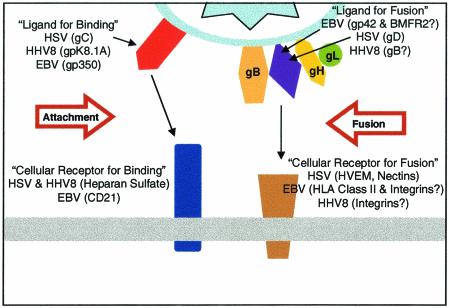
Herpesviruses engage multiple receptors during viral entry. Some are considered binding receptors only, in that engagement with these receptors may be reversible and may serve to concentrate virus on the cell surface without triggering changes required for membrane fusion. Others are considered entry receptors, binding to which triggers events required for membrane fusion. Most herpesviruses probably recognize multiple entry receptors, any one of which may be sufficient for viral entry. The purpose of this review is to summarize recent findings on the entry of alpha- and gammaherpesviruses and on structure-function studies of their entry receptors and viral ligands.
Members of the Herpesviridae form a large and diverse family comprised of three subfamilies designated alpha-, beta-, and gammaherpesviruses. Virions are composed of a large DNA genome encased in an icosahedral capsid, which is in turn coated with a layer of proteins called the tegument and an envelope composed of about a dozen viral proteins and glycoproteins in a lipid bilayer. At least three, sometimes four, of these envelope glycoproteins are absolutely essential for viral entry. The three glycoproteins thought to be essential for the entry of all herpesviruses are designated gB, gH, and gL. The genes for these glycoproteins are conserved, with gB exhibiting the highest degree of sequence similarity. For at least some herpesviruses, gB is a homodimer or homotrimer displayed as a prominent spike. Heterodimerization of gH and gL is a conserved feature, with the addition of another viral protein subunit for some viruses.
Common features of herpesvirus biology include a high incidence of asymptomatic infections and the establishment of latent infections which can be reactivated to cause recurrent or new episodes of disease. The human herpesviruses exhibit these common features as well as diversity in biology and pathogenesis. They include the alphaherpesviruses, herpes simplex virus types 1 and 2 (HSV-1 and HSV-2) and varicella-zoster virus (VZV); the betaherpesviruses, cytomegalovirus (CMV) and human herpesviruses 6 and 7; and the gammaherpesviruses, Epstein-Barr virus (EBV) and human herpesvirus 8 (HHV-8). HSV-1 and HSV-2 are responsible for localized mucocutaneous lesions, most commonly, but can also cause meningitis and encephalitis. VZV causes systemic disease with skin lesions during primary infection (chicken pox) and zoster. All three viruses establish latent infections in neurons and can be reactivated from neurons. The betaherpesviruses cause mostly asymptomatic infections in immunocompetent individuals and can establish latent infections in several cell types, including leukocytes of various lineages. EBV is the major cause of infectious mononucleosis and is causally associated with various malignancies, including Burkitt's lymphoma, Hodgkin's disease, other lymphomas, and nasopharyngeal carcinoma. HHV-8 is associated with Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma.
DIVERSITY OF BINDING AND ENTRY RECEPTORS AND CONSERVED FUSION MACHINERY
Although natural infections with most herpesviruses are restricted to a single species, some of these viruses can infect other species experimentally or accidentally. In general, alphaherpesviruses are viewed as having the broadest host range with respect to species (excepting VZV) and cell type, betaherpesviruses can infect a variety of cell types but are usually restricted to infections of their natural hosts, and gammaherpesviruses are restricted with respect to host and cell type. Much of the data cited in this review will show that the broad host range of HSV and the narrow host range of EBV are consistent with, and perhaps can be explained in part by, the nature of binding and entry receptors used by each virus.
Most, but not all, herpesviruses can make their initial contact with cells by binding to glycosaminoglycans, usually heparan sulfate, on cell surface proteoglycans (37). A notable exception is EBV (Fig. 1), as discussed below. For the majority of herpesviruses that can bind to heparan sulfate, this binding may not be essential for virus infection and can be mediated by viral glycoproteins that are not essential for viral entry. Nevertheless, the presence of cell surface heparan sulfate greatly increases the efficiency of viral entry, probably by concentrating virus on the cell surface so that the appropriate viral ligands for entry receptors can find these receptors. The binding of HSV or other alphaherpesviruses to heparan sulfate is reversible, and eluted virus remains infectious, indicating that fusion activity has not been irreversibly triggered. Interaction with an entry receptor appears to be irreversible, however, perhaps because this binding leads immediately to membrane fusion. This operational definition of binding and entry receptors may also apply to other herpesviruses.
