Abstract
The N-terminal exon of DNA tumor virus T antigens represents a J domain that can direct interaction with the host-encoded Hsp70 chaperones. We have taken advantage of rapid Hsp40 cochaperone assays with Escherichia coli to assess simian virus 40 (SV40)-encoded J-domain loss of function. We found a strong correlation between loss of cochaperone function in E. coli and defective SV40 growth, suggesting that the major role of the J domain in DNA tumor viruses is to provide cochaperone function. We also report the expression of native SV40 virus T antigens in E. coli. Our results show that small t antigen, but not large T antigen (LT) or LT truncation TN125 or TN136, can functionally replace under limited growth conditions DnaJ (Hsp40) function in vivo. In addition, purified small t antigen can efficiently stimulate E. coli DnaK's (Hsp70) ATPase in vitro, thus behaving like a bona fide cochaperone. Furthermore, small t amino acids 83 to 174, which are adjacent to the viral J domain, can replace the E. coli DnaJ J-domain glycine-phenylalanine-rich domain, immediately adjacent to the J-domain sequences, even in the absence of significant amino acid similarity to their DnaJ counterpart. Taken together, our studies demonstrate that functionally related Hsp40 proteins from mammalian viral systems can be rapidly studied in bacteria and exploited to probe the universally conserved Hsp70 chaperone machine mechanism.
The ability of simian virus 40 (SV40) to regulate many aspects of the cell cycle has made it an invaluable tool for probing fundamental biological questions and the molecular origins of human cancer (13). Viral early gene products that include large T antigen (LT), small t antigen (smt), and 17k antigen are coordinately affected by N-terminal exon mutations because this exon is common to all mature spliced early genes (27, 39). Mutations in the N-terminal region of SV40 T antigens have been described that alter viral DNA replication, capsid morphogenesis, cellular transformation, immortalization, transcriptional activation, sensitization to apoptosis, and modulation of growth control signaling pathways (2, 5, 18, 32, 39). The multifunctional viral T antigens and their interactions with host proteins have been the subject of extensive recent reviews (1, 29, 39).
Of particular importance to viral replication is advancing the cell cycle. Viral infection of nonpermissive cells may lead to cellular transformation because of the targeting and sustained inactivation of key regulatory proteins, including the tumor suppressor retinoblastoma (RB) family and p53. SV40 LT specifically interacts with p53, and the retinoblastoma family members pRB, p130, and p107 (39). smt enhances transformation in many cell types, particularly those that are growth arrested (39). smt specifically interacts with protein phosphatase 2A and attenuates its activity, thereby aiding the progression of S phase (43). smt alone can also transactivate the cyclin A and adenovirus E2 promoters independent of smt-dependent inactivation of protein phosphatase 2A (28).
It is now recognized that the N-terminal exon common to SV40 viral early gene products is a J domain that can direct interaction with Hsp70 family members (4, 16, 34). The approximately 70-amino-acid residue-long J domain is the defining signature sequence of the Hsp40 family of molecular chaperones that function via protein-protein interaction to coordinate ATP hydrolysis and substrate selection of the Hsp70 chaperone machine (17, 21). The presence of a J domain in each of the viral early gene products raises the question of whether well-known Hsp70 chaperone machine activities, such as protein refolding, oligomeric state remodeling, or accelerated protein turnover and degradation, are governed by viral early proteins (14). Strong biochemical evidence exists showing that SV40 LT, together with Hsc70, can orchestrate the disassembly of RB family tumor suppressors from the E2F transcription factors and subsequent activation of E2F-dependent genes (35-38, 44).
Up to now, there has been no systematic effort to examine the phenotypes of viral J-domain mutants for cochaperone loss of function. A key question that remains unanswered is whether the large collection of published mutations that map in the SV40 N-terminal domain constitutes loss of function because of impaired J-domain cochaperone activity or whether additional biochemical activities are present in this region. A strong correlation would suggest that the major function of the viral J domain is to uniquely direct interaction with the cognate Hsp70 chaperone of the infected host.
In the present study, we tested a large collection of SV40 J-domain mutants for cochaperone function using our previously established rapid bacterial assays for J domains (16). Our assay exploited l-arabinose-inducible Escherichia coli DnaJ (Hsp40) expression vectors that had been engineered to produce chimeric DnaJ molecules to monitor bacterial and bacteriophage λ growth. In this system, the SV40 J domain can functionally replace the DnaJ J domain in vivo (16). The conditional expression of chimeric DnaJ molecules permits the rapid testing of exogenous J-domain activity. In those cases where multiple viral J-domain mutations gave rise to a particular phenotype, each single or double mutation was engineered and their contributions to cochaperone function were assessed. In parallel, we also examined the intrinsic chaperone activities of viral T antigens expressed in E. coli.
Table 1 summarizes some of the reported phenotypes of nine SV40 J-domain mutants that we examined in detail. The mutants span most of the viral J domain and can be subdivided into three classes. The majority of mutants have been described for their effects arising from the combined LT and smt expression. Two mutants were included that were identified and tested for their effects on smt functional activity alone (P43L/K45N and D44E/G47R). One mutant was included that arose from a virus that could produce only LT and not smt (20). Most mutants have been described as recessive, and their loss-of-function phenotypes can be complemented by coexpression of defective dl1007 virus that produces early, but not late, capsid proteins. Figure 1 shows the collection of SV40 mutants used in this study.
TABLE 1.
