Abstract
We have investigated protein-protein interactions among the respiratory syncytial virus (RSV) RNA polymerase subunits using affinity chromatography. Here we demonstrate a novel interaction of P and M2-1 proteins. Phosphorylation of either M2-1 or P appears to be dispensable for this interaction. Internal deletions within P mapped the M2-1-binding domain to a region between residues 100 and 120. Alanine-scanning mutagenesis within this region of P revealed that substitution of any one of the three residues, L101, Y102, and F109, prevented both M2-1 and P binding and expression of an M2-1-dependent luciferase reporter gene. However, these same mutations did not prevent the activity of an M2-1-independent chloramphenicol acetyltransferase minigenome, suggesting that these residues of P specifically affect M2-1-P interaction. On the basis of these observations, it is possible that the interaction between RSV M2-1 and P proteins is important for viral replication.
Respiratory syncytial virus (RSV) is a nonsegmented negative-strand virus of the family Paramyxoviridae in the genus Pneumovirus (4). The viral genome is composed of 10 genes that are transcribed by the viral RNA polymerase complex (4). The activity of the RSV RNA-dependent RNA polymerase minimally requires the viral proteins N, P, and L, as well as signals in the viral RNA genome (17, 33). For RNA replication, the sequences in the leader and trailer regions function as the promoters for genome and antigenome replication, respectively (11). For mRNA synthesis, the gene-start and gene-end (GE) sequences which frame each RSV gene, in addition to the leader region, are essential (25). Viral transcription occurs sequentially along the genome in a stop-start manner characterized by attenuation at each GE sequence (4). An additional RSV protein, M2-1, is known to be important for transcription by the RSV polymerase, particularly on longer genes. M2-1 functions as an elongation factor, allowing the complete synthesis of RSV mRNAs (6) and as an antitermination factor to permit transit through GE and intergenic regions which allows the RSV polymerase access to promoter-distal genes (10, 18, 19).
The RSV P protein interacts with N (14) and, as shown for bovine RSV (bRSV), with the L protein (23). In both RSV-infected and cotransfected cells, M2-1 has been observed in inclusion bodies, but only in the presence of N and P proteins (14). This colocalization of M2-1 with N and P would be consistent with its function in transcription as part of the ribonucleoprotein (RNP) complex. In that study (14), an interaction between M2-1 and N was observed by coimmunoprecipitation (co-IP). Others have also observed an interaction between M2-1 and N (20), although this interaction is believed to be mediated by RNA (3). Yeast two-hybrid analysis of pairwise combinations of these three RSV proteins did not reveal interacting partners for M2-1 (21).
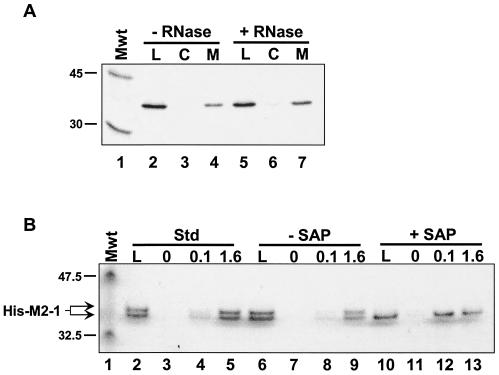
In an effort to identify protein-protein interactions between components of the RSV RNP, we investigated potential interactions between human RSV (hRSV) N, P, and M2-1 proteins by affinity chromatography. Since an interaction between N and P proteins has been extensively investigated, we focused on the M2-1 protein. We cloned, expressed, and purified M2-1 as a histidine-tagged fusion (His-M2-1). To do this, the M2-1 gene was isolated by reverse transcriptase-mediated PCR amplification from the hRSV A2 strain by standard techniques and cloned into pFastBacHT (Invitrogen) for recombination into baculovirus according to the supplier's protocol (details available upon request). His-M2-1 was expressed by infection of a suspension culture of Sf21 cells with a recombinant baculovirus (multiplicity of infection of 10). At 44 h postinfection, the cells were lysed by sonication. The cells were lysed in 5 volumes of extraction buffer (20 mM Tris Cl [pH 7.9], 500 mM NaCl, and 5 mM imidazole containing protease inhibitor cocktail [Sigma]) per gram [wet weight] of cells. Cell debris was removed, first, by centrifugation at 100,000 × g for 30 min (JA30.50Ti rotor in a Beckman J30I centrifuge). The supernatant was precipitated with 40% ammonium sulfate (centrifuged at 100,000 × g for 30 min). The resulting pellet was dissolved in one-fifth the original volume in extraction buffer, and the resulting solution was purified on a 5-ml metal-chelating Hi-Trap column (Amersham) charged with cobalt. The solution was then subjected to buffer exchange using a 10DG desalting column (Bio-Rad) and a second chromatography on a 1-ml cobalt-charged Hi-Trap column. The typical yield was approximately 10 to 15 mg per liter of cells. His-M2-1 purified in this manner ran as a doublet on sodium dodecyl sulfate (SDS)-polyacrylamide gels (see Fig. 3B, lane 2). M2-1 is a phosphoprotein (3, 8, 20, 26), and this doublet most likely represents different phosphorylation states. Indeed, phosphatase treatment eliminated the upper band of the doublet, which suggests that His-M2-1 was phosphorylated during expression in insect cells (see Fig. 3B).