FIG. 1.
Participants in herpesvirus entry and virus-induced cell fusion. For both alphaherpesviruses and gammaherpesviruses, binding to cells can be mediated by a virion glycoprotein that is not essential for entry. The binding receptors are heparan sulfate for HSV gC and HHV-8 K8.1A and CD21 for EBV gp350 (in the case of B cells). Entry requires interaction of a viral ligand with another cell surface receptor. For HSV, virion gD is the ligand for several cell surface receptors (HVEM, nectins, 3-O-sulfated heparan sulfate), any one of which can mediate entry. For EBV entry into B cells, gp42 binds to HLA class II molecules. It should be noted that gp42 is not required for EBV infection of epithelial cells. The entry receptors in epithelial cells have not yet been identified but could include integrins. The viral ligands could be gH and/or BMRF2. For HHV-8 entry, gB can bind to one of the integrins. Any one of these interactions of a viral ligand with an entry receptor is thought to activate the fusion activity of gB and gH-gL.
After the initial binding of virions to cells, entry of herpesviruses occurs by fusion of the virion envelope with a cell membrane (the plasma membrane under certain conditions or the membrane of an endosome under others). Endocytosis and acidification of endosomes may be required in some cell types, but not in others, for entry of certain herpesviruses such as HSV (31, 46). This is somewhat puzzling inasmuch as cell fusion induced by virus infection or by glycoprotein transfection occurs at physiological pH (9, 29, 32) and clearly does not require exposure of any of the glycoproteins to low pH.
It is thought that interactions of one or more viral glycoproteins with cellular receptors can trigger envelope-membrane fusion or cell-cell fusion. The basic fusion machinery for herpesvirus entry is presumed to include gB, gH, and gL, although additional receptor-binding viral glycoproteins may also be required (Fig. 1). For example, HSV requires gD as a ligand for entry receptors. Only alphaherpesviruses (except VZV) encode members of the gD family. EBV requires gp42, a component of the gH-gL-gp42 complex, as a ligand for human leukocyte antigen (HLA) class II molecules on B lymphocytes. Human CMV encodes gO, which is unrelated to gp42 but also forms a complex with gH-gL (14). Similarly, human herpesvirus 6A encodes gQ, which forms a complex with gH-gL (28). It is not yet clear whether gO and gQ are required for entry of these viruses. Alphaherpesviruses do not encode homologs of gp42, gO, or gQ. Thus, it seems likely that the basic membrane-fusing machinery is conserved among the herpesviruses, whereas the viral ligands that bind to cell surface receptors differ among the subfamilies, thereby explaining in part the differences in cell and tissue tropisms.
ALPHAHERPESVIRUS (HSV) ENTRY
The envelope glycoproteins of HSV-1 and HSV-2 number at least a dozen, but to date only five have been shown to have any role in viral entry (Fig. 1). Glycoproteins gB and/or gC can mediate the binding of virus to cell surface heparan sulfate. Although gC is dispensable for the infection of cultured cells, its presence can increase the efficiency of virus binding almost 10-fold, at least for HSV-1. Deletional mutagenesis has shown that gB, gD, gH, and gL are all required for HSV-1 entry, but not for virus binding to cells, provided that gC is present. The four essential glycoproteins are thought to act in concert to induce fusion of the viral envelope with a cell membrane. Cell entry receptors are required to trigger this fusion, and the viral ligand for all known HSV entry receptors is gD (reviewed in reference 38). This summary of requirements for entry applies to animal alphaherpesviruses as well, including pseudorabies virus (PRV) and bovine herpesvirus 1.
