Abstract
Thirty different lymphocryptoviruses (LCV), 26 of them novel, were detected in primates by a panherpesvirus PCR assay. Nineteen LCV from chimpanzees, bonobos, gorillas, and other Old World primates were closely related to Epstein-Barr virus (EBV), the type species of the genus Lymphocryptovirus. Seven LCV originating from New World primates were related to callitrichine herpesvirus 3 (CalHV-3), the first recognized New World LCV. Importantly, a second LCV from gorillas and three LCV from orangutans and gibbons were only distantly related to EBV and CalHV-3. They were tentatively assigned to a novel genogroup of Old World primate LCV. The work described in the paper may also help identify an as yet unknown human LCV.
Alpha-, beta-, and gammaherpesviruses have been found in primates including humans. Old World primates, including great apes, and New World primates have been studied extensively and found to harbor several herpesvirus species, most of them gammaherpesviruses. Some viruses were found in animals suffering from tumors or nonneoplastic diseases, while other viruses were found in systematic investigations of healthy animals (1, 2, 6, 8, 16, 17, 28; reviewed in reference 30). The first gammaherpesvirus identified was Epstein-Barr virus (EBV) (11). It causes infectious mononucleosis and is associated with various tumors in humans (20). It was classified as the type species of the genus Lymphocryptovirus. A human virus member of the genus Rhadinovirus was discovered about 30 years later in AIDS-associated Kaposi's sarcoma. It was named Kaposi's sarcoma herpesvirus or human herpesvirus 8 (HHV-8) (3). Most of the lymphocryptoviruses (LCV) and rhadinoviruses detected in great apes and cercopithecids are closely related to either EBV or HHV-8. However, multiple rhadinoviruses have been found in chimpanzees, gorillas, macaques, and mandrills. They have either been assigned to the HHV-8 genogroup or to a new separate genogroup within the genus Rhadinovirus. This new genogroup was interpreted as an indirect indication of an additional human rhadinovirus (14, 16, 17, 27).
Evidence for LCV from Old World primates was first obtained by serological cross-reactivity to EBV, including LCV of chimpanzees (18), orangutans (23), gorillas (19), baboons (29), and diverse macaque species (12, 15, 22, 24). More recently, PCR-based methods have also been used to detect LCV from New World primates, one virus from the common marmoset (callitrichine herpesvirus 3 [CalHV-3]) (21) and one from the squirrel monkey (saimirine herpesvirus 3 [SaHV-3]) (5).
For initial genetic analyses of herpesviruses, a partial DNA polymerase (DPOL) gene sequence of a few hundred base pairs is generally amplified (9, 10, 14, 16, 17). However, despite the considerable number of recognized LCV, few LCV DPOL gene sequences were available in public databases at the beginning of this study. EBV, cercopithecine herpesvirus 15 (CeHV-15) (25), and CalHV-3 (26) have been completely sequenced (accession no. AY037858, AF091053, and AF091061, respectively). Two short partial DPOL gene sequences have also been published, one of baboon herpesvirus (CeHV-12; accession no. AF091051) (21) and the other of SaHV-3 (accession no. AF229063) (5). In addition, four almost identical DPOL gene sequences of a gorilla LCV have been deposited in the GenBank database (AF250883, AF250884, AF250885, AF290600; V. Lacoste et al., unpublished data). Consequently, this limited sequence data could only provide a fragmented picture of the genetic relationship of LCV.