Selected phenotypes of the viral J-domain mutants used in this study
| Classa | Mutant | SV40 J-domain mutation | Relevant phenotype(s)b | Reference(s) |
|---|---|---|---|---|
| A | dl1135 | Δ(17-27) | Tu, T, V | 16, 26 |
| A | 5002 | L19F, P28S | T, V, VA | 24, 25, 28, 33 |
| A | 5114 | P43F | T, V, R | 25 |
| A | 5110 | D44N | T, V, R, A | 4, 5, 25, 31 |
| A | C6-1 | M301, K51N | R | 11 |
| B | GMSV40 | Δ(67-76) + YK | R | 20 |
| C | 43/45 | P43L, K45N | TA | 23, 28 |
| C | 44/47 | D44E, G47R | TA | 28 |
Class A, all T antigens affected; class B, LT only; class C, smt only.
Defective for the following: Tu, tumor formation in transgenic mice; T, transformation; R, DNA replication; V, viral plaque formation; A, blocking apoptosis in the absence of growth factor; TA, promoter transactivation; VA, viral capsid assembly.
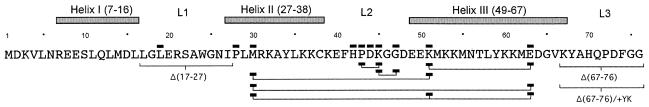
FIG. 1.
Summary of the SV40 J-domain mutations examined in this study. The α-helical boundaries from the SV40 J-domain structure determined in complex with the RB pocket domain (19) (Protein Database accession number 1GH6) are depicted. Loop regions are indicated as L1, L2, and L3. The mutation type is indicated by rectangles above (point mutations) or below (multiple mutations) the sequence. Amino acid residues of T antigens are numbered using the methionine start codon as 1.
To measure cochaperone activity of viral J domains, a sensitive assay was developed using a plasmid vector where the native E. coli J-domain sequence (2 to 70) in DnaJ is excised by EcoRI-KpnI digestion and replaced by PCR-generated viral J domains. Amplification of the viral J domains with appropriate EcoRI and KpnI restriction sites assured the accurate position of the chimeric protein start site and the proper translational reading frame of the heterologous J domain fused to the native DnaJ coding sequence (16). The WKG190 E. coli reporter strain that is used in conjunction with the dnaJ expression plasmid lacks both dnaJ and cbpA genes (16). As a consequence, WKG190 is viable at 30°C but is unable to propagate at both low and high temperatures, nor is it able to support the growth of bacteriophage λ at any temperature (16). Bacterial growth at low and high temperatures and the susceptibility to bacteriophage λ are thus dependent upon conditional expression (i.e., plasmid-borne) of a functional source of wild-type DnaJ, heterologous DnaJ, or a DnaJ chimera harboring an exogenous and functional J domain.
Using PCR, we amplified the SV40 J-domain mutations from viral DNA and cloned the resulting EcoRI-KpnI fragments into pWKG90[H71T-KpnI] DnaJ expression vectors as previously described (16). Alternatively, specific mutations were engineered by site-directed mutagenesis of pWKG92, which contained the wild-type SV40 J domain encoding amino acids 1 to 76. All plasmid constructions used in the study were sequence verified using the appropriate primers. Plasmid DNAs were subsequently transformed into WKG190 and plated at the permissive temperature in the absence of an inducer. Individual colonies were grown overnight, serially diluted, and then tested for complementation of J-domain activity at various temperatures and l-arabinose inducer concentrations. The susceptibility to bacteriophage λ was tested at 30°C using a plaque-forming assay as described previously (9, 16). Whole-cell extracts were prepared from each strain and used in immunoblot analysis of relative steady state protein levels using polyclonal rabbit anti-DnaJ or monoclonal PAb419, which is specific to the SV40 T-antigen J domain. In all assays we used wild-type DnaJ or truncated DnaJ12 as positive controls and empty vector or the mutant DnaJ(H33Q), which abolishes all DnaJ activity, as negative controls (16).
Analysis of DNA replication-defective mutant C6-1 and its revertant, C6-1R.
Gluzman and Ahrens (11) described the C6-1 mutant, which harbors two point mutations in the J domain (M30I, K51N). The mutant C6-1 replicates in permissive CV1 cells, albeit with markedly reduced efficiency and inability to form plaques, but is able to transform COS or established rat cell lines with the same efficiency as wild-type SV40. A revertant of C6-1 was isolated in the same study (C6-1R) by restoring viral plaque formation on CV1 cells. Marker rescue and sequence analyses showed that C6-1R was a pseudorevertant that had acquired a third point mutation (E63K) in the J domain.
We analyzed C6-1 and its revertant C6-1R and tested the hypothesis that the C6-1 mutant had completely eliminated or diminished J-domain cochaperone function and that the revertant had restored J-domain cochaperone function. If the two point mutations in C6-1 had compromised J-domain function, we expected to observe a defective J-domain function in our in vivo chaperone assays with E. coli. Furthermore, if viral plaque formation had been restored in the revertant, we predicted that J-domain function would be concomitantly restored. Since both C6-1 and C6-1R contain multiple point mutations, we also analyzed the phenotypes of each single point mutation and each double mutation.
The results of this study are shown in Fig. 2. We observed that the SV40 C6-1 J domain (M30I, K51N) was unable to complement for bacterial growth at 14 and 40°C. However, the J domain (M30I-K51N) could partially support the growth of bacteriophage λ, albeit at considerably less efficiency (<10−3) than the SV40 J-domain wild-type control. In contrast, the C6-1R J domain (M30I, K51N, E63K) could fully restore bacterial growth at both 14 and 40°C as well as sensitivity to bacteriophage λ at an efficiency similar to that of wild-type SV40 J domain. Immunoblot analysis of whole-cell extracts showed comparable expression of all mutants and wild-type J-domain chimeras (Fig. 2). We conclude that the SV40 C6-1 J domain has greatly reduced J-domain function and that C6-1R fully restores J-domain function to wild-type SV40 J-domain levels.