To date, three classes of HSV entry receptors have been identified (38). They include herpesvirus entry mediator (HVEM), a member of the tumor necrosis factor receptor family; nectin-1 and nectin-2, two members of the immunoglobulin superfamily; and specific sites in heparan sulfate generated by certain isoforms of 3-O-sulfotransferases. Any one of these cell surface molecules can bind to gD to mediate viral entry, with each serotype having somewhat different receptor preferences. Whereas HVEM and nectin-1 are excellent entry receptors for both HSV-1 and HSV-2, nectin-2 is more active for HSV-2 than for HSV-1 and 3-O-sulfated heparan sulfate is probably more active for HSV-1 than for HSV-2. These results were obtained by transfecting cells deficient in endogenous receptors for HSV (for example, Chinese hamster ovary [CHO] cells) with plasmids expressing the entry receptors and then quantifying viral entry using viruses that express reporter genes immediately upon entry. A very useful, if imperfect, surrogate assay for viral entry is a cell fusion assay in which CHO cells expressing HSV gB, gD, gH, and gL are mixed with CHO cells expressing a gD receptor and fusion is assessed by quantifying expression of a reporter gene activated only in heterokaryons (32).
It is not understood how the interaction of HSV gD with an entry receptor leads to viral entry or cell fusion. One hypothesis is that the binding of gD to one of its receptors results in a conformational change in gD, enabling its interaction with gB or gH-gL and activation of fusogenic activity. Possibly, gD is not an integral component of the basic fusion machinery. Also, receptors for gB and/or gH-gL may exist, and binding of either to these receptors could also trigger fusion activity, bypassing the requirement for gD. These ideas emerge in part from results obtained with PRV.
PRV can use several human and animal members of the nectin family as entry receptors (10, 25), as well as other unidentified entry receptors. Although all four PRV glycoproteins, including gD, are required for viral entry, gB, gH, and gL are sufficient for the fusion of rabbit cells (whose receptors have not been identified), suggesting the existence of cell surface receptors for gB or gH-gL (18). Also, one of the gammaherpesviruses, HHV-8, has been shown to require only gB, gH, and gL for cell fusion, although the possible potentiating activity of other viral proteins has not been ruled out (33).
(i) The HSV entry receptors.
HVEM is expressed in a variety of cell types, including lymphocytes, other leukocytes, epithelial cells, and fibroblasts. The natural ligands for HVEM are LIGHT and lymphotoxin-alpha (23). LIGHT can provide a second signal for T-cell activation, and LIGHT-HVEM interactions are the subject of active investigations into the regulation of immune responses (reviewed in reference 19). The nectins are also expressed in a variety of cell types, including epithelial cells, fibroblasts, and neurons. nectin-1 and nectin-2 are related to nectin-3, nectin-4, and the poliovirus receptor, and all appear to be cell adhesion molecules (reviewed in reference 40). The nectins have been shown to colocalize with cadherins in adherens junctions characteristic of various cell types, including epithelial cells and neurons. The nectins, especially nectin-1, are highly conserved among mammalian species with respect to structure, function, and alphaherpesvirus entry (25, 35). Although 3-O-sulfotransferases that can generate gD-binding sites in heparan sulfate are widely expressed, the generation of these sites depends on the expression of other heparan sulfate-modifying enzymes as well (36), complicating the task of determining the distribution of these gD-binding sites without specific probes.
Both the presence and cellular localization of HSV entry receptors are important determinants of viral entry because receptor distribution on polarized cells may or may not permit viral access to the receptors. For example, nectin-1 localizes to adherens junctions and is not accessible for the binding of soluble gD or for service as an HSV entry receptor unless the cell junctions are first disrupted (47).
Since key target cells for HSV infection in vivo may express multiple HSV entry receptors, which are most important for the establishment and spread of virus infection? Use of anti-HVEM antibodies that block viral entry demonstrated that HSV-1 infection of activated human T cells, but not a variety of other cultured human cell types, was principally mediated via HVEM (26). Human epithelial cells express all the known protein entry receptors. As mentioned above, the nectins might not be available for the infection of polarized human epithelial cells unless the epithelium was damaged. HVEM or other receptors might be available, however, even in an intact epithelium. The receptors used by the virus for initial entry into epithelia may differ from those used for cell-to-cell spread of infection within the epithelium. Nectins are good candidates for receptors used in cell-to-cell spread in a polarized epithelium since progeny virus have been shown to exit the cell in the vicinity of adherens junctions on the lateral surfaces of the cells (16). Moreover, ectopic expression of nectins and cadherins in L cells can enhance plaque size (34). Nectins are also good candidates for the infection of neurons (13).