To elucidate the phylogeny of LCV, we analyzed Old World primates including great apes and New World primates for the presence of LCV. Primate species which previously had not been reported to harbor herpesviruses or in which herpesviruses had been found but not genetically defined were investigated. For this purpose, blood and tissue samples were collected from primates housed in several German zoos, in the German Primate Center (Deutsches Primatenzentrum; Göttingen, Germany), and in private households. Blood and tissue samples were also collected from chimpanzees and red colobus monkeys living in the Taï National Park, Ivory Coast, and from gibbons in the Cuc-Phuong National Park, Vietnam. The tissue samples were collected following autopsy of animals which had suffered from lethal diseases, including those with tumors. In summary, 606 samples from more than 30 primate species were tested. The cell lines Austin, LCL278, and 594 (which are infected with chimpanzee, rhesus monkey, and baboon LCV, respectively [7] were also analyzed, in addition to two gorilla cell lines and several primate cell lines of unknown herpesvirus status (European Collection of Cell Cultures, Salisbury, United Kingdom).
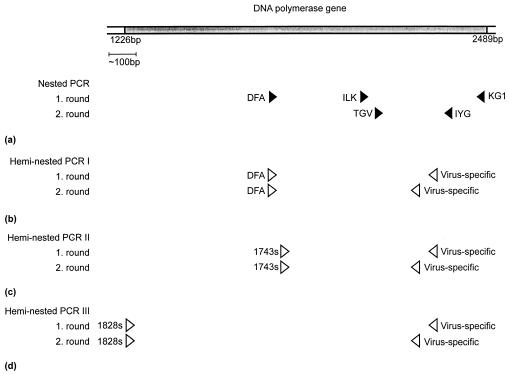
Panherpesvirus consensus PCR was carried out as nested PCR with degenerate and deoxyinosine-substituted primers (4, 9). In first-round PCR, two sense primers and one antisense primer (DFA, ILK, and KG1, respectively) were used. In second-round PCR, one sense and one antisense primer (TGV and IYG, respectively) were used (Fig. 1a). Amplimers of 166 to 175 bp (excluding primer binding sites) were obtained, the length depending on the herpesvirus species amplified. They were purified and sequenced as described previously (13). To extend the sequences in the upstream direction, heminested, hemispecific PCR was performed at an annealing temperature of 46, 50, or 55°C with the degenerate sense primer DFA and two antisense primers specific for each species (Fig. 1b). In cases of insufficient amplification, DFA was used in a 10-fold-higher concentration (31). Alternatively, instead of using DFA, heminested PCR was performed with either the CeHV-15-specific sense primer 1743s (5′-GTTATTCTACCATGATAACGCCGGGAGA-3′) or the EBV-specific sense primer 1828s (5′-GGGGCGTCTGCGAGGTCA-3′). Primer 1743s binds to a conserved region immediately downstream of DFA (Fig. 1c), whereas 1828s binds to a conserved region 0.55 kbp upstream of DFA (Fig. 1d). The final consensus sequences were 0.43 to 0.5 kbp in length.
FIG. 1.
Schematic diagram of the PCR strategy. (a) Initial panherpesvirus nested PCR with degenerate and deoxyinosine-substituted primers, used for amplification of novel herpesvirus DPOL gene sequences. (b) Subsequent heminested PCR with the degenerate sense primer DFA and two virus-specific antisense primers. (c and d) Alternative heminested PCR approaches with a pan-LCV sense primer (1743s or 1828s) and two virus-specific antisense primers. The partial DPOL gene of EBV is shown at the top for demonstration of the primer binding regions. The positions bp 1226 and 2489 refer to the 5′ ends of the binding regions of the primers 1828s and KG1, respectively. Solid arrowheads, degenerate primers; open arrowheads, specific primers. The virus-specific antisense primers have slightly different positions in each individual DPOL gene sequence, but they are always located between primers TGV and IYG.