FIG. 3.
The M2-1-P interaction is not mediated by RNA or M2-1 phosphorylation. (A) Immobilized His-M2-1 and IVT P proteins were both incubated either in the absence (− RNase) or presence (+ RNase) of a mixture of RNase A and RNase T1. Affinity chromatography was performed as described in the legend to Fig. 1. Lanes: L, 1/20 of the material loaded on each column; C, high-salt elutions from the control columns; M, salt elutions from the M2-1 columns. The positions of the 45-kDa (ovalbumin) and 30-kDa (carbonic anhydrase) from 14C-labeled rainbow molecular mass markers (Mwt) (Amersham) are indicated to the left of the gel. (B) His-M2-1 was diluted in affinity column buffer (Std, standard) (lanes 2 to 5) or 1× SAP (Promega) buffer (lanes 6 to 13). In the latter case, the protein was incubated either in the absence or presence of SAP, as indicated, prior to affinity chromatography on columns containing 0, 0.1, or 1.6 mg of immobilized GST-P per ml, as indicated above the gel. High-salt eluates were analyzed on a 4 to 12% NuPAGE (Invitrogen) gel run with 1× MES buffer, as suggested by the manufacturer. The gel was silver stained using the SilverSNAP kit (Pierce). The positions of the 47.5-kDa (aldolase) and 32.5-kDa (triose phosphate isomerase) proteins, which are prestained molecular mass markers (Mwt) (New England BioLabs), and the position of M2-1 are indicated to the left of the gel.
His-M2-1 was prepared for affinity chromatography by buffer exchange in immobilization buffer (20 mM morpholineethanesulfonic acid [MES] [pH 6.1], 500 mM NaCl). The protein was then covalently immobilized on Affi-Gel-10 (Bio-Rad) resin at 0.5 mg/ml (18.5 μM) according to the manufacturer's protocol. M2-1 affinity chromatography was performed with hRSV N and P proteins that were expressed and 35S labeled by in vitro translation (IVT), essentially as described elsewhere (S. W. Mason, L. Lagacé, C. Lawetz, F. Dô, Y. Gaudette, K. Jensen, E. Aberg, M.-J. Massariol, R. DeLong, L. Lamarre, P. Whitehead, and M. Liuzzi, submitted for publication). Lysates containing either N or P were diluted twofold with 2× affinity column buffer (ACB) (20 mM HEPES [pH 7], 0.1 mM EDTA, 10% glycerol [vol/vol], 1 mM dithiothreitol) applied to columns containing immobilized M2-1 or control columns containing no immobilized protein. Columns were washed with 1× ACB containing 100 mM NaCl followed by successive elutions with 1× ACB containing 1 M NaCl or 1% SDS. All fractions were run on SDS-12% polyacrylamide gels and quantified by PhosphorImager analysis (Molecular Dynamics Storm).