An X-ray structure of a truncated form of HSV-1 gD complexed with a truncated form of HVEM revealed that the gD contact sites on HVEM are located in the first two cysteine-rich domains (6). Mutagenic analysis of HVEM identified the subset of amino acid residues in the gD contact regions that were most critical for functional interactions with gD (8). The nectins have three immunoglobulin (Ig)-like domains, including an N-terminal V-like domain. The V-like domain is necessary and sufficient for gD binding. Analysis of hybrid forms of nectin-1 and nectin-2, along with directed mutagenesis, revealed that loops between the C′ and C" beta strands and between the F and G beta strands of the V-like domain are critical for gD binding and for function in viral entry (7, 21, 22, 39). The loop between the F and G beta strands is also critical for homotypic interactions between nectins expressed on adjacent cells. These interactions can be inhibited by soluble forms of gD (40). The fact that nectin-2 appears to be a less effective entry receptor for HSV than nectin-1 might be explained by a lower affinity of interaction of gD with nectin-2, a greater propensity of nectin-2 to engage in competing interactions with other nectins, or both.
(ii) gD structure and binding to receptors.
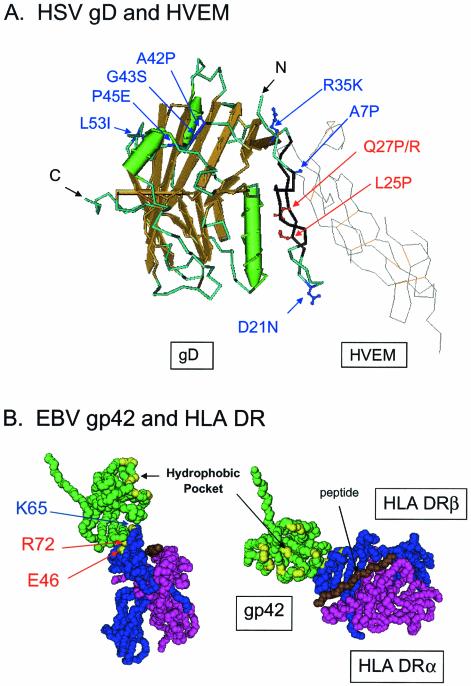
X-ray structures of HSV-1 gD alone and in complex with HVEM revealed, surprisingly, that a portion of gD assumes an Ig-like fold with unconventional disulfide-bonding patterns (6). There is an N-terminal extension from the Ig-like fold that forms a hairpin loop in the complex with HVEM but is disordered in the crystals of gD alone. The contacts in gD for HVEM are localized entirely to amino acids 7 to 15 and 24 to 32 within the N-terminal hairpin. Mutagenic analysis of both HSV-1 and HSV-2 gDs has revealed that the first 32 amino acids of the N-terminal extension have a critical role in functional interactions of gD with all the HSV entry-fusion receptors, except for nectin-1, and that the amino acid sequence within this region governs whether nectin-2 can be recognized as an entry and fusion receptor (48, 49) (Fig. 2A). Deletions in HSV-1 or HSV-2 gD that remove one or both of the contact regions for HVEM eliminated binding to HVEM and cell fusion activity as predicted. Interestingly, cell fusion with other receptors (nectin-2 in the case of HSV-2 gD and 3-O-sulfated heparan sulfate in the case of HSV-1 gD) was also significantly reduced, whereas there was no effect on binding to or fusion with nectin-1. Amino acid substitutions within this region influenced functional interactions with HVEM, nectin-2, or 3-O-sulfated heparan sulfate as described in the legend to Fig. 2A. These results indicate that a major interface in gD for interaction with nectin-1, and probably also nectin-2, must lie downstream of amino acid 32 and outside the N-terminal hairpin. However, the presence of an N-terminal extension with the appropriate amino acid sequence is necessary for functional interactions of gD with nectin-2 and all the other entry receptors except for nectin-1. It is unclear (except for HVEM) whether the N-terminal extension makes direct contact with these other receptors or whether this region influences the conformation of other domains in gD that make the actual contacts.