DPOL gene sequences were detected in 343 blood and tissue samples and seven cell lines. Of these, 322 were more closely related to known LCV sequences than to those of any other herpesviruses. They were therefore regarded as LCV sequences and tentatively assigned to 30 different LCV species. The sequences from 28 samples aligned most closely to those of the DPOL genes of rhadinoviruses and cytomegaloviruses. These will be reported separately. All LCV were provisionally named and are listed in Table 1. Of the 30 LCV species detected, 28 LCV were found in more than one specimen, 20 LCV were found in more than one animal, and 13 LCV were found in animals from different locations. Three LCV were found in specimens from primates that had lived in the wild. Four sequences revealed 95 to 100% identity to DPOL gene sequences of known classified LCV (CeHV-12, CeHV-15, CalHV-3, and SaHV-3). These sequences were found in the same respective primate species and thus were assumed to originate from CeHV-12, CeHV-15, CalHV-3, and SaHV-3. The other 26 sequences were novel, most of them indicating the presence of previously unknown LCV species. Within a primate species, LCV DPOL gene sequences of less than 95% nucleic acid identity were taken to be derived from different LCV species, e.g., LCV1 and LCV2 of gorillas, baboons, mandrills, Japanese macaques, and squirrel monkeys. Sequences of higher identity were assigned to the same LCV species, e.g., LCV1 of rhesus macaques, cynomolgus macaques, wanderoos, and common marmosets (Table 1). For 27 LCV, sequences of more than 0.4 kbp were obtained. For three LCV (MfusLCV2, MtibLCV1, and EpatLCV1 [abbreviations are defined in Table 1]), sequence extension was unsuccessful.
TABLE 1.
Novel primate LCV
| Species | Tentative virus name | Abbre- viation | Accession no. | No. of animalsa/no. of locations (country) and/or cell line | % Identity, with most similar LCVb | % Identity to EBVc
|
|
|---|---|---|---|---|---|---|---|
| na | aa | ||||||
| Old World primates | |||||||
| Chimpanzee | Pan troglodytes LCV1 | PtroLCV1 | AF534226 | 4/2, 18/1 (Ivory Coast); Austin, EB176 | 97, PpanLCV1 | 92 | 93 |
| Bonobo | Pan paniscus LCV1 | PpanLCV1 | AF534220 | 2/1 | 97, PtroLCV1 | 93 | 93 |
| Gorilla | Gorilla gorilla LCV1 | GgorLCV1 | AF534225 | 5/2; EBJC | 100, Gorilla LCVe | 91 | 94 |
| Gorilla | Gorilla gorilla LCV2 | GgorLCV2 | AY129395 | 7/4 | 78, PpygLCV1 and GgorLCV1 | 78 | 82 |
| Orangutan | Pongo pygmaeus LCV1 | PpygLCV1 | AY129398 | 8/5 | 85, MfasLCV1 | 82 | 86 |
| White-cheeked gibbon | Hylobates leucogenys LCV1 | HleuLCV1 | AY174068 | 4/1 (Vietnam) | 82, HlarLCV1 | 73 | 83 |
| White-handed gibbon | Hylobates lar LCV1 | HlarLCV1 | AY196147 | 1/1 | 82, HleuLCV1 | 77 | 84 |
| Hanuman langur | Semnopithecus entellus LCV1 | SentLCV1 | AF534223 | 2/1 | 90, EBVe | 91 | 94 |
| Hamadryas baboon | Papio hamadryas LCV1 | PhamLCV1 | AY174069 | 594 | 100, CeHV12e | 92 | 95 |
| Hamadryas baboon | Papio hamadryas LCV2 | PhamLCV2 | AF534229 | 2/2 | 88, CeHV12 | 90 | 91 |
| Mandrill | Mandrillus sphinx LCV1 | MsphLCV1 | AF534227 | 2/1 | 92, MsphLCV2 | 90 | 91 |
| Mandrill | Mandrillus sphinx LCV2 | MsphLCV2 | AY174066 | 1/1 | 92, MsphLCV1 | 91 | 93 |