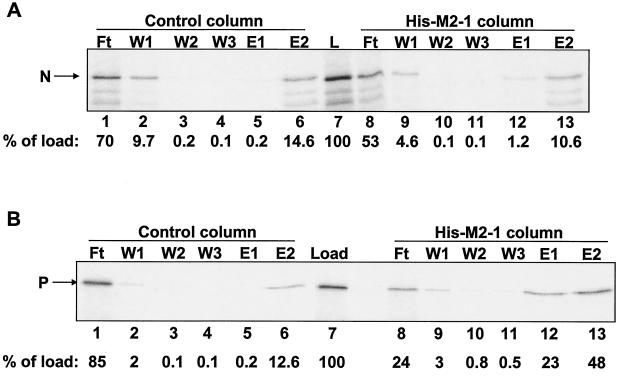
Chromatography of the N protein on immobilized M2-1 (Fig. 1A) revealed that only ∼1% of the N protein that was loaded onto the column eluted with a high level of salt (lane 12). Furthermore, comparable amounts of N protein were present in the SDS eluates from both the M2-1 and control columns (compare lanes 6 and 13). In contrast, almost one-quarter of the IVT P protein that was applied to the M2-1 column was retained and eluted with a high level of salt (Fig. 1B, lane 12). However, only about fourfold-more P protein was eluted with SDS from the M2-1 column than from the control column (Fig. 1B, cf. lanes 6 and 13). Therefore, the presence of P protein in the SDS eluate may represent a mix of specific and nonspecific interactions with immobilized ligand and matrix.
FIG. 1.
M2-1 affinity chromatography with IVT N or P protein. RSV N (A) and P (B) proteins were expressed and labeled with [35S]methionine by IVT. Columns (20 μl each) containing 18.5 μM immobilized M2-1 or control columns containing no immobilized protein were loaded with 20-μl IVT protein lysates. Columns were washed (three times with 40 μl) and eluted first with 40 μl of buffer containing 1 M NaCl (E1) and then with 40 μl of buffer containing 1% SDS (E2). Lanes: L, IVT protein lysate that was loaded onto each column (5 μl per lane); Ft, flowthrough fractions (5 μl per lane); W1 to W3, wash fractions (10 μl per lane). Ten microliters each of E1 and E2 was loaded in each lane. The positions of N and P are indicated to the left of each gel. Phosphorimages of the gels were quantified. The amount of N or P protein present in each fraction was normalized to the amount of the protein in the load sample (percentage of load), as indicated below each gel.
Due to the high background levels of P protein in SDS eluates from the control column, we analyzed only the high-salt eluates in all subsequent experiments. In comparing the results presented in panels A and B of Fig. 1, the amount of N protein that was specifically eluted from the M2-1 column appears to be much less than that observed for P protein. For example, only sixfold-more N eluted with a high level of salt from the M2-1 column than from the control column (Fig. 1A, cf. lanes 5 and 12) compared to 115-fold-more P present in the high-salt eluate from the M2-1 column than from the control column (Fig. 1B, cf. lanes 5 and 12). Therefore, under these conditions, RSV M2-1 binds to P protein but interacts rather weakly or not at all with N protein. As indicated in the literature (3), this N-M2-1 interaction is most likely mediated by RNA and therefore is not direct.
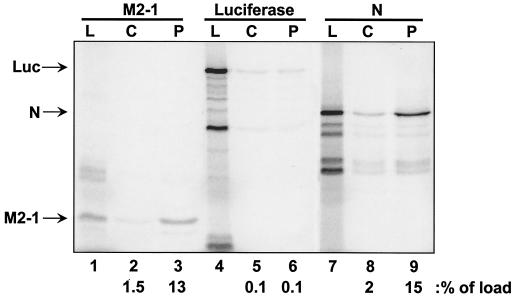
In order to substantiate an interaction between P and M2-1, the reciprocal affinity chromatography was performed with M2-1 and immobilized P protein (Fig. 2). Toward this end, P protein was expressed in Escherichia coli from the plasmid pET-3a-RSVS P (Long strain), kindly provided by S. Barik (University of South Alabama), and purified essentially as described previously (28). IVT lysates containing labeled M2-1, N, or luciferase were subjected to chromatography either on columns containing P protein covalently attached to Affi-Gel-15 resin (Bio-Rad) according to the manufacturer's protocol or on control columns containing no immobilized protein. As shown previously (15, 21, 23, 29), N bound to P protein (Fig. 2, lanes 7 to 9). Consistent with Fig. 1B, M2-1 also bound to immobilized P protein (lanes 1 to 3), but luciferase did not (lanes 4 to 6). Luciferase also did not bind to immobilized M2-1 (data not shown). Thus, the M2-1-P interaction is specific and not dependent upon which protein, either M2-1 or P, is immobilized.
FIG. 2.
