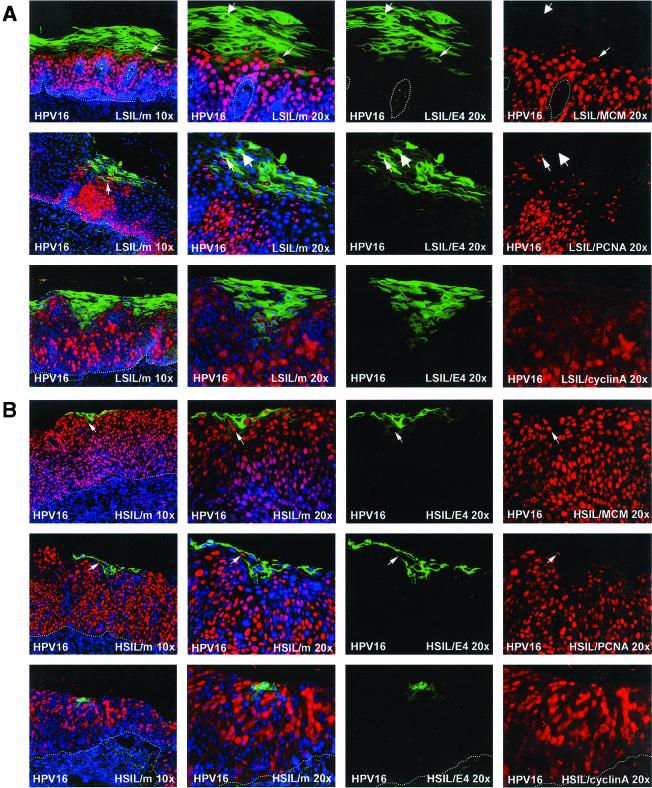
Previous studies have revealed similarities in the life cycles of diverse papillomavirus types by using antibodies to E4, L1, and PCNA (73). PCNA is an E2F-regulated gene product (19, 87, 92) that is induced in papillomas by the presence of E7 (41, 58), and by other viral oncogenes such as E6 and E5. In papillomavirus-infected tissue, PCNA can thus be regarded as a surrogate marker of viral early gene activity. MCM proteins have been suggested to be more effective as surrogate markers of SIL than PCNA (or Ki-67) because of their higher abundance (35, 91). As with MAbs to PCNA (73), antibodies to MCM proteins (MCM 2, 5, and 7) stained the nuclei of cells in the spinous and/or intermediate epithelial layers in productive lesions caused by different HPV types. These included cutaneous papillomas caused by HPV types 1, 2, 63, and 65 (verrucas and warts) and low-grade mucosal lesions caused by HPV11 and HPV16 (Fig. 1A). Although the results obtained with the two MAbs were similar, MCM staining (using MAb 47DC141 [Neomarkers]) was generally more robust than that obtained using monoclonal PC10 (Neomarkers), which detects PCNA. In all the productive lesions examined, MCM staining decreased following the onset of E4 expression, and a region of overlap was apparent where E4-MCM double-positive cells could be found (Fig. 1A). In HPV16-induced lesions, this region of overlap was particularly apparent when signal amplification methods were used to enhance the ability to detect the E4 protein (Fig. 1B).
FIG. 1.
Expression of MCM and E4 in productive lesions caused by different HPV types. (A) Distribution of MCM (red) and E4 (green) in productive lesions caused by HPVs. The HPV16 and HPV11 lesions (LSIL) were obtained from the cervix. The HPV2 lesion was a common hand wart. The lesions caused by HPV1 and HPV65 were verrucas. Cell nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (blue) and are visible in the merged images (leftmost panels). Dotted lines mark the position of the basal layer. Small arrows point to cells expressing both E4 and MCM. Large arrows indicate cells expressing E4 alone. (B) In regions of LSIL where HPV16 E4 expression was abundant, cells expressing both E4 (green) and MCM (red) were clearly visible following signal amplification. The merged image (without DAPI) is shown in the leftmost panel. Images were taken using a 10× objective.
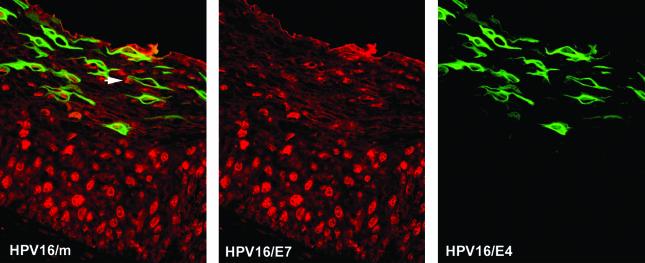
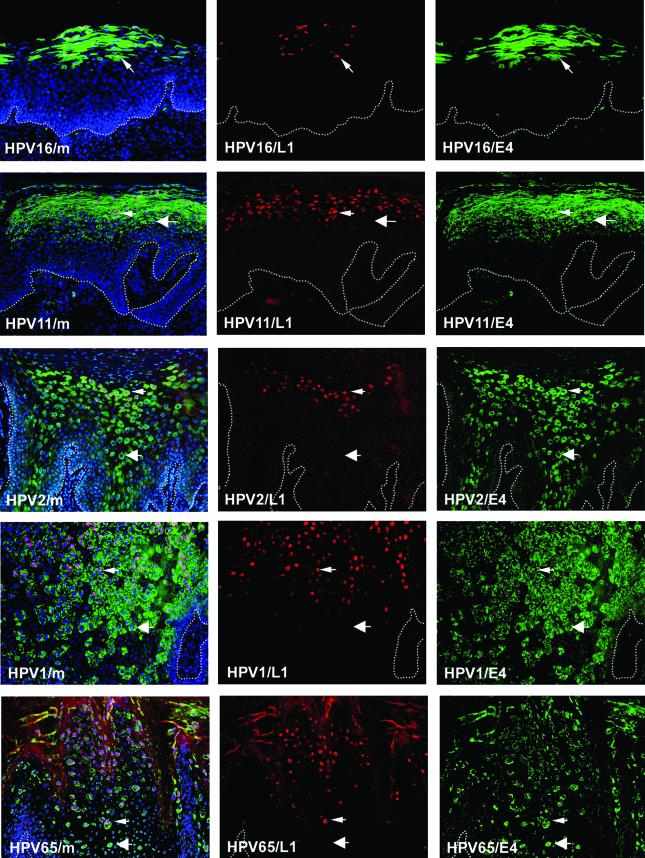
Surrogate markers such as MCM and PCNA are predicted to have an expression pattern that mimics the expression pattern of viral gene products necessary for S-phase entry. E7 is thought to be primarily responsible for stimulating the entry into S phase in most HPV types. To confirm that E7 has a distribution similar to that of E2F-activated genes such as MCM7 and PCNA, double staining was carried out using antibodies to E4 and to E7 (24). E7 is expressed at low levels compared to E4 and can be difficult to detect in archival sections of formalin-fixed, paraffin-embedded material (24, 94). To circumvent this problem, staining was carried out on HPV16-induced epithelial raft tissue prepared using NIKS cells harboring HPV16 episomes (33). Such rafts support the HPV16 productive cycle and are thought to resemble HPV16 infection in vivo. The E7 staining pattern shown in Fig. 2 is representative of that obtained in HPV16 rafts and resembles the pattern seen in HPV16-induced LSIL by use of antibodies to MCM (Fig. 1) or PCNA (73). Cells expressing both E4 and E7 were found in the intermediate epithelial layers (Fig. 2), with a distribution similar to that of E4-MCM-expressing cells in E4-expressing LSIL (Fig. 1). Cells expressing E7 in the absence of E4 were occasionally apparent in the upper epithelial layers (Fig. 2). Raft tissue prepared from NIKS cells that did not contain HPV16 DNA did not show any staining with antibodies to E7. Expression of the major capsid protein L1 always appeared to follow expression of E4, irrespective of the infecting HPV type, and was detectable in cells approaching the epithelial surface (Fig. 3).
FIG. 2.
Expression of E7 and E4 during the HPV16 productive cycle. Shown is the distribution of E7 (red) and E4 (green) in HPV16 raft tissue. The E7 staining pattern resembles that seen with antibodies to E2F-activated gene products such as MCM. The merged image (without 4′,6′-diamidino-2-phenylindole [DAPI]) is shown in the leftmost panel. Arrow points to cells expressing E7 in the absence of E4 in the upper epithelial layers. Images were taken using a 10× objective.
FIG.3.