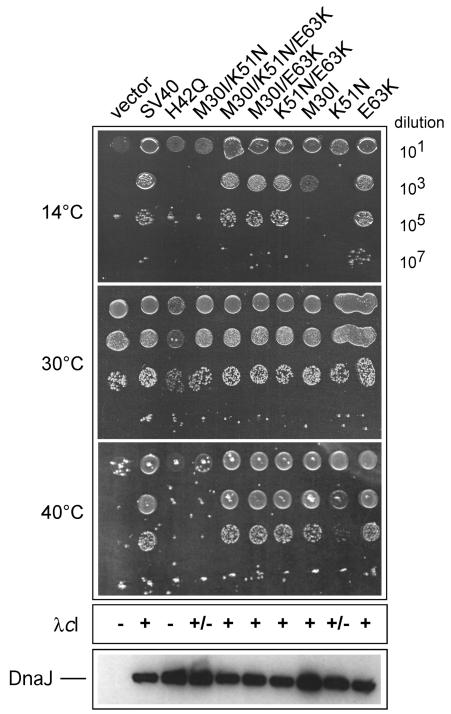
FIG. 2.
Analysis of SV40 J-domain mutant phenotypes in vivo. The WKG190 E. coli reporter strain lacks both dnaJ and cbpA genes, which renders it viable at 30°C but unable to propagate at both low and high temperatures, nor is it able to support the growth of bacteriophage λ at any temperature (16). Complementation of these genetic defects requires a functional source of DnaJ or a DnaJ chimera that has been engineered to contain a heterologous J domain. The l-arabinose-inducible vector pBAD22 or pWKG90 (SV40 J domain inserted in DnaJ in place of the native J domain) and its derivatives with the indicated SV40 J-domain mutations are shown. Fixed aliquots (5 μl) of serial dilutions of bacterial cultures grown at permissive temperature (30°C) were spotted on plates and grown at the indicated temperatures. The relative efficiency of colony formation can be directly determined by counting viable cells. The assay is sensitive for qualitative comparison but cannot be used to conclude relative activity. The efficiency of bacteriophage λcI plaque-forming ability is indicated as + if equivalent to that of wild-type bacteria (i.e., titer on test bacteria divided by titer on wild-type bacteria is ≈1), +/− if partial activity (<10−5 or five orders of magnitude less efficient plaque formation than that for the control), or − if <10−5 compared to control activity. λdnaJ+-transducing bacteriophage was used in every experiment as an infection control. An immunoblot analysis of whole-cell extracts showing the steady-state expression levels of the wild-type and mutant DnaJ proteins is presented at the bottom.
Next, we examined the phenotypes of each single and double mutation combination (Fig. 2). The results showed that the M30I mutation was significantly affected only for bacterial growth at 14°C but could otherwise support bacterial growth at 40°C and propagation of bacteriophage λ. In contrast, the K51N mutation was defective for growth at 14°C and partially defective for growth at 40°C. In addition, the K51N mutation alone resulted in reduced bacteriophage λ plaque formation. The E63K mutation alone showed no discernible phenotype in any assay. Interestingly, our results showed that the M30I and K51N point mutations synergize in C6-1, thus resulting in a more severe phenotype with respect to bacterial growth. The E63K mutation can fully restore J-domain function to both M30I alone and K51N alone. Thus, our results suggest that the K51N mutation resides in a critical region of SV40's J domain. Consistent with this suggestion, when we constructed a charge reversal mutant at this position (K51D), all J-domain function was abolished (data not shown). Structural studies are consistent with our findings. For example, the K51 side chain is solvent exposed in the reported structure of the SV40 N-terminal T-antigen 7-117 fragment in complex with the Rb pocket domain, whereas the side chains of M30 and E63 are buried or less accessible (19). It is possible that the mutations M30I and E63K only minimally perturb the J-domain core packing, whereas substitution at K51 markedly affects an exposed region involved in protein-protein interactions. It is noteworthy that Fewell et al. (7) recently described the mutant K53R, which shows multiple viral defects in vivo, thus underscoring the functional importance of lysine residues 51 and 53 within helix III.
Analysis of multiple classes of SV40 J-domain mutations.
Table 2 shows the composite results of eight additional viral mutants, comprising all three classes, together with an analysis of their constituent point mutants. In almost all cases, we observed that the mutations resulted in loss of J-domain function. Control immunoblots of the mutants showed similar steady-state levels compared to control extracts from cells that express chimeric DnaJ with the wild-type SV40 1-76 J domain (data not shown). We conclude from these analyses that mutations (P43F, P43L, D44N, and D44E) in the highly conserved HPD tripeptide and adjacent loop (K45N, G47E, and G47R) individually abolish J-domain function.
TABLE 2.
SV40 J-domain mutations are defective in bacterial and bacteriophage growth
| SV40 J-domain mutation | Bacterial growtha
|
λ growthb
|
|||
|---|---|---|---|---|---|
| 14°Cc | 30°C | 40°C | λcI dnaJ+ | λcI | |
| pBAD22 (vector) | − | + | − | + | − |
| SV40 (pWKG93) | + | + | + | + | + |
| H42Q | − | + | − | + | − |
| P43F | − | + | − | + | − |
| D44N | − | + | − | + | − |
| G47E | − | + | − | + | − |
| L19F/P28S | − | + | − | + | − |
| L19F | − | + | − | + | − |
| P28S | + | + | ± | + | + |
| P43L/K45N | − | + | − | + | − |
| P43L | − | + | − | + | − |
| K45N | − | + | − | + | − |
| D44E/G47R | − | + | − | + | − |
| D44E | − | + | − | + | − |
| G47R | − | + | − | + | − |
| Δ17-27 | − | + | − | + | − |
| GmSV40 | + | + | ± | + | + |
| GmSV40 (Δ67-82) | + | + | ± | + | + |
Appropriate complementation for bacterial growth was obtained at 0.1% of l-arabinose inducer.