P-affinity chromatography with IVT expressed M2-1, luciferase, and N proteins. Columns containing immobilized P protein or control columns containing no immobilized protein were loaded with IVT protein lysates containing either M2-1 (lanes 1 to 3), luciferase (lanes 4 to 6), or N (lanes 7 to 9) proteins. Lanes: L, load (5 μl per lane); C, high-salt eluate from control column; P, high-salt eluate from P columns (10 μl per lane). The positions of M2-1, luciferase (Luc), and N are indicated to the left of the gel. The amount of each protein present in the high-salt elution fractions from each column and normalized to the load fraction (100%) is indicated at the bottom of the figure (as a percentage of load).
Since M2-1 is an RNA-binding protein (3, 8), it was possible that the interaction between M2-1 and P was mediated through RNA. Therefore, both the immobilized M2-1 and the IVT lysate containing P protein were treated with a mixture of RNase A and T1. The amount of RNase used (2 units of RNase A and 2 μg of RNase T1) was sufficient to degrade 40 μg of tRNA by >95% under identical incubation conditions (data not shown). As shown in Fig. 3A, RNase treatment did not alter the binding of IVT P protein to immobilized M2-1.
M2-1 is a phosphoprotein (3, 8, 26), so we also examined the role of M2-1 phosphorylation in P binding. We treated M2-1 with phosphatase prior to chromatography on P columns. Purified His-M2-1 was diluted to 0.5 mg/ml in either 1× ACB or 1× shrimp alkaline phosphatase (SAP) buffer (50 mM Tris-HCl [pH 9], 10 mM MgCl2) (Promega) supplemented with 1 mg of bovine serum albumin per ml in each case. His-M2-1 in SAP buffer was incubated for 30 min at room temperature either in the absence or presence of 1 U of SAP. As shown in Fig. 3B, untreated His-M2-1 ran as a doublet (lanes 2 and 6), but treatment with SAP eliminated the higher-molecular-weight form of the protein (lane 10), which is consistent with phosphorylation of His-M2-1 in baculovirus-infected cells. These three solutions of His-M2-1 were each chromatographed on Affi-Gel-10 containing either no immobilized protein or glutathione S-transferase (GST) fused to the P protein (GST-P), purified as described elsewhere (Mason et al., submitted), which was immobilized at 0.1 or 1.6 mg/ml (1.8 or 29 μM, respectively). In all three cases, the His-M2-1 protein bound to the immobilized GST-P. It is possible that phosphorylation of M2-1 in insect cells is not necessarily on the same residues as identified previously, namely, Ser 58 and 61 (3). Either way, assuming that the SAP-treated His-M2-1 protein is completely devoid of phosphorylation, it appears that phosphorylation does not contribute to this interaction.
A caveat in examining protein-protein interactions with IVT lysates is that an apparent interaction may be mediated by one or more proteins from the rabbit reticulocyte lysate. However, since the experiment shown in Fig. 3B was performed with two purified pr oteins, we conclude that the interaction between M2-1 and P is direct and not mediated by any other proteins. It can also be concluded that phosphorylation of P is not necessary for this interaction, since the P protein used here and in Fig. 2 was produced in E. coli and, as such, is not phosphorylated.
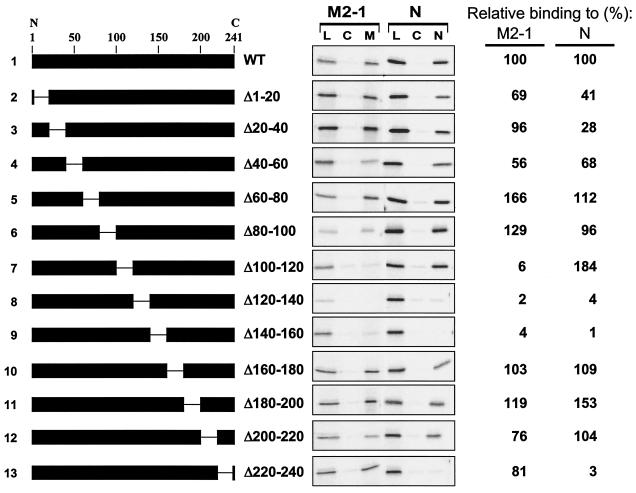
We mapped the M2-1-binding domain on the P protein using a series of in-frame internal deletions that are shown schematically in Fig. 4. Deletions of the P gene were performed using the QuikChange mutagenesis kit (Stratagene) according to the manufacturer's protocols (sequences of mutagenic oligonucleotides available on request). Each mutant P protein was expressed by IVT and tested for binding to immobilized M2-1. Only three mutant proteins with adjacent deletions (PΔ100-120, PΔ120-140, and PΔ140-160) were impaired in binding to M2-1 (Fig. 4, lines 7 to 9). However, when this same panel of P proteins was tested for binding to immobilized N protein, PΔ100-120 (line 7) was retained, but PΔ120-140 and PΔ140-160 (lines 8 and 9) were not. It is possible that the latter two deletion mutants have effects on the secondary or ternary structure of the P protein that prevent its interaction with both M2-1 and N proteins. The deletion of amino acids 100 to 120 in the P protein seems to specifically impair binding of P to M2-1 and does not affect the interaction between N and P. As expected (15, 21, 23, 29), the deletion of amino acid residues 220 to 240 eliminated binding of P to N; however, this mutation did not prevent the interaction between M2-1 and P (Fig. 4, line 13). Thus, P has two distinct domains for interacting with its binding partners: a C-terminal region for binding N and a central domain for binding M2-1.