| Black and white colobus | Colobus guereza LCV1 | CgueLCV1 | AF534219 | 6/2 | 96, MfasLCV1 | 91 | 94 |
| Western red colobus | Piliocolobus badius LCV1 | PbadLCV1 | AF534228 | 2/1 (Ivory Coast) | 96, MfasLCV1 | 86 | 94 |
| Black mangabey | Cercocebus aterrimus LCV1 | CateLCV1 | AY174067 | 1/1 | 89, MfasLCV1 | 88 | 91 |
| Rhesus macaque | Macaca mulatta LCV1 | MmulLCV1d | AY172955 | 24/2; LCL278 | 98-100, CeHV15e | 91-92 | 91-93 |
| Cynomolgus macaque | Macaca fascicularis LCV1 | MfasLCV1d | AF534221 | 1/1 | 95-97, MmulLCV1 | 92-93 | 93-94 |
| Japanese macaque | Macaca fuscata LCV1 | MfusLCV1 | AF534224 | 2/1 | 98-99, MmulLCV1 | 91 | 93 |
| Japanese macaque | Macaca fuscata LCV2 | MfusLCV2 | AY172954 | 1/1 | 93-95, MmulLCV1 | 91 | 91 |
| Wanderoo | Macaca silenus LCV1 | MsilLCV1d | AF534222 | 5/2 | 96-99, MfasLCV1 | 90-92 | 93 |
| Magot | Macaca sylvanus LCV1 | MsylLCV1 | AY172956 | 1/1 | 88, MfasLCV1 | 82 | 91 |
| Tibet macaque | Macaca tibetana LCV1 | MtibLCV1 | AY174065 | 2/2 | 86, MfasLCV1 | 82 | 93 |
| Patas monkey | Erythrocebus patas LCV1 | EpatLCV1 | AY196148 | 1/1 | 91, MmulLCV1 | 90 | 96 |
| New World primates | |||||||
| Common squirrel monkey | Saimiri sciureus LCV1 | SsciLCV1 | AY172953 | 1/1 | 95, SaHV-3e | 67 | 68 |
| Common squirrel monkey | Saimiri sciureus LCV2 | SsciLCV2 | AY139024 | 3/3 | 69, SsciLCV1 | 69 | 70 |
| Saki | Pithecia pithecia LCV1 | PpitLCV1 | AY139025 | 3/2 | 72, CjacLCV1 | 64 | 68 |
| White-fronted capuchin | Cebus albifrons LCV1 | CalbLCV1 | AY139027 | 2/2 | 62, SsciLCV1 | 59 | 62 |
| Black spider monkey | Ateles paniscus LCV1 | ApanLCV1 | AY139028 | 1/1 | 69, PpitLCV1 | 65 | 72 |
| Black-pencilled marmoset | Callithrix penicillata LCV1 | CpenLCV1 | AY139026 | 2/1 | 96, CjacLCV1 | 68 | 68 |
| Common marmoset | Callithrix jacchus LCV1 | CjacLCV1d | AY174064 | 11/2 | 96-100, CalHV-3e | 67-68 | 67-68 |
The number of animals positive for the indicated LCV species in panherpesvirus consensus PCR. For GoLCV2, PpygLCV1, HlarLCV1, and HleuLCV1, the data are from consensus PCR and specific PCR.
Determined from pairwise alignments of each novel DPOL gene sequence (166 to 175 bp) with the DPOL gene sequences of known LCV and with the novel DPOL gene sequences determined in this study.
Determined from pairwise alignments of 166 to 175 bp (left column) and 53 to 58 amino acids (right column).
In Macaca mulatta, Macaca fascicularis, Macaca silenus, and Callithrix jacchus, sequences of 96 to 100% nucleic acid (na) identity were found, respectively. They were assigned to the same virus species (MmulLCV1, MfasLCV1, MsilLCV1, and CjacLCV1, respectively).
Accession number is given in the text.
In pairwise nucleic acid and amino acid sequence comparisons, 19 of the 23 viruses detected in Old World primates were more than 90% identical to EBV (Table 1). Moreover, phylogenetic analysis (see below) revealed a clade with EBV (Fig. 2). On the other hand, the seven viruses detected in New World primates were related to CalHV-3. For the first time this indicates unequivocally that New World primate LCV form a group which is clearly distinct from Old World primate LCV (Table 1; Fig. 2).