Expression of E4 and L1 in productive lesions caused by different HPV types. Shown is the distribution of L1 (red) and E4 (green) in lesions (described in the legend to Fig. 1) caused by different HPV types. Cell nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (blue) and are visible in the merged images (leftmost panels). Dotted lines mark the position of the basal layer. Small arrows point to cells expressing both E4 and L1. Large arrows indicate cells expressing E4 alone. L1 is expressed in a subset of the cells that express E4. Images were taken using a 10× objective.
The E4 protein is first detected in S-phase competent cells that support viral genome amplification.
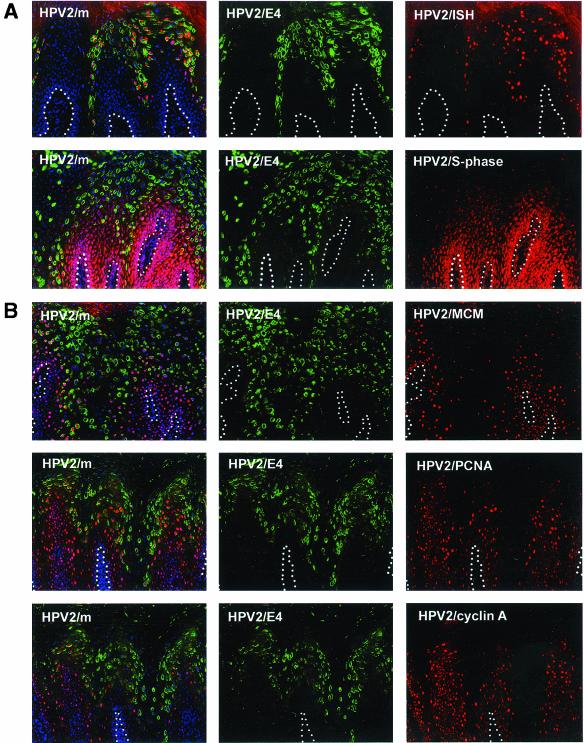
E4 expression results from the activation of a differentiation-dependent promoter which is situated within the E7 open reading frame (ORF) (12, 13, 38, 43, 70, 71). Proteins expressed from this promoter are either known (E1 and E2) or thought (E4 and E5) to be involved in genome amplification, and it has been suggested that E4 may be a marker of cells in which such proteins have been up-regulated (27, 73). In an attempt to understand the conserved expression pattern seen in productive lesions caused by different HPV types (Fig. 1 and 3) (73), DNA in situ hybridization and S-phase analysis (by in situ DNA replication) were carried out. As in situ DNA replication staining requires snap-frozen clinical material (60), the analysis was restricted to common warts caused by HPV2 (Fig. 4A) and mucosal lesions caused by rabbit oral papillomavirus. HPV2 is a supergroup A virus like HPV11 and HPV16, while rabbit oral papillomavirus is more closely related to HPV1 and falls into supergroup E (23, 64). In both lesions, S-phase competent cells (Fig. 4A) had a distribution that resembled that of replication proteins such as MCM, PCNA, and cyclin A (Fig. 4B). Since viral genome amplification coincides with E4 expression (27, 73) and occurs in cells in which the cellular replication machinery is active, it was not surprising to find that some of the replication-competent (and MCM- and/or PCNA-positive) cells contained abundant E4 protein (Fig. 4A). Our results suggest that the increase in expression of viral replication proteins that results from the activation of the differentiation-dependent promoter (such as E1, E2, and E4) occurs prior to the down-regulation of proteins necessary for S-phase entry (such as E6 and E7).
FIG. 4.
E4 expression begins in cells that express S-phase markers and are S-phase competent. (A) (Upper panels) Distribution of amplified viral DNA (red) and E4 (green) in an HPV2-induced wart. (Lower panels) Distribution of cells in S phase (red) and cells expressing E4 (green). Cell nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (blue) and are apparent in the merged images shown on the left. Dotted lines mark the position of the basal layer. (B) The distribution of three different markers necessary for S phase (MCM, PCNA, and cyclin A) (red) is compared with the distribution of E4 (green) in lesions caused by HPV2. In each case, the first appearance of E4 occurs before the loss of such markers. The merged images including DAPI staining (blue) are shown on the left. Dotted lines mark the position of the basal layer. Images were taken using a 10× objective.
Immunostaining studies suggest a model of productive HPV infection.
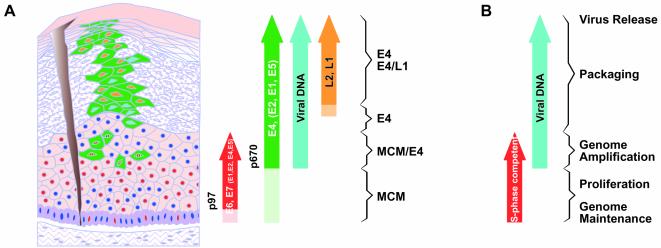
The results described here and previously (73) suggest a pattern of viral protein expression that is preserved across a diverse range of papillomavirus types including HPV16 (shown diagrammatically in Fig. 5). During normal productive infection, MCM expression, and presumably therefore the expression of viral oncogenes (Fig. 1 and 2), extends from the basal layer upward (Fig. 5A). E4 expression begins in these MCM-positive cells and usually persists to the epithelial surface (Fig. 5A). It is in these cells (i.e., those that express E4 and S-phase markers) that viral genome amplification begins (Fig. 5A, viral DNA) (8, 27, 73). Cells that are S-phase competent overlap those that contain amplified viral DNA (Fig. 5B). Expression of the papillomavirus major coat protein (L1) occurs in a subset of E4-positive cells in the upper epithelial layers (Fig. 5A) (27, 73). It appears from these studies that a productively infected cell expresses each of these proteins (E7, E4, L1) in turn during its migration from the basal layer to the epithelial surface, irrespective of the infecting virus type.
FIG. 5.
Expression data suggest a model of productive HPV infection. (A) The differentiated epithelium is represented diagrammatically on the left, and the markers expressed are represented as arrows on the right. Following access to the basal layer, the virus is thought to establish itself as a low-copy-number episome. The E7 protein (red circles) is expressed in the lower epithelial layers as determined by the presence of E2F-activated gene products such as MCMs (Fig. 1) and direct detection of the E7 protein (Fig. 2). Dark blue circles represent nuclei of uninfected or nonpermissive cells. It is not clear whether E7 is expressed in cells of the basal layer or only in cells of the parabasal and intermediate cell layers (Fig. 2). The expression of E4, and presumably that of other proteins whose genes lie downstream of the differentiation-dependent promoter (p670 in HPV16) (green arrow), is triggered before the expression of E7 ceases. In lesions caused by HPV16, E7 is expressed from the early promoter (p97), which also directs the expression of E6 and may direct the low-level expression of other early region proteins (red arrow). The E4-expressing cells (green) that also express E7 are predicted to contain all the early viral gene products in order to facilitate genome amplification. Expression of the virus capsid proteins (L1 and L2) (orange arrow) follows the completion of genome amplification and occurs in a subset of cells that express E4. The precise stage in the virus life cycle supported at any particular point is apparent from the combination of markers expressed. (B) Based on the distribution of cells in S phase (red arrow) and the distribution of amplified viral DNA by in situ DNA hybridization (turquoise arrow), it is apparent that genome amplification is limited to the region where E4 expression overlaps the expression of proteins necessary for DNA synthesis such as MCMs. Stages in the papillomavirus productive cycle that are revealed by use of life cycle markers are given on the right.
The order of life cycle events is preserved in HPV-associated cervical neoplasia.
Given this basic understanding of how viral proteins are expressed during productive papillomavirus infection, we next examined the changes in expression patterns that occur during the progression from productive infection to cervical cancer. Cervical cancer represents a “dead end” for the virus, because viral DNA becomes integrated into the host cell chromosome (often with large deletions), and the ability to produce infectious virions is abolished. This happens rarely in lesions caused by HPV1, -2, or -65 but occurs more commonly in cervical lesions caused by high-risk papillomavirus types such as HPV16.