FIG. 4.
Analysis of P deletion mutants by M2-1 and N-affinity chromatography. (Left) Schematic representations of P deletion mutants used to program IVT protein lysates loaded onto M2-1 and N columns. The numbers above line 1 give the positions of amino acid residues in the P protein (from the N to C terminus, as indicated). The gap in the black rectangle represents the region deleted in each protein. The amino acid residues that flank the deleted residues in each protein are indicated to the right of the schematic representation. (Center) M2-1 and N-affinity chromatography, as described in the legend to Fig. 1, except that each column was eluted only with a high level of salt (50 μl). Lanes: L, 1 μl of the load material that was applied to each column; C, high-salt eluate from control column (10 μl per lane); M and N, high-salt eluates from M2-1 and N column, respectively (10 μl per lane). (Right) The M2-1- and N-binding activity of each mutant, relative to the WT (set at 100%), as determined by quantification of the gels in the center panel, is shown.
The individual residues within this central region of the P protein were investigated for their contribution to the M2-1-P interaction. Single or double amino acid substitutions to alanine were made, expressed by IVT, and tested for binding to immobilized M2-1 protein (Table 1). Most of the substitutions did not have any significant effect on binding of P to M2-1 (e.g., PTI105/106AA or PNN111/112AA). For unknown reasons, some substitutions apparently stimulated binding (e.g., PKE103/104AA and PEE113/114AA). However, two double-amino-acid substitutions, PLY101/102AA and PTF108/109AA, greatly reduced binding of P to M2-1 (3 and 11% of wild-type [WT] P activity, respectively) but retained the ability to interact with N. To determine which of these four residues of the P protein, L101, Y102, T108, and F109, were important for the interaction, each amino acid was individually changed to alanine. As shown in Table 1, any one of the three amino acid substitutions L101A, Y102A, and F109A prevented binding of P to immobilized M2-1, whereas T108A affected binding only modestly. All four of these mutant proteins retained the ability to bind to N protein.
TABLE 1.
Effects of point mutations in P protein on binding to M2-1 and N proteins and minigenome activities
| Mutation in P protein | Bindinga to protein:
|
Minigenome activitya
|
||
|---|---|---|---|---|
| M2-1 | N | Luciferaseb | CATc | |
| None (WT) | 100 | 100 | 100 | 100 |
| Δ100-120 | 6 | 184 | <2 | ND |
| LY101/102AA | 3 | 126 | <2 | 142 |
| KE103/104AA | 263 | ND | 38 | 40 |
| TI105/106AA | 147 | ND | 109 | ND |
| E107A | 84 | ND | ND | ND |
| TF108/109AA | 11 | 165 | 25 | 32 |
| D110A | 58 | ND | ND | ND |
| NN111/112AA | 111 | ND | ND | ND |
| EE113/114AA | 207 | ND | 171 | ND |
| EE114/115AA | 45 | ND | 110 | ND |
| SS116/117AA | 102 | ND | 104 | 70 |
| YS118/119AA | 57 | 64 | <2 | 7 |
| S120A | 103 | 124 | ND | ND |
| EE121/122AA | 300 | 103 | 2 | 7 |
| L101A | 4 | 227 | <2 | 173 |
| Y102A | 4 | 144 | 3 | 223 |
| T108A | 77 | 91 | 60 | 64 |
| F109A | 3 | 105 | 4 | 97 |
| F241A | 125 | 4 | <2 | <2 |
| No P | NA | NA | <2 | <2 |
| No M2-1 | NA | NA | <2 | 100 |
All values are relative to WT P, which was set at 100%. ND, not determined; NA, not applicable.