Appropriate complementation for bacteriophage λ growth was obtained at 0.05% of l-arabinose inducer.
+ indicates full complementation for bacterial and λ growth when compared to the wild-type SV40 J-domain, i.e., efficiency of approximately one in colony or plaque formation, ± indicates partial complementation (with efficiency of colony or plaque formation of <10−2), and − indicates no complementation (efficiency of colony or plaque formation of <10−5). Additional definition of efficiency is shown in the legend to Fig. 2.
The mutation dl1135 (Δ17-27), which results in the deletion of 11 amino acid residues comprising most of the L1 loop joining helices I and II of the J domain, also abolished all J-domain activity. The L19F/P28S mutation, which alters one amino acid in the L2 loop joining helices I and II and one at the start of helix II, also abolishes J-domain function. The point mutation P28S alone has nearly wild-type activity, indicating that the defect in the double mutant is most likely due to the mutation L19F. It is likely that L19F disrupts J-domain stability by perturbing a hydrophobic pocket that stabilizes the J-domain fold (19). Collectively, our results reveal that the majority of SV40 J-domain loss-of-function mutants with measurable phenotypes in the virus concomitantly lose J-domain function. We conclude that the major function of the J domain is to provide cochaperone function for the viral T antigens.
Analysis of GMSV40, a mutant strongly defective in vivo but encoding LT that has improved activity in vitro.
Maulbecker et al. (20) described the isolation of the GMSV40 virus from a human skin fibroblast line, GM637, stably transformed with SV40, curiously carrying persistent multicopy extrachromosomal viral plasmids. The GMSV40 virus replicates with reduced efficiency in human cells but is markedly defective for replication and plaque formation in normally permissive monkey cells and cannot transform Rat-2 cells. Interestingly, in in vitro replication assays, the purified LT from GMSV40 was at least 25-fold more active than wild-type LT and showed reduced replication lag time. This striking effect of the mutant LT was observed irrespective of the source of the cellular replication factors or the method of preparation of T antigen. A 322-bp deletion in GMSV40 destroys smt production and produces an LT with a J-domain deletion of amino acid residues 67 to 82 and a breakpoint that changes the LT residues 83/84 from KY to YK. The replication defect of GMSV40 could not be rescued by expression of smt antigen in trans, which suggested that the observed phenotype was due solely to a defect in LT.
The three-dimensional structure of the LT 7-117 fragment in complex with the RB pocket domain shows that this Δ67-82 deletion results in the removal of part of the L3 loop outside the three-helix J-domain fold (19). The L3 loop directs some protein-protein interaction with the A-B box of the RB pocket domain. Portions of the loop also contact the J-domain core and may help to stabilize it (19). Deletion of residues 67 to 82 may not alter the essential J-domain fold but would likely relocate helix IV and perturb RB-LT interaction. Compared to the three-dimensional structure of the E. coli J domain, the SV40 J domain possesses a longer alpha helix III, and the region encompassing the GMSV40 deletion comprises a fourth helix in the DnaJ J domain. We have recently shown in the context of E. coli DnaJ that this helix IV has no detectable function in vivo, and multiple mutants in this region, or its complete deletion, behave like wild-type DnaJ (9).
We generated the GMSV40 J domain by PCR using primers that encoded the breakpoint change as found in the original virus (Δ67-82/YK). We also made a variant without the 83/84 KY residues reversal that contained only the Δ67-82 deletion. Both plasmids were tested for cochaperone activity (Table 2). We observed that the GMSV40 deletion as originally isolated with the added YK junction, as well as our own engineered deletion of Δ67-82 without the YK junction, did not abolish J-domain function in either assay, except for partial complementation at elevated temperature. Both mutants fully supported bacteriophage λ growth. We conclude that this region of the viral J domain is not necessary for J-domain function. In the context of virus, this region may help direct interaction with partner proteins, such as RB.
Expression of native T antigens and tests for cochaperone function.
The sensitivity of our E. coli assay in detecting J-domain function was also applied to detect the presence of intrinsic DnaJ cochaperone function of native LT and smt proteins. To do this, arabinose-inducible expression plasmids containing the cloned cDNA coding for native SV40 LT, LT truncation variants TN136 and TN125, and native smt or various truncations of smt were constructed by PCR and cloned in pBAD22 (12). All plasmid constructions were sequence verified. Plasmids were transformed into WKG190 at 30°C and then tested for DnaJ-like cochaperone activities as described above (Fig. 3).
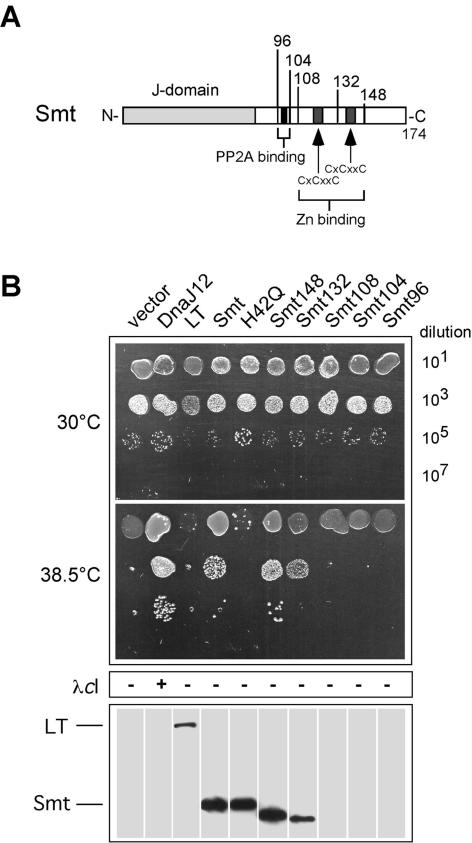
FIG. 3.
smt but not LT can functionally replace DnaJ for E. coli growth at elevated temperature. (A) The boundaries of the C-terminal smt truncation mutations used in the study are shown, together with the locations of previously mapped physical features of smt (28). (B) The assays for DnaJ cochaperone activity were performed as described in the legend to Fig. 2. Western blots of whole-cell extracts were performed using the PAb419 monoclonal antibody that specifically recognizes the SV40 N-terminal J domain.