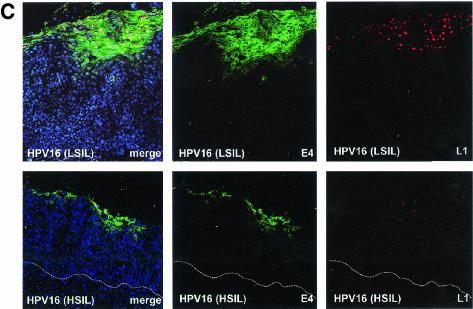
HPV16 is the primary cause of HSIL and accounts for around 55% of all invasive cervical cancer cases (53). In LSIL, HPV16 and HPV6 or -11 are equally prevalent (53). The patterns of protein expression in productive LSIL caused by the two virus types were very similar (see Fig. 1) and resembled that seen in productive papillomas caused by other virus types (Fig. 1). Similar results were obtained by using the E2F-activated markers (MCM, PCNA, and cyclin A) (Fig. 6A) used to examine expression patterns in warts caused by HPV2 (Fig. 5A). Expression of MCM, PCNA, and cyclin A was abundant in the lower epithelial layers and was lost after the appearance of E4 (Fig. 6A). In HSIL, which originate from LSIL during cancer progression, the extent of E4 expression was considerably reduced, and E4 expression was restricted to small pockets of cells close to the epithelial surface (Fig. 6B). This loss of E4 expression was accompanied by a corresponding increase in the prevalence of cells expressing surrogate markers of viral oncogene activity such as MCM, PCNA, and cyclin A (Fig. 6B). While the expression of these markers appeared to correlate directly with the severity of SIL, the expression of E4 correlated inversely with severity. In HSIL and LSIL that supported late gene expression, the appearance of E4 followed the expression of oncogene markers and was first detected in cells that were expressing these proteins (Fig. 6). Although the full productive cycle of the virus is not supported during cancer progression (i.e., in HSIL), the order of life cycle events is preserved. This was also apparent when antibodies to the major virus coat protein L1 were used (Fig. 6C). As in productive infections, the expression of L1 in HSIL followed the expression of E4 and could be detected only in cells that were E4 positive (Fig. 6C). L1 expression appeared to be less extensive than that of E4 and could not be detected in many high-grade cervical lesions (data not shown).
FIG. 6.
Distribution of surrogate markers, HPV16 E4, and L1 in regions of LSIL and HSIL caused by HPV16. (A) Distribution of surrogate markers (MCM, PCNA, and cyclin A) (red) and HPV16 E4 (green) in a region of LSIL where E4 staining was extensive. Merged images (including 4′,6′-diamidino-2-phenylindole [DAPI] staining [blue]) are shown on the left at high (20× objective) and low (10× objective) magnifications. Small arrows indicate cells expressing both markers; large arrows indicate cells expressing E4 only. Dotted lines mark the position of the basal layer. (B) Distribution of surrogate markers and HPV16 E4 (as above) in a region of HSIL where E4 expression was apparent close to the epithelial surface. Small arrows point to cells expressing both markers. Although an overlap between the expression of the two proteins is apparent, the expression of E4 in HSIL is less extensive than that in LSIL. Dotted lines mark the position of the basal layer. (C) Distribution of HPV16 E4 (green) and L1 (red) in regions of LSIL (upper panels) and HSIL (lower panels) associated with HPV16. L1 expression follows the expression of E4 in LSIL and HSIL but is less extensive in HSIL. Dotted lines mark the position of the basal layer. Merged images, which include a DAPI counterstain (blue), are shown on the left. Images were taken using a 10× objective.
The distribution of life cycle markers in infected tissue changes significantly during neoplastic progression.
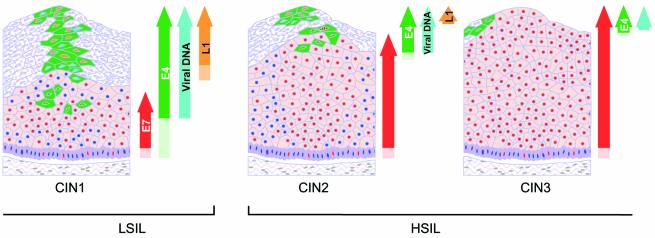
Changes in the expression pattern of viral gene products during cancer progression suggest that HSIL may sometimes represent an abortive infection for the virus. While the order of life cycle events remains unaltered, their timing changes considerably, and in HSIL, L1 expression is only poorly supported (Fig. 7). As the expression of viral gene products reveals the extent to which the virus life cycle is supported (Fig. 5), we wondered whether the severity of cervical disease could be established from the distribution of such markers at the epithelial surface (Fig. 7). To examine this further, productive lesions caused by HPV11 (mucosal) and HPV2 (cutaneous) were compared with LSIL and HSIL caused by HPV16. Although these viruses are all contained within supergroup A, HPV2 causes warts and verrucas whereas HPV11 causes low-grade cervical lesions. In addition to their classification as either LSIL or HSIL, cervical lesions caused by HPV16 were also classified (where possible) as HPVI, CIN1, CIN2, or CIN3, because CIN2 lesions differ from CIN3 in the extent to which undifferentiated cells occupy the full thickness of the epithelium (77). We suspected that such changes may be important in regulating the extent to which the virus life cycle is completed.
FIG. 7.
The expression of viral proteins changes in a predictable way during cancer progression. The timing and order of events in regions of LSIL can resemble those seen in productive infections caused by other papillomavirus types (Fig. 5). In HSIL caused by HPV16, the order of events is preserved, but the timing is disturbed and the number of cells expressing surrogate markers of viral oncogene activity is increased. In HSIL (CIN3) a total failure to complete the papillomavirus life cycle can occur, resulting in an abortive infection. Red circles, cells expressing E7; dark blue circles, nuclei of uninfected or nonpermissive cells. Cells expressing E4 are depicted as green. Nuclei of cells expressing L1 are shown in orange, whereas those that contain only amplified viral DNA are turquoise.
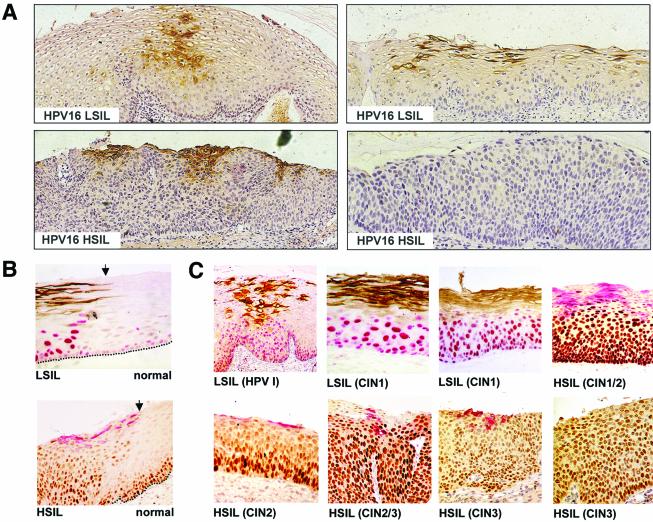
The E4 protein was visualized by immunoperoxidase staining and DAB detection in order to allow protein expression and tissue morphology to be assessed simultaneously (Fig. 8A). The HPV16 E4 protein could not be detected in lesions that were negative for HPV16 DNA by PCR and was not detected in regions of normal tissue immediately adjacent to areas of dysplasia (Fig. 8B). When HPV16 E4 was present in LSIL (CIN1), it was usually found in the intermediate and superficial layers in cells that showed some evidence of morphological differentiation but did not exactly correlate with the appearance of koilocytes (Fig. 8A). When HPV16 E4 expression was apparent in HSIL, it was restricted to areas of cell differentiation close to the epithelial surface (Fig. 8A).
FIG. 8.
Distribution of HPV16 E4 and MCM in precancerous cervical epithelium infected with HPV16. (A) Detection of HPV16 E4 (brown) by immunoenzymatic staining in a region of HPV16 LSIL (upper panels) and HSIL (lower panels). Lesions were counterstained with hematoxylin to enable grading. Images were taken using a 4× objective. (B) Detection of HPV16 E4 and MCM at the edge of an HPV16-induced LSIL (upper panel [brown, E4; red, MCM]) or HSIL (lower panel [red, E4; brown, MCM]). E4 staining is not apparent in normal epithelial tissue. In normal cervical epithelium, MCM expression is confined to cells of the basal and parabasal cell layers. Images were taken using a 10× objective. (C) Patterns of HPV16 E4 and MCM expression in cervical lesions showing some evidence of HPV16 late gene expression. The lesions examined included HPVI and CIN1 (brown, E4; red, MCM) and CIN1/2, CIN2 and CIN3 (red, E4; brown, MCM). The progressive loss of HPV16 E4 staining is accompanied by an increase in the abundance of cells that express MCM. Images were taken using a 10× objective.