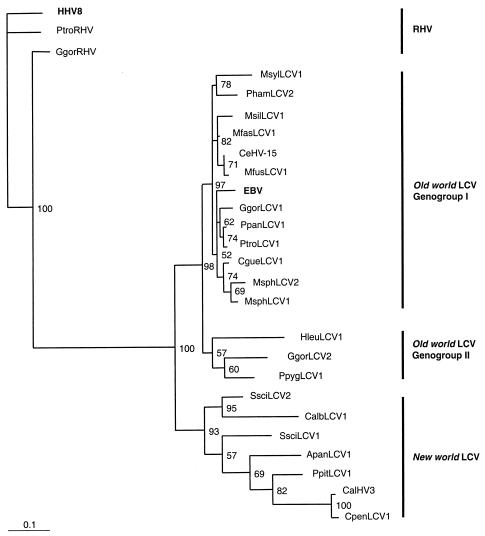
FIG. 2.
Phylogenetic analysis of novel LCV. A maximum-likelihood tree was constructed for the novel DPOL gene sequences listed in Table 1 and DPOL gene sequences of LCV available in GenBank (accession numbers are given in the text). Primate rhadinoviruses (RHV) were also included (PtroRHV, Pan troglodytes RHV; accession no. AF250879; GgorRHV, Gorilla gorilla RHV; accession no. Af250886). Sequences of 435 bp were aligned with ClustalW. The multiple alignment (with gaps removed) was analyzed with the TREE-PUZZLE, version 5.0, program. A rooted phylogram is shown, with HHV-8 as the outgroup. Support values >50% are indicated at the nodes of the tree. RHV, Old World primate LCV (genogroups I and II), and New World primate LCV are indicated. LCV virus abbreviations are defined in Table 1.
One New World primate species (squirrel monkeys) and four Old World primate species (gorillas, mandrills, baboons, and Japanese macaques) appeared each to be infected with two different LCV. In the last three primates, nucleic acid sequence comparisons of the LCV pairs (MsphLCV1-MsphLCV2,PhamLCV1-PhamLCV2, and MfusLCV1-MfusLCV2) re-vealed pair identities of 88 to 92%. All six viruses had 90 to 91% identity to EBV. In the gorilla, the situation was clearly different. GgorLCV1 and GgorLCV2 were only 78% identical to each other. Furthermore, GgorLCV1 had a 91% nucleic acid and 94% amino acid sequence identity to EBV but GgorLCV2 was much more distantly related. It had only 78% nucleic acid and 82% amino acid sequence identity to EBV. Similar genetic distances to EBV was found for the LCV of orangutans (PpygLCV1) and gibbons (HleuLCV1 and HlarLCV1). These viruses showed 73 to 82% nucleic acid identity and 82 to 86% amino acid identity to EBV (Table 1). Furthermore, in the partial DPOL gene sequences of GgorLCV2, HleuLCV1, and HlarLCV1, a base triplet is missing, a unique observation among the Old World primate LCV. Phylogenetic trees were constructed by the maximum-likelihood method and the neighbor-joining method with TREE-PUZZLE, version 5.0, and PHYLIP, version 3.6, software and sequence alignments of 435 bp or 145 amino acids (aa). In all trees, GgorLCV2, PpygLCV1, and HleuLCV1 branched separately from EBV and all other Old World primate LCV. Figure 2 shows a representative tree, in which GgorLCV2, PpygLCV1, and HleuLCV1 form a separate clade. Trees with all LCV sequences including those of 175 bp and 58 aa showed a very similar topology. However, the limited lengths of the latter sequences resulted in lower probability values (not shown).