Each luciferase minigenome activity value is the average of two to four separate assays, each performed in duplicate (with the exception of P Δ100-120 and with no M2-1 which were each tested only once in duplicate).
The CAT minigenome determinations were each performed twice. The raw data obtained in each of the two assays were similar. The data shown are from only one of the two experiments and the values shown were normalized for relative levels of N and P to account for the decreased levels of WT P (three- to fivefold) observed in Western blots with anti-RSV antibody (BioDesign) and K-109 (polyclonal anti-P). All CAT minigenome determinations were performed in the absence of the M2-1 plasmid.
Some of the same substitutions in the P protein were tested for effects on expression of minigenome reporters. Two separate minigenome reporter plasmids were constructed with the required RSV cis-acting sequences positioned exactly as described previously (7, 33). Reconstitution of RSV RNA polymerase activity in transfected cells would result in the expression of either a luciferase or chloramphenicol acetyltransferase (CAT) reporter gene. Constructs expressing RSV L, N, P, and M2-1 proteins and transfection procedures for minigenome assays will be described elsewhere (Mason et al., submitted). Detection of luciferase activity was performed using the Promega luciferase assay system per the manufacturer's protocol. The level of CAT activity was assessed with a CAT enzyme-linked immunosorbent assay kit, following the manufacturer's protocol (Roche). Expression of the luciferase gene in this system is absolutely dependent on the presence of M2-1, whereas the expression of the much shorter CAT gene does not require M2-1 (10). In fact, the M2-1 expression plasmid is not present in any experiment assessing CAT activity. As shown in Table 1, either double (PLY101/102AA) or single (PL101A, PY102A, and PF109A) amino acid substitutions that prevented P binding to M2-1 also reduced the luciferase minigenome activity to near background levels. However, these same mutations in P did not eliminate CAT minigenome activity. In particular, CAT levels were either equal to (PF109A) or as much as twofold greater (L101A and PY102A) than that produced by WT P. The reason for the elevated level of CAT expression with L101A and PY102A is not clear but may require further investigation. As a control, PF241A, which has been shown to prevent the N-P interaction (22), but not the M2-1-P interaction (Table 1), also prevented the reconstitution of RSV RNA-dependent RNA polymerase activity of both minigenome reporters. Thus, the mutations that prevent M2-1-P interaction impaired P protein function only in M2-1-dependent transcription by the RSV polymerase.
The results presented above demonstrate a previously uncharacterized interaction between two components of the RSV polymerase complex, the M2-1 and P proteins. Though both of these proteins on their own are essential for RSV replication (2, 5, 31, 33), that an interaction between them is important for RSV polymerase activity was not previously suggested. We have shown that single amino acid mutations of leucine 101, tyrosine 102, and phenylalanine 109 within the P protein each impaired the binding of P to M2-1 and also eliminated (L101A) or greatly decreased (Y102A and F109A) M2-1-dependent RSV polymerase activity in a minigenome system. Although effects on the secondary or ternary structure of P by these single amino acid mutations cannot be totally ruled out, the absence of effects of these same mutations on the N-P interaction and expression of the M2-1-independent CAT reporter would suggest that their overall structure is indeed intact. These observations are consistent with the interpretation that the M2-1-P interaction is important for viral replication, which is dependent on the complete transcription of all 10 RSV genes.
We have also found that deletion of residues 120 to 140 or 140 to 160 in the P protein impaired both N-P and M2-1-P interactions. The same deletions in bRSV P also prevented its interaction with L (23). Although we cannot rule out the possibility that this region of P also directly contributes to binding to M2-1, the multiple effects that deletions in the region from amino acids 120 to 160 appear to have on P activities could be mediated indirectly through changes in the oligomerization state of P. If oligomerization were required for interaction of P with each of its partners, M2-1, N, and L, then changes in P that alter its oligomerization state would be expected to influence proper folding of P and thus prevent these interactions. Consistent with this, it has been found that changing seven serine residues within this region (between amino acids 99 and 161) in hRSV P abolished P oligomerization (1).
Interestingly, we have also found that two of our P mutations (PYS118/119AA and PEE121/122AA) resulted in greatly reduced minigenome-directed luciferase and CAT activities, but were still able to interact with M2-1 and N. Since the region of bRSV P between amino acids 120 and 161 has been implicated in binding to L (23), we speculate that certain amino acid substitutions neighboring (e.g., PYS1118/119AA) and within (PEE121/122AA) this region may be influencing an interaction between the P and L proteins of hRSV without affecting oligomerization. This remains to be investigated.