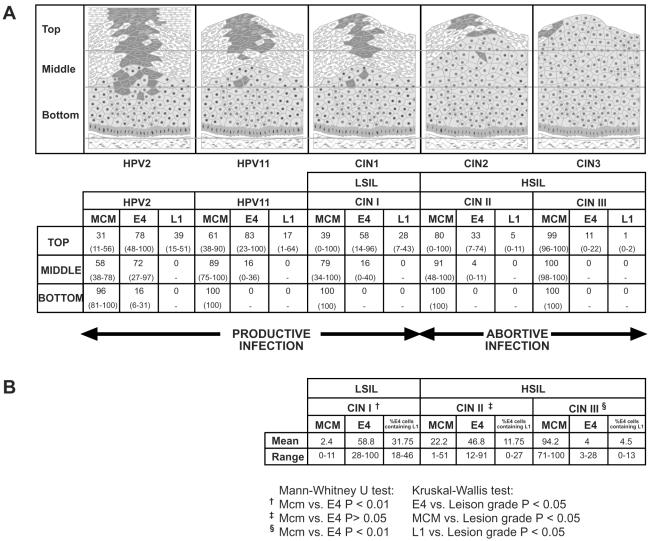
In order to examine the potential of combining life cycle markers during diagnosis, the percentages of cells expressing MCM, E4, and L1 in the top, middle, and lower thirds of cervical neoplastic lesions of differing grades (Fig. 8C) were established. Counting was carried out on at least five individual epithelial foci classified as CIN1 (LSIL), CIN2, or CIN3 (HSIL) (Fig. 8C) following immunofluorescence staining. As suggested from the data shown in Fig. 1 and 6, E4 expression was confined to the upper epithelial layers (Fig. 9A) and showed a significant inverse correlation with lesion grade (P < 0.01 by the Kruskal-Wallis test) (Fig. 9A). E4 could be detected in 100% of the surface cells in some regions of LSIL (Fig. 8C). L1 expression was always less extensive than that of E4, but a similar inverse correlation was apparent (P < 0.01 by the Kruskal-Wallis test) (Fig. 9A). E4-L1 double staining was achieved by using either horseradish peroxidase- or AP-conjugated antibodies, followed by visualization with DAB (brown) or fast red. When such staining was carried out using antibodies to MCM (Fig. 8C), a significant positive correlation with lesion grade was observed (P < 0.01 by the Kruskal-Wallis test) (data shown in Fig. 9A). Both E4 and MCM were easily detected by immunoenzymatic methods in formalin-fixed, paraffin-embedded tissue (Fig. 8).
FIG. 9.
E4, MCM, and L1 as markers of HPV-associated cervical disease. (A) Lesions caused by HPV2, HPV11, or HPV16 were double stained with antibodies to E4 and L1 or to E4 and MCM, and the mean number of cells expressing each protein was established after counting cells in the upper, middle, or lower third of the epithelium. The counting data are shown in the table and illustrated diagrammatically above it. The range of values obtained after counting five or more lesions of each type is given in parentheses beneath the mean. In productive lesions caused by HPV2, HPV11, and HPV16 (i.e., LSIL), cells expressing E4 are more abundant in the upper epithelial layers than cells expressing MCM. In CIN2 and CIN3 lesions (HSIL) caused by HPV16, cells expressing MCM greatly outnumber those expressing E4. The ratio of cells expressing E4, MCM, or L1 in the upper epithelial third is symptomatic of the severity of cervical disease. (B) Counting was performed on cells in the uppermost five cell layers in cervical lesions classified as CIN1, CIN2, or CIN3 and was restricted to regions in which some evidence of late gene activity was apparent as determined by the expression of E4. Where HPV16 E4 expression was most extensive, such as LSIL (CIN1), MCM was only rarely found at the epithelial surface. In regions where E4 was expressed at very low levels, such as in HSIL (CIN3), MCM expression was widespread. The extent of L1 expression at the epithelial surface decreased as E4 expression was retarded into the more differentiated epithelial cell layers, and L1 expression was not widespread in HSIL. The ratio of cells expressing E4, MCM, and L1 at the epithelial surface can indicate the grade of neoplasia in the underlying cervix.
Cells in the lower epithelial third were usually positive for MCM, irrespective of disease status (Fig. 8B and C and 9A). By contrast, the percentage of positive cells in the middle and upper epithelial layers increased with increasing disease severity and usually extended to the epithelial surface in regions of CIN3 (Fig. 8B and C and 9A). In CIN3 lesions, but not in CIN1 lesions that expressed the HPV16 E4 protein, the number of MCM-positive cells was larger than the number of cells that contained E4 (P < 0.01 by the Mann-Whitney U test) (Fig. 9A). MCM staining did not reach the epithelial surface in areas of E4 expression in low-grade lesions or productive papillomas (Fig. 5, 6, and 8), even though MCM-positive cells were apparent in the upper third (Fig. 9). E4-expressing cells were present in the lower epithelial layers of lesions caused by HPV2 but not in those caused by HPV11 or -16, which may reflect differences in infection site and transmission route between HPV2 and genital papillomavirus types (73). Apart from this, however, no significant difference (P < 0.01 by the Kruskal-Wallis test) (data presented in Fig. 9) was apparent in the distribution of MCM, E4, or L1 between productive lesions caused by HPV2, productive lesions caused by HPV11, and regions of LSIL in which HPV16 late gene expression could be observed (CIN1). This suggests that productive infections caused by different papillomaviruses share an overall organizational similarity. A significant difference was observed, however, in the distribution of all markers when CIN1 (LSIL) and CIN2 or CIN3 (HSIL) were compared (P < 0.05 by the Mann-Whitney U test). No such difference was apparent between CIN2 and CIN3.
Potential of life cycle markers for the diagnosis of cervical neoplasia.
The use of antibodies for cervical screening requires their evaluation in cells taken from the surface of the cervix. Although the problems associated with staining such cells are likely to be different from those associated with staining tissue sections (described here), our results suggest that antibodies to viral gene products and/or antibodies to surrogate markers of viral protein expression may be used in combination to assess disease status. In our hands, however, efforts at detection of viral gene products other than E7, E4, and L1, such as E6 (MAb C1P5 [39]), E2 (MAbs TVG 261 and TVG 271 [40]), or E1 (MAbs E1-N1 and E1-C1 [55]) in infected tissue were not successful. While it may be possible to detect these proteins by using appropriate antibodies and staining protocols (72, 84), the ease with which E4, L1, and MCM (as a surrogate marker) can be detected suggests that they may be particularly useful for diagnostic purposes.
As cervical screening specifically investigates cells taken from the epithelial surface, we decided to examine the uppermost cell layers of cervical lesions for the presence of the three markers described above. Epithelial foci that were classified as CIN1 (LSIL), CIN2, or CIN3 (HSIL) and which showed some evidence of life cycle completion (as determined by the expression of E4 and/or L1 [shown diagrammatically in Fig. 9A]) were selected. Five foci of each type were counted, and the mean percentage of cells expressing E4, L1, or MCM in the upper five cell layers was determined (Fig. 9B). CIN3/HSIL that lacked detectable levels of E4 were not included, as they usually lacked signs of epithelial differentiation and contained only MCM-expressing cells at their surfaces. Such lesions may represent a more severe form of CIN3/HSIL than those that still express E4. The expression of E4 and L1 in the superficial cell layers showed an inverse correlation with lesion grade (P < 0.05 by the Kruskal-Wallis test), whereas the expression of MCM showed a significant positive correlation (P < 0.05 by the Kruskal-Wallis test). In the CIN1/LSIL foci selected here, cells expressing E4 in the upper epithelial layers were significantly more abundant than those expressing MCM (P < 0.01 by the Mann-Whitney U test). In foci of CIN3/HSIL, the situation was reversed, with the number of MCM-positive cells becoming significantly larger than the number of cells expressing E4 (P < 0.01 by the Mann-Whitney test). The combined detection of such markers, which are present at the epithelial surface, and which are present at different levels in lesions of different grades, may improve the accuracy of cervical screening.