GgorLCV2, PpygLCV1, HleuLCV1, and HlarLCV1 were detected with specific PCR solely in their species of origin. This included detection of GgorLCV2, PpygLCV1, and HlarLCV1 in animals from various zoological gardens and detection of HleuLCV1 in gibbons living in the wild (Table 1). These viruses were therefore regarded as genuine gorilla, orangutan, and gibbon LCV, respectively.
Based on the results of pairwise nucleotide and amino acid sequence comparisons (Table 1) and phylogenetic analyses (Fig. 2), we put forward the hypothesis that GgorLCV2, PpygLCV1, HleuLCV1, and HlarLCV1 are members of a second, new genogroup of Old World primate LCV (genogroup II; Fig. 2). So far, this new genogroup II comprises LCV of great apes (gorillas and orangutans) and lesser apes (gibbons), while genogroup I is made up of EBV (humans) and LCV of great apes (chimpanzees and gorillas) and several nonhominid Old World primates. Further studies will show whether, in addition to gorillas, other Old World primates harbor LCV of both genogroups I and II.
Since great apes are closest to humans in the context of evolution, the novel genogroup raises the question of whether a second human LCV exists. EBV was discovered about 40 years ago and is now the only known human member of the genus Lymphocryptovirus. However, three HHVs (HHV-6, HHV-7, and HHV-8) were discovered quite recently. Therefore it is possible that an as yet unknown human LCV may exist. Possible reasons for difficulties in detection include nonpathogenicity, low prevalence, serological non-cross-reactivity with EBV, and insufficient amplifiability with universal PCR-based methods (like the panherpesvirus PCR used in this study). On the other hand, this putative human LCV may have already been extinguished during evolution.
In summary, this study is the first comprehensive search for LCV in primates. It describes a large number of new LCV and allows the first detailed insight into their genetic relationships. The characterization of the complete genomes of EBV and CeHV-15 on the one hand and of CalHV-3 on the other has revealed considerable differences in the repertoires of LCV-specific genes (25, 26). More detailed analysis of the great ape LCV of genogroup II and of the New World monkey LCV, which are only distantly related to CalHV-3, will allow a better understanding of these interviral relationships. Furthermore, the availability of LCV sequences from 30 different monkey species may provide a solid basis for diagnosis of LCV-induced diseases in primates. Ultimately, the sequences of the genogroup II viruses may be an exciting lead in the search for an additional human LCV.
ADDENDUM IN PROOF
After acceptance of the manuscript, de Thoisy et al. (B. de Thoisy, J.-F. Pouliquen, V. Lacoste, A. Gessain, and M. Kazanji, J. Virol., 77:9099-9105, 2003) reported three novel LCV species of New World monkeys. Two of them (named SscLCV1 and PpiLCV1) are nearly identical to SsciLCV2 and PpitLCV1, respectively, at the nucleic acid level.
Acknowledgments
We thank Tatjana Franz and Siegfried Pociuli for excellent technical assistance, Stewart Lowden for helpful discussions, and Ursula Erikli for copy editing the manuscript. The supply of cell lines and primate samples by Andrew Bell, Thomas Haaf, Tilo Nadler, Werner Schempp, and Sabine Burckhardt is kindly acknowledged.