We observed that LT could not complement for E. coli growth at the nonpermissive temperature and for the growth of bacteriophage λ at 30°C. The same observations were made for the LT truncation variants TN136 and TN125 (data not shown). In addition, no complementation for bacterial growth was observed for any LT variant at low temperatures (data not shown). Control immunoblots using whole-cell extracts and the monoclonal antibody PAb419 (Oncogene Science), which specifically recognizes the T-antigen J domain, showed that all LTs were expressed at equivalent steady-state levels (Fig. 3). We conclude that native LT and the truncation variants TN136 and TN125 cannot substitute for DnaJ in this assay under all experimental conditions tested.
Strikingly, and in contrast to our results with LT, we observed that smt could functionally replace DnaJ for bacterial growth at 38.5°C (Fig. 3) but could not functionally substitute for DnaJ at higher temperatures or at low temperature (data not shown). The ability to partially complement for DnaJ was abolished when we introduced the H42Q J-domain inactivating mutation into smt (16). We observed that smt, however, could not support the growth of bacteriophage λ. We next prepared a series of smt C-terminal truncation mutations to map the limits of J-domain flanking sequences necessary for the observed cochaperone function. PCR primers were designed to amplify the indicated regions of the smt cDNA, and the end points of each truncation are shown in Fig. 3. Expression plasmids were prepared, sequence verified, and tested as described above. We observed that smt148 behaved essentially like wild-type smt. Further deletion up to residue 132, which removed the last zinc binding motif, reduced but did not entirely abolish cochaperone activity. Deletion beyond residue 108 completely abolished cochaperone activity. Control immunoblots with PAb419 showed comparable steady-state protein expression of smt, smt(H42Q), and truncations smt148 and smt132 (Fig. 3). Immunoblots did not detect truncated smt proteins truncated beyond smt132. Most likely these deletions resulted in unstable proteins, and we did not pursue further studies with them. We conclude that smt encodes a detectable DnaJ cochaperone function that can support bacterial growth at elevated temperature. The propagation of bacteriophage λ apparently requires additional sequences, or particular tertiary structures, adjacent to the J domain that are not present in smt. It is noteworthy that residues 82 to 132 in smt have no significant homology to the DnaJ glycine-phenylalanine-rich (G/F-rich) domain encompassing residues 77 to 108.
smt can stimulate DnaK's ATPase and acts as a cochaperone in vitro.
To confirm that smt acted as a cochaperone, we analyzed the ability of purified smt to stimulate the ATPase activity of DnaK in vitro. Smt was purified as described previously (22) and used in a steady-state ATPase assay (8). The results are shown in Fig. 4. In contrast to the DnaJ(H33Q) mutant, which shows no cochaperone function, we observed that wild-type smt could stimulate DnaK's ATPase threefold to fourfold. Compared to control reactions performed in parallel, smt stimulation of DnaK's ATPase represented 30 to 40% of the maximal stimulation observed with purified DnaJ. These results are comparable to the fourfold smt-dependent stimulation of Ssa1p's ATPase, a yeast cytosolic Hsc70 homolog (34). We next tested smt for its intrinsic ability to function as a bona fide chaperone by testing its ability to protect luciferase from aggregation following chemical or thermal denaturation (8, 40). This assay has previously been used to define whether Hsp40 or other proteins possess chaperone activity alone. We observed that smt could not by itself protect luciferase from aggregation, in contrast to control experiments conducted in parallel using purified DnaJ (data not shown). We conclude from these analyses that smt possesses cochaperone activity but does not possess detectable intrinsic chaperone activity.
FIG. 4.
smt can stimulate DnaK's ATPase activity. Stimulation of the steady-state ATPase activity of DnaK by purified DnaJ, DnaJ H33Q, or smt is shown. The percentage of hydrolyzed ATP/min is plotted as a function of the final DnaJ or smt concentration in the reaction mixture. DnaK was constant in each assay and was used at a final concentration of 1 μM. The ATPase assay was performed as described previously (8).
Our results suggest that smt can be exploited in E. coli to probe fundamental questions about cochaperone function and its interaction with its cognate Hsp70 partner (DnaK). Since there is no significant amino acid homology between smt residues (82 to 174) that flank the viral J domain and the DnaJ G/F-rich 72-108 region adjacent to the DnaJ J domain, we conclude that neither E. coli nor DnaK itself requires a specific J-domain adjacent sequence for cochaperone function. This conclusion is in agreement with our finding that the E. coli cytoplasmic DjlA protein fragment can functionally complement for DnaJ (8). In DjlA, the J domain is positioned at the extreme C-terminal portion of the protein. DjlA J-domain flanking sequences, which are thus in inverted position relative to DnaJ or smt, have no homology either to the DnaJ G/F-rich sequence or to the smt unique sequence. We therefore propose that the fundamental DnaJ cochaperone function requires a J domain and flanking amino acid sequences positioned N terminally or C terminally that may adopt a particular tertiary structure that permits correct interaction with DnaK.
smt flanking sequence fused to DnaJ J domain can provide cochaperone function.