DISCUSSION
The life cycle of papillomaviruses can be divided into three phases that can be distinguished by using antibodies to PCNA, E4, and L1 (73). PCNA is one of a number of cellular proteins that are up-regulated during papillomavirus infection as a result of viral early gene expression. Such proteins include MCM, Ki-67, cyclin A, and cyclin E, which are normally active during S-phase entry as a result of the presence of external growth factors. During the normal cell cycle, the phosphorylation of Rb by G1/S cyclins and the subsequent release of the E2F transcription factor are the trigger for the activation of these genes. In HPV-infected tissue, their activation is regulated by the viral protein E7, which binds to Rb and stimulates the release of the E2F transcription factor irrespective of Rb phosphorylation. E2F-activated genes are expressed above the basal and parabasal layers in papillomavirus-infected tissue but are not found in these cell layers in uninfected epithelium. As a result, such proteins can be regarded as surrogate markers of HPV infection (49).
While MCM, PCNA, Ki-67, and the G1/S cyclins may all be useful as markers, differences in their abundance and stability make them more or less appropriate for screening purposes. Both PCNA and MCM persist into G2 (19, 69), but the greater abundance of MCM (63) allows it to be detected more easily. Antibodies to MCM have recently been used to identify cycling cells in several different types of cancer including urinary tract cancer, renal cancer, and colon cancer (17, 35, 79, 85) and have also been used to identify proliferating cells during cervical screening (91). Although we have focused on MCM in this study, other markers of viral oncogene activity, such as cyclin E or p16 (68), may have the additional advantage of allowing neoplasia to be distinguished from metaplasia or wound repair. Both proteins are present at increased levels in lesions caused by papillomaviruses but are usually absent or only transiently expressed during normal cell proliferation. Despite differences in the utility of the different markers, a common pattern emerges. In high-grade lesions and cervical cancers, such markers extend through the full thickness of the epithelium, whereas in low-grade lesions they are confined to the lower epithelial layers and reach the epithelial surface less frequently. Several studies have suggested that the abundance of these proteins in cells taken during cervical screening may be used to predict the severity of cervical disease (20, 21, 75, 76, 81, 86), although reservations regarding their specificity as markers of papillomavirus infection exist (78). The ability to detect E7 directly, as is possible for E4 and L1, would simplify the situation and may be possible with appropriate antibodies and staining procedures.
As a marker of papillomavirus infection, E4 differs from the markers described above, which depend for their expression on the presence of E7. Antibodies to E4 detect a population of cells that is largely separate from that which expresses E7, irrespective of whether the E7 protein is detected directly or indirectly (Fig. 1 and 2). The E4 protein is present in cells at the epithelial surface in productive lesions caused by most (if not all) papillomavirus types (73), whereas oncogene markers are expressed below this level. In some lesions, such as those caused by HPV1, E4 accounts for as much as 30% of total cell protein (8, 25), and the general ease with which it can be detected in lesions caused by other virus types (73) suggests that it is similarly abundant. For HPV16, the availability of high-affinity MAbs (27) that can detect E4 in paraffin-embedded, formalin-fixed tissue simplifies its detection, and in this study, the E4 protein was visualized directly using an Alexa 488-conjugated monoclonal Fab (TVG 405 [27]). TVG 405 was derived from a synthetic immunoglobulin library displayed on phage (27) and is not recognized by the anti-mouse and anti-rabbit secondary antibodies used for the detection of MCM and L1 during immunostaining. In contrast to surrogate markers of E7, which increase in abundance in high-grade neoplasia and cancer, E4 is primarily a marker of HPV-associated LSIL, and its prevalence decreases during cervical cancer progression.
In cervical smear preparations, the presence of oncogene-expressing cells (as determined using surrogate markers) cannot necessarily distinguish between HSIL and LSIL. Both may be expected to give rise to smears that contain cells expressing surrogate markers of HPV oncogene activity, such as MCM, p16, or cyclin E. In the traditional Pap test, cytology provides information that allows the diagnosis of LSIL, and it seems likely that surrogate markers of viral oncogene expression (MCM, PCNA, G1/S cyclins, p16) will need to be considered along with cytological data for accurate diagnosis. The inclusion of LSIL markers such as E4 (and possibly also L1) may improve diagnostic accuracy and reveal the extent of abortive infection. As E4 antibodies generally have limited cross-reactivity (73), such an approach may, at the same time, allow high-risk infections to be distinguished from those caused by low-risk papillomavirus types.
The molecular changes that govern progression from productive infection to HSIL are not fully understood, and the prognostic significance of the failure to express E4 (or to express it in higher epithelial layers) is not known. Productive papillomas such as those caused by HPV11 or HPV2 contain only episomal viral DNA (reviewed in reference 42). In cervical cancers the viral DNA is nearly always integrated into the host cell chromosomes (11, 16, 89, 93), leading to an elevation in the levels of viral oncogene expression (44, 45) as a result of the disruption of the E2 ORF. In precursor lesions caused by HPV16 (LSIL and HSIL), episomal and integrated genomes coexist (2, 31, 74), and it has been suggested that integration may occur early during neoplastic progression (74). The changes in life cycle organization that accompany cervical cancer progression may have several possible explanations and may in fact be linked to changes in the levels of E6 and E7 that occur following integration. While HPV16 episomes can clearly persist in cells harboring integrated viral genomes, the onset of genome amplification may be delayed in cells expressing higher levels of E6 and E7, or be inhibited altogether (CIN3). Many CIN2 and CIN3 lesions, which support productive infection only poorly, contain both episomal and integrated HPV16 DNA (74). Although the effect of deregulated E6 and E7 expression on the timing of initiation of late events remains to be established, work from several groups suggests that the expression of these proteins does in fact increase during the progression from LSIL to HSIL (36, 56, 88), whereas cervical cancer originates from HSIL following the accumulation of secondary mutations in the host cell genome.
The failure of HPV16 to complete its productive life cycle in HSIL may have other explanations, however, and may be related to the specific site at which HPV16-associated precancerous lesions develop. Most cervical intraepithelial lesions originate from cells in the transformation zone, the region of cervical epithelium that changes from columnar to squamous in response to hormonal changes such as those occurring at puberty or during pregnancy. This region of the cervix is particularly sensitive to estrogen (1) and to estrogen-induced carcinogenesis (29), and the prolonged use of estrogen-containing oral contraceptives has been reported to double the incidence of cervical cancer (9) and affect the level of viral gene expression in vivo (6, 59) and in vitro (10, 50, 61). Although the effect of hormones on late gene expression has not yet been characterized, it is clear from the data presented here that the timing, but not the order, of late events changes in a marked but predictable way during the progression from CIN1 to CIN3. Such variation has previously been reported for lesions caused by different papillomavirus types but is not usually apparent when lesions caused by the same papillomavirus type are compared (73).
The loss of the E2 ORF and the subsequent deregulation of E7 expression is considered a major factor in the development of cervical cancer (reviewed in reference 42). The expression of E6 and E7 from the p97 promoter increases in the absence of E2, which can act as a transcriptional repressor. Integration also leads to the loss of a negative regulatory element that is thought to limit the abundance of E6 and E7 mRNAs during normal productive infection (45). The loss of E4, which usually accompanies the disruption of E2, has not previously been implicated in cancer progression but may also be a predisposing factor. The E4 proteins of both HPV16 and HPV18 have recently been shown to induce G2 arrest in cervical epithelial cells (18, 65) and are thus incompatible with cell proliferation. It appears that the reduced expression of E4 seen in HSIL may be necessary for cell proliferation to extend into the upper epithelial layers.
A general feature of all DNA tumor viruses is their ability to stimulate quiescent cells to enter the cell cycle and to provide an environment that favors viral genome amplification (67). In cells that are nonpermissive for productive infection, the integration of viral DNA into the host cell chromosome can lead to cellular transformation. For most papillomaviruses, and indeed for DNA tumor viruses in general, natural infection usually leads to the formation of lesions that allow the production of new virus particles. Abortive infections result from the infection of a tissue or cell type in which the virus has not evolved to complete its productive life cycle successfully. The different papillomavirus types cause lesions at different epithelial sites, with the extent of productive infection reflecting a balance between the need to evade immune detection and the need to produce sufficient infectious progeny to allow the successful infection of a new host (73). As papillomavirus entry is mediated through a receptor that is present on many cell types (30, 37, 46, 82), viruses that have evolved to do well at a particular epithelial site can in principle gain access to inappropriate cell types, where their ability to complete their productive cycle may be compromised. Natural infection of cattle by bovine papillomavirus type 1 (BPV1) causes productive fibropapillomas, whereas in horses BPV1 infection leads to the formation of nonproductive equine sarcoids (66). Similarly, cottontail rabbit papillomavirus (CRPV) produces productive lesions in cottontail rabbits but causes nonproductive lesions in New Zealand White rabbits (14). Such lesions, which are in effect abortive CRPV infections, progress to cancer at a higher frequency than do the productive papillomas induced in cottontails (14). As many HPV16-induced cervical lesions are in fact abortive infections in which the full life cycle of the virus is not supported, it seems possible that the virus may have evolved to complete its productive cycle at a primary site other than the cervix. Although the extent to which HPV16 can undergo productive infection at other epithelial sites remains to be established, it would now be interesting, since HPV16 is a sexually transmitted virus, to examine the lesions it causes in men, as well as lesions at other sites in the female genital tract. In contrast to cancer of the cervix, penile cancer and cancers at other genital sites in women are relatively uncommon (7, 80).