REFERENCES
- 1.Ablashi, D. V., J. M. Easton, and J. H. Guegan. 1976. Herpesviruses and cancer in man and subhuman primates. Biomedicine 24:286-305. [PubMed] [Google Scholar]
- 2.Barahona, H., L. V. Melendez, and J. L. Melnick. 1974. A compendium of herpesviruses from non-human primates. Intervirology 3:175-192. [DOI] [PubMed] [Google Scholar]
- 3.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 4.Chmielewicz, B., M. Goltz, and B. Ehlers. 2001. Detection and multigenic characterization of a novel gammaherpesvirus in goats. Virus Res. 75:87-94. [DOI] [PubMed] [Google Scholar]
- 5.Cho, Y., J. Ramer, P. Rivailler, C. Quink, R. L. Garber, D. R. Beier, and F. Wang. 2001. An Epstein-Barr-related herpesvirus from marmoset lymphomas. Proc. Natl. Acad. Sci. USA 98:1224-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damiana, B., and R. C. Desrosiers. 2001. Simian homologues of human herpesvirus 8. Phil. Trans. R. Soc. Lond. B. Biol. Sci. 356:535-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dillner, J., H. Rabin, N. Letvin, W. Henle, G. Henle, and G. Klein. 1987. Nuclear DNA-binding proteins determined by the Epstein-Barr virus-related simian-lymphotropic herpesviruses H. gorilla, H. pan, H. pongo and H. papio. J. Gen. Virol. 68:1587-1596. [DOI] [PubMed] [Google Scholar]
- 8.Eberle, R., and J. Hilliard. 1995. The simian herpesviruses. Infect. Agents Dis. 4:55-70. [PubMed] [Google Scholar]
- 9.Ehlers, B., K. Borchers, C. Grund, K. Frölich, H. Ludwig, and H.-J. Buhk. 1999. Detection of new DNA polymerase genes of known and potentially novel herpesviruses by PCR with degenerate and deoxyinosine-substituted primers. Virus Genes 18:211-220. [DOI] [PubMed] [Google Scholar]
- 10.Ehlers, B., S. Ulrich, and M. Goltz. 1999. Detection of two novel porcine herpesviruses with high similarity to gammaherpesviruses. J. Gen. Virol. 80:971-978. [DOI] [PubMed] [Google Scholar]
- 11.Epstein, M. A., B. G. Achong, and Y. M. Barr. 1964. Virus particles in cultured lymphoblasts from Burkitt′s lymphoma. Lancet i:702-703. [DOI] [PubMed] [Google Scholar]
- 12.Fujimoto, K., K. Terato, J. Miyamoto, H. Ishiko, M. Fujisaki, F. Cho, and S. Honjo. 1990. Establishment of a B-lymphoblastoid cell line infected with Epstein-Barr-related virus from a cynomolgus monkey (Macaca fascicularis). J. Med. Primatol. 19:21-30. [PubMed] [Google Scholar]
- 13.Goltz, M., T. Ericsson, C. Patience, C. A. Huang, S. Noack, D. H. Sachs, and B. Ehlers. 2002. Sequence analysis of the genome of porcine lymphotropic herpesvirus 1 and gene expression during post-transplant lymphoproliferative disease of pigs. Virology 294:383-393. [DOI] [PubMed] [Google Scholar]
- 14.Greensill, J., J. A. Sheldon, N. M. Renwick, B. E. Beer, S. Norley, J. Goudsmit, and T. F. Schulz. 2000. Two distinct gamma-2 herpesviruses in African green monkeys: a second gamma-2 herpesvirus lineage among Old World primates? J. Virol. 74:1572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi, K., H. L. Chen, H. Yanai, T. R. Koirala, N. Ohara, N. Teramoto, T. Oka, T. Yoshino, K. Takahashi, K. Miyamoto, K. Fujimoto, Y. Yoshikawa, and T. Akagi. 1999. Cyno-EBV (EBV-related herpesvirus from cynomolgus macaques) induces rabbit malignant lymphomas and their tumor cell lines frequently show specific chromosomal abnormalities. Lab. Investig. 79:823-835. [PubMed] [Google Scholar]
- 16.Lacoste, V., P. Mauclere, G. Dubreuil, J. Lewis, M. C. Georges-Courbot, and A. Gessain. 2000. KSHV-like herpesviruses in chimps and gorillas. Nature 407:151-152. [DOI] [PubMed] [Google Scholar]