Several attempts have been made in the past to detect M2-1-interacting partners among the RSV RNP proteins; mostly through co-IP (13) or yeast two-hybrid analysis (21). We have successfully used affinity chromatography to demonstrate a direct interaction between M2-1 and P. We have not been able to observe this interaction through co-IP (data not shown), but the inability to detect this interaction using co-IP could be due to weak or transient interactions. In addition, technical limitations potentially exist if the antibodies used for IP actually displace or compete with the interaction domain on either of the proteins. Affinity chromatography tends to be a much more sensitive approach and may detect interactions that are not sufficiently stable for co-IP (12). With covalent attachment of proteins on Affi-Gel resin, immobilization occurs at random lysine residues within the immobilized protein. Therefore, affinity chromatography should be free of epitope effects which may prevent co-IP.
Our demonstration of an interaction between P, a transcription factor for the RSV polymerase which is presumably involved in the initiation of transcription (9), and M2-1, an elongation or antitermination factor for RSV polymerase (10, 19), may not be surprising. Many protein-protein interactions mediate the assembly and function of transcription complexes in the cell (for example, the assembly of RNA polymerase II transcription initiation and elongation complexes). In addition, multiprotein complexes are known to be important in the regulation of viral (e.g., human immunodeficiency virus Tat [16, 24, 27]) and bacteriophage (e.g., lambda N-mediated antitermination [16, 32]) transcription elongation. A cascade of interactions, built upon the scaffold of N-encapsidated RNA which binds to P followed by binding of P to M2-1, may be important for delivery of M2-1 to the RSV transcription elongation complex. Though we have shown that the M2-1-P interaction is important for M2-1-dependent RSV polymerase function, the mechanism of M2-1-mediated transcription remains unclear. M2-1 most likely interacts with L protein to execute this function. Full elucidation of other potential interactions with M2-1 may be a significant step toward an understanding of the stop-start transcription that occurs on the RSV genome (18, 30).
Acknowledgments
We thank S. Barik (University of South Alabama) for the pET-3a-RSVS P (Long strain) plasmid and M.-J. Massariol and L. Lagacé for the CAT minigenome construct. We also thank Jacques Archambault and George Kukolj for comments on the manuscript.
REFERENCES
- 1.Asenjo, A., and N. Villanueva. 2000. Regulated but not constitutive human respiratory syncytial virus (HRSV) P protein phosphorylation is essential for oligomerization. FEBS Lett. 467:279-284. [DOI] [PubMed] [Google Scholar]
- 2.Bitko, V., and S. Barik. 2001. Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol. 1:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cartee, T. L., and G. W. Wertz. 2001. Respiratory syncytial virus M2-1 protein requires phosphorylation for efficient function and binds viral RNA during infection. J. Virol. 75:12188-12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1443-1485. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, New York, N.Y.
- 5.Collins, P. L., E. Camargo, and M. G. Hill. 1999. Support plasmids and support proteins required for recovery of recombinant respiratory syncytial virus. Virology 259:251-255. [DOI] [PubMed] [Google Scholar]
- 6.Collins, P. L., M. G. Hill, J. Cristina, and H. Grosfeld. 1996. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc. Natl. Acad. Sci. USA 93:81-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, P. L., M. A. Mink, and D. S. Stec. 1991. Rescue of synthetic analogs of respiratory syncytial virus genomic RNA and effect of truncations and mutations on the expression of a foreign reporter gene. Proc. Natl. Acad. Sci. USA 88:9663-9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuesta, I., X. Geng, A. Asenjo, and N. Villanueva. 2000. Structural phosphoprotein M2-1 of the human respiratory syncytial virus is an RNA binding protein. J. Virol. 74:9858-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupuy, L. C., S. Dobson, V. Bitko, and S. Barik. 1999. Casein kinase 2-mediated phosphorylation of respiratory syncytial virus phosphoprotein P is essential for the transcription elongation activity of the viral polymerase: phosphorylation by casein kinase 1 occurs mainly at Ser215 and is without effect. J. Virol. 73:8384-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fearns, R., and P. L. Collins. 1999. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 73:5852-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fearns, R., M. E. Peeples, and P. L. Collins. 2002. Mapping the transcription and replication promoters of respiratory syncytial virus. J. Virol. 76:1663-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Formosa, T., J. Barry, B. M. Alberts, and J. Greenblatt. 1991. Using protein affinity chromatography to probe structure of protein machines. Methods Enzymol. 208:24-45. [DOI] [PubMed] [Google Scholar]