Acknowledgments
Funding for this work was provided largely by the United Kingdom Medical Research Council (to K.M., H.G., N.C., and J.D.), the Association for International Cancer Research (to S.S.), and the Royal Society (to J.D.).
This report was written by J.D.
We thank John Skehel and Jonathan Stoye for advice and encouragement during the course of this work.
REFERENCES
- 1.Autier, P., M. Coibion, F. Huet, and A. R. Grivegnee. 1996. Transformation zone location and intraepithelial neoplasia of the cervix uteri. Br. J. Cancer 74:488-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badaracco, G., A. Venuti, A. Sedati, and M. L. Marcante. 2002. HPV16 and HPV18 in genital tumors: significantly different levels of viral integration and correlation to tumor invasiveness. J. Med. Virol. 67:574-582. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin, P., R. Laskey, and N. Coleman. 2003. Translational approaches to improving cervical screening. Nat. Rev. Cancer 3:217-226. [DOI] [PubMed] [Google Scholar]
- 4.Banks, L., P. Spence, E. Androphy, N. Hubbert, G. Matlashewski, A. Murray, and L. Crawford. 1987. Identification of human papillomavirus type 18 E6 polypeptide in cells derived from human cervical carcinomas. J. Gen. Virol. 68:1351-1359. [DOI] [PubMed] [Google Scholar]
- 5.Barrera-Oro, J. G., K. O. Smith, and J. L. Melnick. 1962. Quantitation of papova virus particles in human warts. J. Natl. Cancer Inst. 29:583-595. [PubMed] [Google Scholar]
- 6.Bhattacharya, D., A. Redkar, I. Mittra, U. Sutaria, and K. D. MacRae. 1997. Oestrogen increases S-phase fraction and oestrogen and progesterone receptors in human cervical cancer in vivo. Br. J. Cancer 75:554-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleeker, M. C., C. J. Hogewoning, A. J. Van Den Brule, F. J. Voorhorst, R. E. Van Andel, E. K. Risse, T. M. Starink, and C. J. Meijer. 2002. Penile lesions and human papillomavirus in male sexual partners of women with cervical intraepithelial neoplasia. J. Am. Acad. Dermatol. 47:351-357. [DOI] [PubMed] [Google Scholar]
- 8.Breitburd, F., O. Croissant, and G. Orth. 1987. Expression of human papillomavirus type-1 E4 gene products in warts, p. 115-122. In Cancer cells 5. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.Brisson, J., C. Morin, M. Fortier, M. Roy, C. Bouchard, J. Leclerc, A. Christen, C. Guimont, F. Penault, and A. Meisels. 1994. Risk factors for cervical intraepithelial neoplasia: differences between low- and high-grade lesions. Am. J. Epidemiol. 140:700-710. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y. H., L. H. Huang, and T. M. Chen. 1996. Differential effects of progestins and estrogens on long control regions of human papillomavirus types 16 and 18. Biochem. Biophys. Res. Commun. 224:651-659. [DOI] [PubMed] [Google Scholar]
- 11.Choo, K. B., C. C. Pan, and S. H. Han. 1987. Integration of human papillomavirus type 16 into cellular DNA of cervical carcinoma: preferential deletion of the E2 gene and invariable retention of the long control region and the E6/E7 open reading frames. Virology 161:259-261. [DOI] [PubMed] [Google Scholar]
- 12.Chow, L. T., M. Nasseri, S. M. Wolinsky, and T. R. Broker. 1987. Human papillomavirus types 6 and 11 mRNAs from genital condylomata acuminata. J. Virol. 61:2581-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow, L. T., S. S. Reilly, T. R. Broker, and L. B. Taichman. 1987. Identification and mapping of human papillomavirus type 1 RNA transcripts recovered from plantar warts and infected epithelial cell cultures. J. Virol. 61:1913-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen, N. D., R. Han, and J. W. Kreider. 2000. Cottontail rabbit papillomavirus (CRPV), p. 485-502. In R. Ahmed and I. Chen (ed.), Persistent viral infections. John Wiley & Sons Ltd., Sussex, England.
- 15.Crum, C. P., S. Barber, M. Symbula, K. Snyder, A. M. Saleh, and J. K. Roche. 1990. Coexpression of the human papillomavirus type 16 E4 and L1 open reading frames in early cervical neoplasia. Virology 178:238-246. [DOI] [PubMed] [Google Scholar]
- 16.Cullen, A. P., R. Reid, M. Campion, and A. T. Lorincs. 1991. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasms. J. Virol. 65:606-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies, R. J., A. Freeman, L. S. Morris, S. Bingham, S. Dilworth, I. Scott, R. A. Laskey, R. Miller, and N. Coleman. 2002. Analysis of minichromosome maintenance proteins as a novel method for detection of colorectal cancer in stool. Lancet 359:1917-1919. [DOI] [PubMed] [Google Scholar]
- 18.Davy, C. E., D. J. Jackson, Q. Wang, K. Raj, P. J. Masterson, N. F. Fenner, S. Southern, S. Cuthill, J. B. A. Millar, and J. Doorbar. 2002. Identification of a G2 arrest domain in the E1^E4 protein of human papillomavirus type 16. J. Virol. 76:9806-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeGregori, J., T. Kowalik, and J. R. Nevins. 1995. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol. Cell. Biol. 15:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dellas, A., E. Schultheiss, M. R. Leivas, et al. 1998. Association of p27Kip1, cyclin E and c-myc expression with progression and prognosis in HPV-positive cervical neoplasms. Anticancer Res. 18:3991-3998. [PubMed] [Google Scholar]
- 21.Demeter, L. M., M. H. Stoler, T. R. Broker, and L. T. Chow. 1994. Induction of proliferating cell nuclear antigen in differentiated keratinocytes of human papillomavirus-infected lesions. Hum. Pathol. 25:343-348. [DOI] [PubMed] [Google Scholar]
- 22.de Villiers, E. M. 1999. Human papillomavirus. Introduction. Semin. Cancer Biol. 9:377. [DOI] [PubMed] [Google Scholar]
- 23.de Villiers, E. M. 2001. Taxonomic classification of papillomaviruses. Papillomavirus Rep. 12:57-63. [Google Scholar]
- 24.Di Lonardo, A. D., M. L. Marcante, F. Poggiali, E. Hamsoikova, and A. Venuti. 2001. Egg yolk antibodies against the E7 oncogenic protein of human papillomavirus type 16. Arch. Virol. 146:117-125. [DOI] [PubMed] [Google Scholar]
- 25.Doorbar, J., D. Campbell, R. J. A. Grand, and P. H. Gallimore. 1986. Identification of the human papillomavirus-1a E4 gene products. EMBO J. 5:355-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doorbar, J., I. Coneron, and P. H. Gallimore. 1989. Sequence divergence yet conserved physical characteristics among the E4 proteins of cutaneous human papillomaviruses. Virology 172:51-62. [DOI] [PubMed] [Google Scholar]
- 27.Doorbar, J., C. Foo, N. Coleman, E. Medcalf, O. Hartley, T. Prospero, S. Napthine, J. Sterling, G. Winter, and H. Griffin. 1997. Characterisation of events during the late stages of HPV16 infection in vivo using high affinity synthetic Fabs to E4. Virology 238:40-52. [DOI] [PubMed] [Google Scholar]
- 28.Doorbar, J., and P. H. Gallimore. 1987. Identification of proteins encoded by the L1 and L2 open reading frames of human papillomavirus type 1a. J. Virol. 61:2793-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elson, D. A., R. R. Riley, A. Lacey, G. Thordarson, F. J. Talamantes, and J. M. Arbeit. 2000. Sensitivity of the cervical transformation zone to estrogen-induced squamous carcinogenesis. Cancer Res. 60:1267-1275. [PubMed] [Google Scholar]
- 30.Evander, M., I. H. Frazer, E. Payne, Y. Mei Qi, K. Hengst, and N. A. J. McMillan. 1997. Identification of the α6 integrin as a candidate receptor for papillomaviruses. J. Virol. 71:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans, M. F., S. L. Mount, B. G. Beatty, and K. Cooper. 2002. Biotinyl-tyramide-based in situ hybridization signal patterns distinguish human papillomavirus type and grade of cervical intraepithelial neoplasia. Mod. Pathol. 15:1339-1347. [DOI] [PubMed] [Google Scholar]
- 32.Ferlay, J., F. Bray, P. Pisani, and D. M. Parkin. 2001. GLOBOCAN 2000: cancer incidence, mortality and prevalence worldwide, version 1.0, vol. 5. IARC CancerBase. IARCPress, Lyon, France.