FIG. 2.
Mutational analysis of HSV and EBV entry receptors and ligands. (A) Mutations in HSV gD that influence functional interactions of gD with the various entry receptors. The features of HSV-1 gD (left; in color) and HVEM (right; in gray) shown are based on the crystal structure of gD-HVEM complexes (6). The peptide backbone of gD is shown as a tube in light blue with the beta strands and alpha-helices shown in gold and green, respectively. The peptide backbone of HVEM is shown as a wire in gray with the disulfide bonds in orange. Amino acids 7 to 15 and 24 to 32, the contact regions with HVEM, are shown in black; deletion of one or both of these regions significantly reduces the number of functional interactions of HSV-1 or HSV-2 gD with all known entry-fusion receptors except nectin-1 (48). The amino acid substitutions shown in red (Q27P/R and L25P) enhance functional interactions of HSV-1 gD with nectin-2 but do not have this effect in HSV-2 gD since the latter already has significant activity with nectin-2. The substitution Q27P/R significantly reduces functional interactions of either HSV-1 or HSV-2 gD with HVEM, whereas L25P has much less effect. However, both substitutions reduce the activity of HSV-1 gD with 3-O-sulfated heparan sulfate. The substitutions shown in dark blue represent the amino acid differences found in HSV-2 gD within the first 66 amino acids. These amino acid differences are principally responsible for the greater activity of HSV-2 gD with nectin-2 (49). Thus, the combination of these amino acid substitutions or either of two single amino acid substitutions (Q27P/R or L25P) can render HSV-1 gD more active with nectin-2. (B) Mutations in an HLA class II molecule that influence interactions of gp42 with HLA class II molecules. The structure of gp42 (green) bound to an HLA class II molecule, based on X-ray crystallography of the complex (30), is shown in side view (left) or from above (right). The HLA class II α chain (purple) and β chain (blue) are illustrated, with bound peptide (brown). The residues E46 and R72 (red labels) within HLA DRβ, which areessential for gp42 binding and EBV entry, are indicated by arrows (24). K65 (blue label) mutations can have differential effects on gp42 binding and EBV entry. When the residue is changed to alanine, there is little effect on gp42 binding and EBV entry, whereas mutation to glutamic acid completely abolishes gp42 binding and EBV entry. K65 and R72 are within the alpha helix of the β chain that forms one side of the peptide-binding groove. E46 is within a loop of the β chain that extends from the base of the peptide-binding groove. The hydrophobic pocket of gp42, consisting of I159, V184, Y185, I187, F188, Y194, F198, V201, F210, and L211, is labeled and highlighted.
GAMMAHERPESVIRUS (EBV AND HHV-8) ENTRY
Like alphaherpesvirus entry, entry of EBV and HHV-8 into target cells involves interactions of multiple viral glycoproteins with multiple cell surface determinants. Understanding these interactions is important for determining gammaherpesvirus cell tropism.
(i) EBV entry.
The initial interaction of EBV with target B cells is mediated by binding of the EBV major outer envelope glycoprotein gp350/220 to complement receptor 2 (CR2; also called CD21) (reviewed in reference 17) (Fig. 1). This binding may be considered analogous to the interaction of alphaherpesvirus gC with heparan sulfate. Recombinant viruses with deletions of gp350/220 retain the ability to infect cells, although the efficiency is reduced (15). Also, gp350 is not required for EBV-induced membrane fusion in a cell-based fusion assay (11).
After gp350/220 interaction with CD21, gp42, a C-type lectin family member, binds to HLA class II molecules that are expressed abundantly on B lymphocytes (20). Like the interaction of HSV gD with its receptors, binding of gp42 to HLA class II molecules depends on multiple amino acid interactions. Initial studies identified the essential nature of a negative charge at amino acid 46 of the HLA class II sequence (12). Interestingly, this amino acid is conserved as glutamic acid in only one HLA class II DQ allele, whereas it is conserved in all HLA class II DR and DP alleles. An HLA class II molecule is required for antigen presentation and as such is very polymorphic in the human population. This observation may provide clues to understanding the association of EBV pathologies in the human host with particular HLA subtypes.