The failure of native LT and its truncated variants TN125 and TN136 to complement for DnaJ in our assays further suggested that not just any adjacent polypeptide sequence, or even a sequence of comparable length, could function together with a J domain to provide cochaperone function. To examine the role of flanking sequences further, we constructed a series of chimeric DnaJ molecules where the DnaJ J domain was linked to adjacent sequences from smt or LT. It has been shown that E. coli DnaJ12, a truncation mutant of DnaJ with only 108 residues, of which the first 72 residues represent the J domain, is sufficient to provide cochaperone function for partial bacterial growth and propagation of bacteriophage λ (41). Since the E. coli J domain alone has been shown to have no detectable biological function alone (15, 16, 40), a certain number of residues adjacent to the J domain must be necessary to provide biological function as a cochaperone.
The LT residues 82 to 125, 82 to 136, or smt 82 to 174 were fused to E. coli's DnaJ J domain in the vector pWKG190[H71T-KpnI] using PCR primers that introduced appropriate KpnI-BglII restriction sites for the flanking sequence replacement and tested in our in vivo assays. The vector pWKG190 served as a control to express DnaJ12 (16). The results are shown in Fig. 5. The chimeric DnaJ J domain fused to smt residues 82 to 174 could provide cochaperone function at elevated temperature comparable to that of DnaJ12. The chimeric DnaJ-smt protein could function up to 40°C, unlike native smt, which functions only up to 38.5°C in this assay. These results indicated that the E. coli J domain can function with heterologous flanking sequence and that some thermal stability, or activity, may be gained by the chimeric protein compared to native smt alone. We also observed that neither chimera with J-domain LT-derived flanking amino acid residues 76 to 125 or 76 to 136 could function in this assay. This result shows that sequences specific to smt can cooperate in cis with a bacterial J domain to provide cochaperone function but that LT sequences of the same approximate length cannot provide this function. In contrast to DnaJ12, none of the chimeric proteins could function for bacteriophage λ growth, and thus, we conclude that λ requires specific sequences, or tertiary structures, outside the J domain that remain to be identified. Mechanistically, it remains to be determined if flanking amino acid sequences themselves act through direct protein-protein interaction with a cognate Hsp70 or, alternatively, whether these adjacent sequences promote alterations in the cis J domain that lead to productive and reversible J-domain interaction with its cognate Hsp70 partner(s).
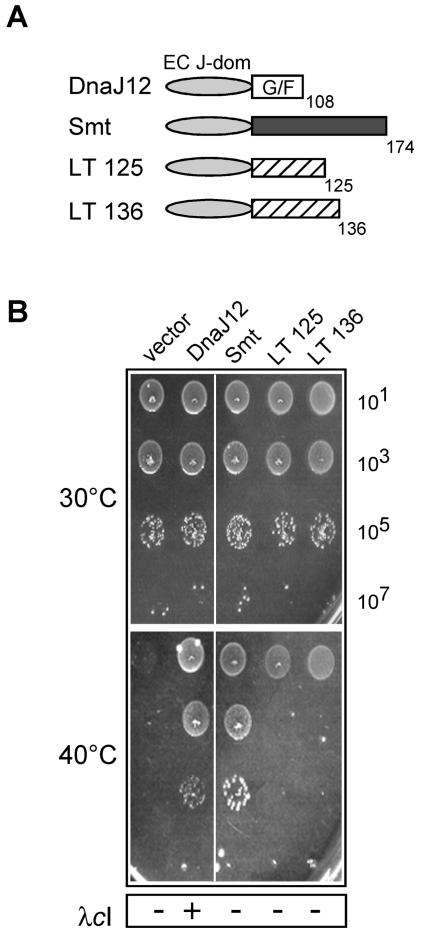
FIG. 5.
Analysis of J-domain flanking regions in T antigens for their ability to functionally reconstitute DnaJ cochaperone activity when linked to the E. coli J domain. (A) Schematic representation of the various chimeric proteins used in the assay and constructed in the vector pBAD22. (B) Ability to complement for bacterial growth in the E. coli WKG190 reporter strain. The assay conditions for DnaJ cochaperone activity were as described in the legend to Fig. 2.
Conclusions.
The following original observations have been made in this study. (i) There exists a correlation between loss-of-function phenotypes observed in multiple T-antigen J-domain mutants and scored in mammalian cells and viral J-domain cochaperone loss of function when scored in E. coli. The wide variety of both type and dispersion of mutations strongly suggests that the major role of the viral J domain is to direct molecular chaperones (Hsp70s) to multiple steps in the viral life cycle. (ii) We have identified SV40 smt as a bona fide cochaperone and provided the first demonstration that a native mammalian DNA tumor virus protein can functionally replace a bacterial Hsp40 chaperone in vivo. (iii) Amino acid sequences adjacent to the DnaJ J domain that are necessary for cochaperone function activity in vivo in E. coli can be functionally replaced by smt residues that possess no significant amino acid homology. Thus, fundamental questions underlying the Hsp70 chaperone machine mechanism can be addressed using interchangeable parts from distantly related mammalian DNA tumor virus T antigens and bacterial Hsp40 proteins.
We recently reported an extensive scanning mutagenesis study of the E. coli DnaJ J domain that identified a small set of essential amino acids located on the same solvent-exposed face that were necessary for J-domain function in vivo (9). Two high-resolution structures have been reported for the SV40 LT N-terminal 7-117 polypeptide in complex with the RB pocket domain and the murine polyomavirus T-antigen J-domain 1-79 (3, 19). Both structures show a similar overall J-domain fold, and the polyomavirus T-antigen J domain was also shown to functionally substitute for the DnaJ J domain in our assay system (3). Because of conserved J-domain function, a similar interaction surface in the viral J domains should map in the same region. An examination of the SV40 T-antigen loss-of-function point mutations used in the present study shows that some map on the same solvent-exposed face that we identified. In particular, mutations spanning the 42HPDKGG47 region affect J-domain function and coincide closely with a portion of the essential contact surface that we identified in DnaJ. Other mutations that map outside the region that we defined may perturb local structure, leading to loss of function. Extensive mutagenesis of the SV40 J domain will resolve exactly which residues are essential for J-domain function.