- 13.Garcia, J., B. Garcia-Barreno, I. Martinez, and J. A. Melero. 1993. Mapping of monoclonal antibody epitopes of the human respiratory syncytial virus P protein. Virology 195:239-242. [DOI] [PubMed] [Google Scholar]
- 14.Garcia, J., B. Garcia-Barreno, A. Vivo, and J. A. Melero. 1993. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195:243-247. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Barreno, B., T. Delgado, and J. A. Melero. 1996. Identification of protein regions involved in the interaction of human respiratory syncytial virus phosphoprotein and nucleoprotein: significance for nucleocapsid assembly and formation of cytoplasmic inclusions. J. Virol. 70:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenblatt, J., J. R. Nodwell, and S. W. Mason. 1993. Transcriptional antitermination. Nature 364:401-406. [DOI] [PubMed] [Google Scholar]
- 17.Grosfeld, H., M. G. Hill, and P. L. Collins. 1995. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins: transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol. 69:5677-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, R. W., S. B. Harmon, and G. W. Wertz. 1999. Diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J. Virol. 73:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy, R. W., and G. W. Wertz. 1998. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J. Virol. 72:520-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardy, R. W., and G. W. Wertz. 2000. The Cys3-His1 motif of the respiratory syncytial virus M2-1 protein is essential for protein function. J. Virol. 74:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hengst, U., and P. Kiefer. 2000. Domains of human respiratory syncytial virus P protein essential for homodimerization and for binding to N and NS1 protein. Virus Genes 20:221-225. [DOI] [PubMed] [Google Scholar]
- 22.Khattar, S. K., A. S. Yunus, P. L. Collins, and S. K. Samal. 2001. Deletion and substitution analysis defines regions and residues within the phosphoprotein of bovine respiratory syncytial virus that affect transcription, RNA replication, and interaction with the nucleoprotein. Virology 285:253-269. [DOI] [PubMed] [Google Scholar]
- 23.Khattar, S. K., A. S. Yunus, and S. K. Samal. 2001. Mapping the domains on the phosphoprotein of bovine respiratory syncytial virus required for N-P and P-L interactions using a minigenome system. J. Gen. Virol. 82:775-779. [DOI] [PubMed] [Google Scholar]
- 24.Kobor, M. S., and J. Greenblatt. 2002. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577:261-275. [DOI] [PubMed] [Google Scholar]
- 25.Kuo, L., H. Grosfeld, J. Cristina, M. G. Hill, and P. L. Collins. 1996. Effects of mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J. Virol. 70:6892-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert, D. M., J. Hambor, M. Diebold, and B. Galinski. 1988. Kinetics of synthesis and phosphorylation of respiratory syncytial virus polypeptides. J. Gen. Virol. 69:313-323. [DOI] [PubMed] [Google Scholar]
- 27.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 28.Mazumder, B., G. Adhikary, and S. Barik. 1994. Bacterial expression of human respiratory syncytial viral phosphoprotein P and identification of Ser237 as the site of phosphorylation by cellular casein kinase II. Virology 205:93-103. [DOI] [PubMed] [Google Scholar]
- 29.Slack, M. S., and A. J. Easton. 1998. Characterization of the interaction of the human respiratory syncytial virus phosphoprotein and nucleocapsid protein using the two-hybrid system. Virus Res. 55:167-176. [DOI] [PubMed] [Google Scholar]
- 30.Sutherland, K. A., P. L. Collins, and M. E. Peeples. 2001. Synergistic effects of gene-end signal mutations and the M2-1 protein on transcription termination by respiratory syncytial virus. Virology 288:295-307. [DOI] [PubMed] [Google Scholar]
- 31.Tang, R. S., N. Nguyen, X. Cheng, and H. Jin. 2001. Requirement of cysteines and length of the human respiratory syncytial virus M2-1 protein for protein function and virus viability. J. Virol. 75:11328-11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weisberg, R. A., and M. E. Gottesman. 1999. Processive antitermination. J. Bacteriol. 181:359-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu, Q., R. W. Hardy, and G. W. Wertz. 1995. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J. Virol. 69:2412-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]