- 33.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, C. A. Sattler, and P. F. Lambert. 1999. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology 262:344-354. [DOI] [PubMed] [Google Scholar]
- 34.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74:6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freeman, A., L. S. Morris, A. D. Mills, K. Stoeber, R. A. Laskey, G. H. Williams, and N. Coleman. 1999. Minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin. Cancer Res. 5:2121-2132. [PubMed] [Google Scholar]
- 36.Giannoudis, A., M. Duin, P. J. Snijders, and C. S. Herrington. 2001. Variation in the E2-binding domain of HPV 16 is associated with high-grade squamous intraepithelial lesions of the cervix. Br. J. Cancer 84:1058-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giroglou, T., L. Florin, F. Schafer, R. E. Streeck, and M. Sapp. 2001. Human papillomavirus infection requires cell surface heparan sulfate. J. Virol. 75:1565-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grassmann, K., B. Rapp, H. Maschek, K. U. Petry, and T. Iftner. 1996. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J. Virol. 70:2339-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu, Z., D. Pim, S. Labrecque, L. Banks, and G. Matlashewski. 1994. DNA damage induced p53 mediated transcription is inhibited by human papillomavirus type 18 E6. Oncogene 9:629-633. [PubMed] [Google Scholar]
- 40.Hibma, M. H., K. Raj, S. J. Ely, M. Stanley, and L. Crawford. 1995. The interaction between human papillomavirus type 16 E1 and E2 proteins is blocked by an antibody to the N-terminal region of E2. Eur. J. Biochem. 229:517-525. [DOI] [PubMed] [Google Scholar]
- 41.Hiraiwa, A., T. Kiyono, S. Suzuki, M. Ohashi, and M. Ishibashi. 1996. E7 proteins of four groups of human papillomaviruses, irrespective of their tissue tropism or cancer association, possess the ability to transactivate transcriptional promoters E2F site dependently. Virus Genes 12:27-35. [DOI] [PubMed] [Google Scholar]
- 42.Howley, P. M., and D. R. Lowy. 2001. Papillomaviruses and their replication, p. 2197-2229. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 43.Hummel, M., H. B. Lim, and L. A. Laimins. 1995. Human papillomavirus type 31b late gene expression is regulated through protein kinase C-mediated changes in RNA processing. J. Virol. 69:3381-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeon, S., B. L. Allen-Hoffmann, and P. F. Lambert. 1995. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 69:2989-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeon, S., and P. F. Lambert. 1995. Integration of HPV16 DNA into the human genome leads to increased stability of E6/E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. USA 92:1654-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joyce, J. G., J. S. Tung, C. T. Przysiecki, J. C. Cook, E. D. Lehman, J. A. Sands, K. U. Jansen, and P. M. Keller. 1999. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J. Biol. Chem. 274:5810-5822. [DOI] [PubMed] [Google Scholar]
- 47.Kanda, T., S. Watanabe, S. Zanma, H. Sato, A. Furuno, and K. Yoshiike. 1991. Human papillomavirus type 16 E6 proteins with glycine substitution for cysteine in the metal-binding motif. Virology 185:536-543. [DOI] [PubMed] [Google Scholar]
- 48.Keating, J. T., A. Cviko, S. Riethdorf, L. Riethdorf, B. J. Quade, D. Sun, S. Duensing, E. E. Sheets, K. Munger, and C. P. Crum. 2001. Ki-67, cyclin E, and p16INK4 are complementary surrogate biomarkers for human papilloma virus-related cervical neoplasia. Am. J. Surg. Pathol. 25:884-891. [DOI] [PubMed] [Google Scholar]
- 49.Keating, J. T., T. Ince, and C. P. Crum. 2001. Surrogate biomarkers of HPV infection in cervical neoplasia screening and diagnosis. Adv. Anat. Pathol. 8:83-92. [DOI] [PubMed] [Google Scholar]
- 50.Kim, C. J., S. J. Um, T. Y. Kim, E. J. Kim, T. C. Park, S. J. Kim, S. E. Namkoong, and J. S. Park. 2002. Regulation of cell growth and HPV genes by exogenous estrogen in cervical cancer cells. Int. J. Gynecol. Cancer 10:157-164. [DOI] [PubMed] [Google Scholar]
- 51.Klaes, R., A. Benner, T. Friedrich, R. Ridder, S. Herrington, D. Jenkins, R. J. Kurman, D. Schmidt, M. Stoler, and M. von Knebel Doeberitz. 2002. p16INK4a immunohistochemistry improves interobserver agreement in the diagnosis of cervical intraepithelial neoplasia. Am. J. Surg. Pathol. 26:1389-1399. [DOI] [PubMed] [Google Scholar]
- 52.Klaes, R., T. Friedrich, D. Spitkovsky, R. Ridder, W. Rudy, U. Petry, G. Dallenbach-Hellweg, D. Schmidt, and M. von Knebel Doeberitz. 2001. Overexpression of p16INK4A as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int. J. Cancer 92:276-284. [DOI] [PubMed] [Google Scholar]
- 53.Lorincz, A. T., R. Reid, A. B. Jenson, M. D. Greenberg, W. D. Lancaster, and R. J. Kurman. 1992. Human papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types. Obstet. Gynecol. 79:328-337. [DOI] [PubMed] [Google Scholar]
- 54.Lowy, D. R., and P. M. Howley. 2001. Papillomaviruses, p. 2231-2264. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 55.Masterson, P. J., M. A. Stanley, A. P. Lewis, and M. A. Romanos. 1998. A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit. J. Virol. 72:7407-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathur, S. P., R. S. Mathur, P. F. Rust, and R. C. Young. 2001. Human papilloma virus (HPV)-E6/E7 and epidermal growth factor receptor (EGF-R) protein levels in cervical cancer and cervical intraepithelial neoplasia (CIN). Am. J. Reprod. Immunol. 46:280-287. [DOI] [PubMed] [Google Scholar]
- 57.McClean, C. S., M. J. Churcher, J. Meinke, G. L. Smith, G. Higgins, M. A. Stanley, and M. C. Minson. 1990. Production and characterisation of a monoclonal antibody to human papillomavirus type 16 using a recombinant vaccinia virus. J. Clin. Pathol. 43:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melillo, R. M., K. Helin, D. R. Lowy, and J. T. Schiller. 1994. Positive and negative regulation of cell proliferation by E2F-1: influence of protein level and human papillomavirus oncoproteins. Mol. Cell. Biol. 14:8241-8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michelin, D., L. Gissmann, D. Street, R. K. Potkul, S. Fisher, A. M. Kaufmann, L. Qiao, and C. Schreckenberger. 1997. Regulation of human papillomavirus type 18 in vivo: effects of estrogen and progesterone in transgenic mice. Gynecol. Oncol. 66:202-208. [DOI] [PubMed] [Google Scholar]
- 60.Mills, A. D., N. Coleman, L. S. Morris, and R. A. Laskey. 2000. Detection of S-phase cells in tissue sections by in situ DNA replication. Nat. Cell Biol. 2:244-245. [DOI] [PubMed] [Google Scholar]
- 61.Mitrani-Rosenbaum, S., R. Tsvieli, and R. Tur-Kaspa. 1989. Oestrogen stimulates differential transcription of human papillomavirus type 16 in SiHa cervical carcinoma cells. J. Gen. Virol. 70:2227-2232. [DOI] [PubMed] [Google Scholar]
- 62.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518-527. [DOI] [PubMed] [Google Scholar]
- 63.Musahal, C., H. P. Holthoff, R. Lesch, and R. Knippers. 1998. Stability of the replicative MCM3 protein in proliferating and differentiating human cells. Exp. Cell Res. 241:260-264. [DOI] [PubMed] [Google Scholar]
- 64.Myers, G. 1997. HPV and animal papillomavirus nucleotide sequences, p. I1-I9. In G. Myers, C. C. Baker, K. Munger, F. Sverdrup, A. McBride, and H.-U. Bernard (ed.), Human papillomaviruses, part 1. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 65.Nakahara, T., A. Nishimura, M. Tanaka, T. Ueno, A. Ishimoto, and H. Sakai. 2002. Modulation of the cell division cycle by human papillomavirus type 18 E4. J. Virol. 76:10914-10920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nasir, L., and S. W. Reid. 1999. Bovine papillomaviral gene expression in equine sarcoid tumours. Virus Res. 61:171-175. [DOI] [PubMed] [Google Scholar]
- 67.Nevins, J. R., and P. K. Vogt. 1996. Cell transformation by viruses, p. 301-343. In B. N. Fields, D. N. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 1. Lippincott-Raven, Philadelphia, Pa.