It is worthwhile to note that experimental evidence exists to support a role for defective Hsp70 chaperone machines that retain some biological activity. It is possible in the context of SV40 viral T antigens that certain viral functions require a productive chaperone machine and a J domain that can target and stimulate the cognate Hsp70 ATPase, while other viral functions may simply require the J domain together with perhaps other proteins as a protein scaffold. This may explain why certain SV40 mutants, such as D44N, P43L/K51N, D44E/G47R, or even mutations in other J-domain family proteins, such as HPD-AAA in p58IPK (42), may be interpreted as biologically active in certain assays despite an apparent loss of J-domain cochaperone function (5, 10, 30, 39). Additional studies are needed to address this possibility in depth.
Recently, Fewell and colleagues reported the development of a genetic screen for SV40 J-domain loss of function using Saccharomyces cerevisiae (7). Their method, the reverse of the approach used here, uncovered new LT J-domain loss-of-function mutations, some of which were subsequently reengineered into SV40 and shown to be defective in plaque formation and cellular transformation. These authors concluded that a correlation existed between the ability of a given viral J domain to stimulate Hsc70's ATPase in vitro and its ability to successfully support viral infection in vivo. Our overall conclusions are in agreement with this study. The successful use of yeast or bacteria to probe complex questions in mammalian tumor virus biology underscores the power of simple systems to shed light on the universally conserved chaperone mechanisms.
Acknowledgments
This work was supported by grant 3100-065403-01 from the Swiss National Science Foundation to C.G. and W.L.K., the Canton of Geneva, and U.S. National Institutes of Health grants CA40586 to J.M.P. and CA21327 to K.R.
REFERENCES
- 1.Ali, S. H., and J. A. DeCaprio. 2001. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin. Cancer Biol. 11:15-22. [DOI] [PubMed] [Google Scholar]
- 2.Beachy, T. M., S. L. Cole, J. F. Cavender, and M. J. Tevethia. 2002. Regions and activities of simian virus 40 T antigen that cooperate with an activated ras oncogene in transforming primary rat embryo fibroblasts. J. Virol. 76:3145-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berjanskii, M. V., M. I. Riley, A. Xie, V. Semenchenko, W. R. Folk, and S. R. Van Doren. 2000. NMR structure of the N-terminal J. domain of murine polyomavirus T antigens. J. Biol. Chem. 275:36094-36103. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, K. S., K. P. Mullane, I. A. Aksoy, H. Stubdal, J. Zalvide, J. M. Pipas, P. A. Silver, T. M. Roberts, B. S. Schaffhausen, and J. A. DeCaprio. 1997. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 11:1098-1110. [DOI] [PubMed] [Google Scholar]
- 5.Cole, S. L., and M. J. Tevethia. 2002. Simian virus 40 large T antigen and two independent T-antigen segments sensitize cells to apoptosis following genotoxic damage. J. Virol. 76:8420-8434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, B. S., and J. M. Pipas. 1995. T antigens encoded by replication-defective simian virus 40 mutants dl1135 and 5080. J. Biol. Chem. 270:15377-15384. [DOI] [PubMed] [Google Scholar]
- 7.Fewell, S. W., J. M. Pipas, and J. L. Brodsky. 2002. Mutagenesis of a functional chimeric gene in yeast identifies mutations in the simian virus 40 large T antigen J. domain. Proc. Natl. Acad. Sci. USA 99:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genevaux, P., A. Wawrzynow, M. Zylicz, C. Georgopoulos, and W. L. Kelley. 2001. DjlA is a third DnaK co-chaperone of Escherichia coli, and DjlA-mediated induction of colanic acid capsule requires DjlA-DnaK interaction. J. Biol. Chem. 276:7906-7912. [DOI] [PubMed] [Google Scholar]
- 9.Genevaux, P., F. Schwager, C. Georgopoulos, and W. L. Kelley. 2002. Scanning mutagenesis identifies amino acid residues essential for the in vivo activity of the Escherichia coli DnaJ J-domain. Genetics 162:1045-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gjoerup, O., D. Zaverni, and T. M. Roberts. 2001. Induction of p53-independent apoptosis by simian virus 40 small t antigen. J. Virol. 75:9142-9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gluzman, Y., and B. Ahrens. 1982. SV40 early mutants that are defective for viral DNA synthesis but competent for transformation of cultured rat and simian cells. Virology 123:78-92. [DOI] [PubMed] [Google Scholar]
- 12.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation and high level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn, W. C., S. K. Dessain, M. W. Brooks, J. E. King, B. Elenbaas, D. M. Sabatini, J. M. DeCaprio, and R. A. Weinberg. 2002. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 22:2111-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartl, F. U. 1996. Molecular chaperones in cellular protein folding. Nature 381:571-579. [DOI] [PubMed] [Google Scholar]
- 15.Karzai, A. W., and R. McMacken. 1996. A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J. Biol. Chem. 271:11236-11246. [DOI] [PubMed] [Google Scholar]
- 16.Kelley, W. L., and C. Georgopoulos. 1997. The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc. Natl. Acad. Sci. USA 94:3679-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley, W. L. 1998. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 23:222-227. [DOI] [PubMed] [Google Scholar]