- 17.Lacoste, V., P. Mauclere, G. Dubreuil, J. Lewis, M. C. Georges-Courbot, J. Rigoulet, T. Petit, and A. Gessain. 2000. Simian homologues of human gamma-2 and betaherpesviruses in mandrill and drill monkeys. J. Virol. 74:11993-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landon, J. C., L. B. Ellis, V. H. Zeve, and D. P. Fabrizio. 1980. Herpes-type virus in cultured leukocytes from chimpanzees. J. Natl. Cancer Inst. 40:181-192. [PubMed] [Google Scholar]
- 19.Neubauer, R. H., H. Rabin, B. C. Strand, M. Nonoyama, and W. A. Nelson-Rees. 1979. Establishment of a lymphoblastoid cell line and isolation of an Epstein-Barr-related virus of gorilla origin. J. Virol. 31:845-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niedobitek, G., G. Meru, and H. J. Delecluse. 2001. Epstein-Barr virus infection and human malignancies. Int. J. Exp. Pathol. 82:149-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramer, J. C., R. L. Garber, K. E. Steele, J. F. Boyson, C. O'Rourke, and J. A. Thomson. 2000. Fatal lymphoproliferative disease associated with a novel gammaherpesvirus in a captive population of common marmosets. Comp. Med. 50:59-68. [PubMed] [Google Scholar]
- 22.Rangan, S. R., L. N. Martin, B. E. Bozelka, N. Wang, and B. J. Gormus. 1986. Epstein-Barr virus-related herpesvirus from a rhesus monkey (Macaca mulatta) with malignant lymphoma. Int. J. Cancer 38:425-432. [DOI] [PubMed] [Google Scholar]
- 23.Rasheed, S., R. W. Rongey, J. Bruszweski, W. A. Nelson-Rees, H. Rabin, R. H. Neubauer, G. Esra, and M. B. Gardner. 1977. Establishment of a cell line with associated Epstein-Barr-like virus from a leukemic orangutan. Science 198:407-409. [DOI] [PubMed] [Google Scholar]
- 24.Rivadeneira, E. D., M. G. Ferrari, R. F. Jarrett, A. A. Armstrong, P. Markham, T. Birkebak, S. Takemoto, C. Johnson-Delaney, J. Pecon-Slattery, E. A. Clark, and G. Franchini. 1999. A novel Epstein-Barr-virus-like virus, HVMNE, in a Macaca nemestrina with mycosis fungoides. Blood 94:2090-2101. [PubMed] [Google Scholar]
- 25.Rivailler, P., H. Jiang, Y. G. Cho, C. Quink, and F. Wang. 2002. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J. Virol. 76:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivailler, P., Y. G. Cho, and F. Wang. 2002. Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a new world primate: a defining point in the evolution of oncogenic lymphocryptoviruses. J. Virol. 76:12055-12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz, E. R., G. W. Rankin, Jr., M. P. Blanc, B. W. Raden, C. C. Tsai, and T. M. Rose. 2000. Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:4919-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strand, K., E. Harper, S. Thormahlen, M. E. Thouless, C. C. Tsai, T. Rose, and M. L. Bosch. 2000. Two distinct lineages of macaque γ herpesviruses related to the Kaposi′s sarcoma associated herpesvirus. J. Clin. Virol. 16:253-269. [DOI] [PubMed] [Google Scholar]
- 29.Vasiljeva, V. A., D. S. Markarjan, B. A. Lapin, L. A. Yakovleva, M. T. Ivanov, V. F. Schekolodkin, and E. K. Dzikidze. 1974. Establishment of continuous cell lines from leukocytes: culture of a hamadryas baboon with leukosis-reticulosis. Neoplasma 21:537-544. [PubMed] [Google Scholar]
- 30.Wang, F., P. Rivailler, P. Rao, and Y. Cho. 2001. Simian homologues of Epstein-Barr virus. Phil. Trans. R. Soc. Lond. B. Biol. Sci. 356:489-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, X., and J. E. Marchand. 2002. Optimal ratio of degenerate primer pairs improves specificity and sensitivity of PCR. BioTechniques 32:1002-1006. [DOI] [PubMed] [Google Scholar]