- 68.Noya, F., W.-M. Chien, T. R. Broker, and L. T. Chow. 2001. p21cip1 degradation in differentiated keratinocytes is abrogated by costabilization with cyclin E induced by human papillomavirus E7. J. Virol. 75:6121-6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohtani, K., R. Iwanaga, M. Nakamura, M. Ikeda, N. Yabuta, H. Tsuruga, and H. Nojima. 1999. Cell growth-regulated expression of mammalian MCM5 and MCM6 genes mediated by the transcription factor E2F. Oncogene 18:2299-2309. [DOI] [PubMed] [Google Scholar]
- 70.Ozbun, M. A., and C. Meyers. 1997. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J. Virol. 71:5161-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ozbun, M. A., and C. Meyers. 1998. Human papillomavirus type 31b E1 and E2 transcript expression correlates with vegetative viral genome amplification. Virology 248:218-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Padmanathan, A., M. Yadav, A. R. Gregory, S. Kumar, and A. W. Norhanum. 1997. Human papillomavirus DNA and virus-encoded antigens in cervical carcinoma. Med. J. Malaysia 52:108-116. [PubMed] [Google Scholar]
- 73.Peh, W. L., K. Middleton, N. Christensen, P. Nicholls, K. Egawa, K. Sotlar, J. Brandsma, A. Percival, J. Lewis, W. J. Liu, and J. Doorbar. 2002. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J. Virol. 76:10401-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peitsaro, P., B. Johansson, and S. Syrjanen. 2002. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J. Clin. Microbiol. 40:886-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ragu, G. C. 1994. Expression of the proliferating cell nuclear antigen in cervical neoplasia. Int. J. Gynecol. Pathol. 13:337-341. [DOI] [PubMed] [Google Scholar]
- 76.Resnick, M., S. Lester, J. E. Tate, E. E. Sheets, C. Sparks, and C. P. Crum. 1996. Viral and histopathologic correlates of MN and MIB-1 expression in cervical intraepithelial neoplasia. Hum. Pathol. 27:234-239. [DOI] [PubMed] [Google Scholar]
- 77.Richart, R. M. 1973. Cervical intraepithelial neoplasia. Pathol. Annu. 8:301-328. [PubMed] [Google Scholar]
- 78.Riethdorf, L., S. Riethdorf, K. R. Lee, A. Cviko, T. Loning, and C. P. Crum. 2002. Human papillomaviruses, expression of p16, and early endocervical glandular neoplasia. Hum. Pathol. 33:899-904. [DOI] [PubMed] [Google Scholar]
- 79.Rodins, K., M. Cheale, N. Coleman, and S. B. Fox. 2002. Minichromosome maintenance protein 2 expression in normal kidney and renal cell carcinomas: relationship to tumor dormancy and potential clinical utility. Clin. Cancer Res. 8:1075-1081. [PubMed] [Google Scholar]
- 80.Rubin, M. A., B. Kleter, M. Zhou, G. Ayala, A. L. Cubilla, W. G. Quint, and E. C. Pirog. 2001. Detection and typing of human papillomavirus DNA in penile carcinoma: evidence for multiple independent pathways of penile carcinogenesis. Am. J. Pathol. 159:1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sano, T., T. Oyama, K. Kashiwabara, T. Fukuda, and T. Nakajima. 1998. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am. J. Pathol. 153:1741-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Selinka, H. C., T. Giroglou, and M. Sapp. 2002. Analysis of the infectious entry pathway of human papillomavirus type 33 pseudovirions. Virology 299:279-287. [DOI] [PubMed] [Google Scholar]
- 83.Snijders, P. J. F., A. J. F. van den Brule, H. J. F. Schrijnemakers, G. Snow, C. J. L. M. Meijer, and J. M. M. Walboomers. 1990. The use of general primers in the polymerase chain reaction permits the detection of a broad spectrum of human papillomavirus genotypes. J. Gen. Virol. 71:173-181. [DOI] [PubMed] [Google Scholar]
- 84.Stevenson, M., L. C. Hudson, J. E. Burns, R. L. Stewart, M. Wells, and N. J. Maitland. 2000. Inverse relationship between the expression of the human papillomavirus type 16 transcription factor E2 and virus DNA copy number during the progression of cervical intraepithelial neoplasia. J. Gen. Virol. 81:1825-1832. [DOI] [PubMed] [Google Scholar]
- 85.Stoeber, K., R. Swinn, A. T. Prevost, P. de Clive-Lowe, I. Halsall, S. M. Dilworth, J. Marr, W. H. Turner, N. Bullock, A. Doble, C. N. Hales, and G. H. Williams. 2002. Diagnosis of genito-urinary tract cancer by detection of minichromosome maintenance 5 protein in urine sediments. J. Natl. Cancer Inst. 94:1071-1079. [DOI] [PubMed] [Google Scholar]
- 86.Tae Kim, Y., E. Kyoung Choi, N. Hoon Cho, et al. 2000. Expression of cyclin E and p27KIP1 in cervical carcinoma. Cancer Lett. 153:41-50. [DOI] [PubMed] [Google Scholar]
- 87.Tommasi, S., and G. P. Pfeifer. 1999. In vivo structure of two divergent promoters at the human PCNA locus. Synthesis of antisense RNA and S phase-dependent binding of E2F complexes in intron 1. J. Biol. Chem. 274:27829-27839. [DOI] [PubMed] [Google Scholar]
- 88.Tonon, S. A., M. A. Picconi, P. D. Bos, J. B. Zinovich, J. Galuppo, L. V. Alonio, and A. R. Teyssie. 2001. Physical status of the E2 human papilloma virus 16 viral gene in cervical preneoplastic and neoplastic lesions. J. Clin. Virol. 21:129-134. [DOI] [PubMed] [Google Scholar]
- 89.Vernon, S. D., E. R. Unger, D. L. Miller, D. R. Lee, and W. C. Reeves. 1997. Association of human papillomavirus type 16 integration in the E2 gene with poor survival from cervical cancer. Int. J. Cancer 74:50-56. [DOI] [PubMed] [Google Scholar]
- 90.Walboomers, J., M. Jacobs, M. M. Manos, F. Bosch, J. Kummer, K. Shah, P. Snijders, J. Peto, C. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 91.Williams, G. H., P. Romanowski, L. S. Morris, M. Madine, A. D. Mills, K. Stoeber, J. Marr, R. A. Laskey, and N. Coleman. 1998. Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc. Natl. Acad. Sci. USA 95:14932-14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yamaguchi, M., Y. Hayashi, and A. Matsukage. 1995. Essential role of E2F recognition sites in regulation of the proliferating cell nuclear antigen gene promoter during Drosophila development. J. Biol. Chem. 270:25159-25165. [DOI] [PubMed] [Google Scholar]
- 93.Yee, C., I. Krishnan-Hewlett, C. C. Baker, R. Schlegel, and P. M. Howley. 1985. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am. J. Pathol. 119:361-366. [PMC free article] [PubMed] [Google Scholar]
- 94.Zatsepina, O., J. Braspenning, D. Robberson, M. A. Hajibagheri, K. J. Blight, S. Ely, M. Hibma, D. Spitkovsky, M. Trendelenburg, L. Crawford, and M. Tommasino. 1997. The human papillomavirus type 16 E7 protein is associated with the nucleolus in mammalian and yeast cells. Oncogene 14:1137-1145. [DOI] [PubMed] [Google Scholar]