X-ray crystallography of the gp42-HLA class II complex (Fig. 2B) confirmed that glutamic acid 46 in the HLA class II molecule is critical for the interaction with gp42 (30). Other sites of interaction were also identified. Site-specific mutational analysis using the structural studies as a guide confirmed that arginine 72 of the HLA class II molecule is required for gp42 binding and EBV entry (24). Interestingly, a substitution at lysine 63 had variable effects on viral entry and gp42 binding. When the lysine residue was switched to glutamic acid, EBV entry and gp42 binding were abolished, whereas mutation to an alanine did not affect either EBV entry or binding. This residue may be important for the overall structure of the HLA class II molecule, or the electrostatic switch from a negatively to a positively charged amino acid may alter gp42 binding. Most surprising from the structural studies was the finding that gp42 has a distinct interaction site with the HLA class II molecule, in comparison to NK receptor supercomplexes such as the Ly49A-major histocompatibility complex (MHC) class I complex. Ly49A, also a member of the C-type lectin family, interacts with MHC class I molecules in an entirely different manner than the interaction of gp42 with HLA class II molecules. The homodimerization domain within Ly49A corresponds to the interaction site of gp42 with HLA DR. As a result of the binding site differences, gp42 binds much higher and closer to the peptide-binding groove of the HLA class II molecule.
Subsequent to the binding of gp42 to HLA class II molecules, fusion of the virion envelope to the plasma membrane is triggered by the concerted action of gB, gH, and gL (11). By functioning in this manner, gp42 appears to serve a role in triggering fusion similar to that of HSV gD, despite the absence of any sequence homology (Fig. 1). How the interaction of gp42 with HLA class II molecules triggers fusion is unknown, but functional as well as structural studies may offer some clues. Previous studies have shown that gp42 interacts with gH and gL via a domain contained within the amino terminus of gp42 (44). This interaction may be important for linking gp42 binding with the induction of fusion by gB, gH, and gL. The structure of gp42 complexed with an HLA class II molecule identified a large unoccupied hydrophobic pocket on the surface of gp42 (30). A similar pocket was not found on the structure of gD (6). Typically, external hydrophobic pockets are complexed with other proteins. Since this region does not contain the amino-terminal amino acids required for interaction of gp42 with gH and gL, gp42 may interact with another viral protein such as gB, or possibly a host protein, and this interaction may be crucial for triggering the fusion mediated by gB, gH, and gL. Thus, EBV may offer a special opportunity to understand the mechanism of herpesvirus-induced membrane fusion.
Entry of EBV into epithelial cells has different requirements than entry of EBV into B lymphocytes (4, 45). At least for some epithelial cells, gp42 and gp350/220 are not required. Instead, initial attachment of EBV to epithelial cells not expressing CD21 or HLA class II molecules may be mediated by gH. This alternative mode of entry does not function for EBV entry into B cells since viruses lacking gp42 cannot infect B lymphocytes (44). This observation highlights an interesting strategy that EBV has evolved to enhance viral tropism for specific cell types. EBV virions produced in B cells are more efficient for infection of epithelial cells than of B cells, whereas the converse is true for virus produced in epithelial cells (4). In B cells, gp42 is sequestered by HLA class II molecules, resulting in the production of virions that are depleted of gp42. This virus infects epithelial cells very efficiently since gp42 is not required. When virus is produced in epithelial cells, which typically do not express HLA class II molecules, gp42 is not sequestered and is found in the virion, allowing entry into B cells through binding of gp42 to HLA class II molecules.
In addition, other EBV-encoded glycoproteins appear to influence the infection of epithelial cells. EBV gp150 is a highly glycosylated protein that does not have a homolog in the alpha- or betaherpesviruses. Recombinant viruses which lack gp150 have no defects in assembly, egress, binding, or infectivity for B cells or epithelial cells; however, infection of epithelial cells is enhanced (5). Finally, EBV BMRF2, which contains an RGD motif and can interact with integrins, is thought to be important for the infection of polarized epithelial cells via basolateral surfaces (41).
(ii) HHV-8 entry.