- 18.Kiersted, T. D., and J. M. Pipas. 1996. Database of mutations that alter the large tumor antigen in simian virus 40. Nucleic Acids Res. 24:125-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, H.-Y., A. Byung-Yoon, and Y. Cho. 2001. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 20:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maulbecker, C., I. Mohr, Y. Gluzman, J. Bartholomew, and M. Botchan. 1992. A deletion in the simian virus 40 large T antigen impairs lytic replication in monkey cells in vivo but enhances DNA replication in vitro: new complementation function of T antigen. J. Virol. 66:2195-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer, M. P., D. Brehmer, C. S. Gassler, and B. Bukau. 2001. Hsp70 chaperone machines. Adv. Protein Chem. 59:1-44. [DOI] [PubMed] [Google Scholar]
- 22.Montano, X., and K. Rundell. 2001. Role of SV40 small t in cell lysis and transformation. Methods Mol. Biol. 165:229-242. [DOI] [PubMed] [Google Scholar]
- 23.Mungre, S., K. Enderle, B. Turk, A. Porras, Y.-Q. Wu, M. Mumby, and K. Rundell. 1994. Mutations which affect the inhibition of protein phosphatase 2A by simian virus 40 small-t antigen in vitro decrease viral transformation. J. Virol. 68:1675-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peden, K. W., S. L. Spence, L. C. Tack, C. A. Cartwright, A. Srinivasan, and J. M. Pipas. 1990. A DNA replication-positive mutant of simian virus 40 that is defective for transformation and the production of infectious virus. J. Virol. 64:2912-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peden, K. W. C., and J. M. Pipas. 1992. Simian virus 40 mutants with amino-acid substitutions near the amino terminus of large T antigen. Virus Genes 6:107-118. [DOI] [PubMed] [Google Scholar]
- 26.Pipas, J. M., K. W. Peden, and D. Nathans. 1983. Mutational analysis of simian virus 40 T antigen: isolation and characterization of mutants with deletions in the T-antigen gene. Mol. Cell. Biol. 3:203-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pipas, J. M. 1992. Common and unique features of T antigens encoded by the polyomavirus group. J. Virol. 66:3979-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porras, A., J. Bennett, A. Howe, K. Tokos, N. Bouck, B. Henglein, S. Sathamangalam, B. Thimmapaya, and K. Rundell. 1996. A novel simian virus 40 early-region domain mediates transactivation of the cylin A promoter by small-t antigen and is required for transformation in small-t antigen-dependent assays. J. Virol. 70:6902-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rundell, K., and R. Parakati. 2001. The role of the SV40 ST antigen in cell growth promotion and transformation. Semin. Cancer Biol. 11:5-13. [DOI] [PubMed] [Google Scholar]
- 30.Sheng, Q., T. M. Love, and B. Schaffhausen. 2000. J domain-independent regulation of the Rb family by polyomavirus t antigen. J. Virol. 74:5280-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slinsky, A., D. Barnes, and J. M. Pipas. 1999. Simian virus 40 large T antigen J domain and Rb-binding motif are sufficient to block apoptosis induced by growth factor withdrawal in a neural stem cell line. J. Virol. 73:6791-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sock, E., J. Enderich, and M. Wegner. 1999. The J. domain of papovaviral large tumor antigen is required for synergistic interaction with the POU-domain protein Tst-1/Oct6/SCIP. Mol. Cell. Biol. 19:2455-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spence, S. L., and J. M. Pipas. 1994. SV40 large T antigen functions at two distinct steps in virion assembly. Virology 204:200-209. [DOI] [PubMed] [Google Scholar]
- 34.Srinivasan, A., A. J. McClellan, J. Vartikar, I. Marks, P. Cantalupo, Y. Li, P. Whyte, K. Rundell, J. L. Brodsky, and J. M. Pipas. 1997. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol. Cell. Biol. 17:4761-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stubdal, H., J. Zalvide, K. S. Campbell, C. Schweizer, T. M. Roberts, and J. S. DeCaprio. 1997. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol. Cell. Biol. 17:4979-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan, C. S., P. Cantalupo, and J. M. Pipas. 2000. The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol. Cell. Biol. 20:6233-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan, C. S., J. D. Tremblay, S. W. Fewell, J. A. Lewis, J. L. Brodsky, and J. M. Pipas. 2000. Species-specific elements in the large T-antigen J domain are required for cellular transformation and DNA replication by simian virus 40. Mol. Cell. Biol. 20:5749-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan, C. S., S. P. Gilbert, and J. M. Pipas. 2001. ATP-dependent simian virus T-antigen-Hsc70 complex formation. J. Virol. 75:1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan, C. S., and J. M. Pipas. 2002. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol. Mol. Biol. Rev. 66:179-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szabo, A., T. Langer, H. Schröder, J. Flanagan, M. K. Hayer, and F. U. Hartl. 1994. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc. Natl. Acad. Sci. USA 91:10345-10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wall, D., M. Zylicz, and C. Georgopoulos. 1994. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J. Biol. Chem. 269:5446-5451. [PubMed] [Google Scholar]
- 42.Yan, W., M. G. Gale, Jr., S.-L. Tan, and M. G. Katze. 2002. Inactivation of the PKR protein kinase and stimulation of mRNA translation by the cellular co-chaperone p58IPK does not require J. domain function. Biochemistry 41:4938-4945. [DOI] [PubMed] [Google Scholar]
- 43.Yang, S.-I., R. L. Lickteig, R. Estes, K. Rundell, G. Walter, and M. Mumby. 1991. Control of protein phosphatase 2A by simian virus 40 small-t antigen. Mol. Cell. Biol. 11:1988-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zalvide, J., H. Stubdal, and J. A. DeCaprio. 1998. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol. Cell. Biol. 18:1408-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]