HHV-8 has been shown to infect a variety of cell types both in vivo and in vitro, including B cells, endothelial cells, keratinocytes, and macrophages (reviewed in reference 27). This broad cellular host range suggests that HHV-8 may interact with ubiquitous host cell molecules to gain access to target cells. In part, this appears to be the case. K8.1A, an HHV-8-encoded virion envelope glycoprotein that is a positional homologue of EBV gp350/220, may be responsible for the initial binding of HHV-8 to target cells. K8.1A has been shown to bind to heparan sulfate with an affinity similar to that shown for HSV gC (3, 42). HHV-8 gB also binds heparan sulfate through a conserved region found in most of the herpesvirus gBs (1). In contrast, heparan sulfate binding by EBV gB has not been reported. Finally, HHV-8 gB contains an RDG motif which binds specifically to integrin α3β1 (CD49c/29) (2, 43). Binding to integrins may not be a common feature of herpesvirus gBs; at least, no other gB has a similar conserved RDG motif. The integrin α3β1 is broadly expressed and has been detected on all cells susceptible to infection by HHV-8, including human foreskin fibroblasts and B, epithelial, endothelial, and 293 cells (2). Finally, HHV-8-induced membrane fusion in a cell-based assay with susceptible cells as targets requires only gB, gH, and gL (33).
OTHER IMPORTANT TOPICS
There are at least three important topics not addressed in this minireview, including (i) the actual mechanism of herpesvirus-induced membrane fusion, (ii) the identities and activities of viral proteins that may modulate herpesvirus entry and cell fusion, and (iii) the potential for signal transduction following the interactions of herpesvirus ligands with their binding and entry receptors. These topics are the subjects of active study and will deserve reviews in the not-too-distant future.
SUMMARY
The entry of herpesviruses into cells depends upon interactions of several viral glycoproteins with multiple cell surface receptors. Also, each herpesvirus may have evolved multiple pathways for entry into different cell types, as is evident for HSV and EBV. The broad host range of HSV is consistent with its use of cell surface heparan sulfate as a binding receptor and both 3-O-sulfated heparan sulfate and multiple conserved and widely expressed proteins as entry receptors. The more limited host range of EBV (at least in the case of its B-cell target) is also consistent with its use of binding and entry receptors that are found together on very few cell types, chief of which are B cells. Much remains to be learned about the actual requirements for entry of these viruses into the target cell types that are critical for disease and about the mechanisms of virus-induced membrane fusion.
ADDENDUM IN PROOF
A recent publication (S. A. Connolly, D. J. Landsburg, A. Carfi, D. C. Wiley, G. H. Cohen, and R. J. Eisenberg, J. Virol. 77:8127-8140, 2003) describes substitution of most of the gD residues that make contact with HVEM and the effects of these substitutions on functional interactions of mutated gD with HVEM and other receptors.
Acknowledgments
Research by P.G.S. is supported by Public Health Service grants CA21776 from the National Cancer Institute and AI36293, AI31494, AI49394, and AI53774 from the National Institute of Allergy and Infectious Diseases. R.L. is supported by Public Health Service grants CA62234, CA73507, and CA93444 from the National Cancer Institute and DE13127 from the National Institute of Dental and Craniofacial Research. R.L. is a Scholar of the Leukemia Society of America.
We thank current and former members of the Longnecker, Spear, and Jardetzky laboratories for their contributions to the work described.
REFERENCES
- 1.Akula, S. M., N. P. Pramod, F. Z. Wang, and B. Chandran. 2001. Human herpesvirus 8 envelope-associated glycoprotein B interacts with heparan sulfate-like moieties. Virology 284:235-249. [DOI] [PubMed] [Google Scholar]
- 2.Akula, S. M., N. P. Pramod, F.-Z. Wang, and B. Chandran. 2002. Integrin α3β1 (CD49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108:407-419. [DOI] [PubMed] [Google Scholar]
- 3.Birkmann, A., K. Mahr, A. Ensser, S. Yaguboglu, F. Titgemeyer, B. Fleckenstein, and F. Neipel. 2001. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 75:11583-11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borza, C. M., and L. M. Hutt-Fletcher. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594-599. [DOI] [PubMed] [Google Scholar]
- 5.Borza, C. M., and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus recombinant lacking expression of glycoprotein gp150 infects B cells normally but is enhanced for infection of epithelial cells. J. Virol. 72:7577-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 7.Cocchi, F., M. Lopez, P. Dubreuil, G. Campadelli-Fiume, and L. Menotti. 2001. Chimeric nectin 1-poliovirus receptor molecules identify a nectin 1 region functional in herpes simplex virus entry. J. Virol. 75:7987-7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, R. J. Eisenberg, and G. H. Cohen. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 76:10894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis-Poynter, N., S. Bell, T. Minson, and H. Browne. 1994. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J. Virol. 68:7586-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 11.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106-114. [DOI] [PubMed] [Google Scholar]
- 12.Haan, K. M., and R. Longnecker. 2000. Coreceptor restriction within the HLA-DQ locus for Epstein-Barr virus infection. Proc. Natl. Acad. Sci. USA 97:9252-9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haarr, L., D. Shukla, E. Rødahl, M. C. Dal Canto, and P. G. Spear. 2001. Transcription from the gene encoding the herpesvirus entry receptor nectin-1 (HveC) in nervous tissue of adult mouse. Virology 287:301-309. [DOI] [PubMed] [Google Scholar]
- 14.Huber, M. T., and T. Compton. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J. Virol. 72:8191-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janz, A., M. Oezel, C. Kurzeder, J. Mautner, D. Pich, M. Kost, W. Hammerschmidt, and H. J. Delecluse. 2000. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol. 74:10142-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, D. C., M. Webb, T. W. Wisner, and C. Brunetti. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J. Virol. 75:821-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In D. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 18.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon, B., B.-S. Kim, H. R. Cho, J.-E. Park, and B. S. Kon. 2003. Involvement of tumor necrosis factor receptor superfamily (TNFRSF) members in the pathogenesis of inflammatory diseases. Exp. Mol. Med. 35:8-16. [DOI] [PubMed] [Google Scholar]
- 20.Li, Q., M. K. Spriggs, S. Kovats, S. M. Turk, M. R. Comeau, B. Nepom, and L. M. Hutt-Fletcher. 1997. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 71:4657-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez, W. M., and P. G. Spear. 2002. Amino acid substitutions in the V domain of nectin-1 (HveC) that impair entry activity for herpes simplex viruses 1 and 2 but not for pseudorabies virus or bovine herpesvirus 1. J. Virol. 76:7255-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez, W. M., and P. G. Spear. 2001. Structural features of nectin-2 (HveB) required for herpes simplex virus entry. J. Virol. 75:11185-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mauri, D. N., R. Ebner, R. I. Montgomery, K. D. Kochel, T. C. Cheung, G.-L. Yu, S. Ruben, M. Murphy, R. J. Eisenberg, G. H. Cohen, P. G. Spear, and C. F. Ware. 1998. LIGHT, a new member of the TNF superfamily, and lymphotoxin α are ligands for herpesvirus entry mediator. Immunity 8:21-30. [DOI] [PubMed] [Google Scholar]
- 24.McShane, M. P., M. M. Mullen, K. M. Haan, T. S. Jardetzky, and R. Longnecker. 2003. Mutational analysis of the HLA class II interaction with the Epstein-Barr virus glycoprotein 42. J. Virol. 77:7655-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milne, R. S. B., S. A. Connolly, C. Krummenacher, R. J. Eisenberg, and G. H. Cohen. 2001. Porcine HveC, a member of the highly conserved HveC/nectin 1 family, is a functional alphaherpesvirus receptor. Virology 281:315-328. [DOI] [PubMed] [Google Scholar]
- 26.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 27.Moore, P. S., and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus, p. 2803-2833. In D. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 28.Mori, Y., X. Yang, P. Akkapaiboon, T. Okuno, and K. Yamanishi. 2003. Human herpesvirus 6 variant A glycoprotein H-glycoprotein L-glycoprotein Q complex associates with human CD46. J. Virol. 77:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muggeridge, M. I. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 81:2017-2027. [DOI] [PubMed] [Google Scholar]
- 30.Mullen, M. M., K. M. Haan, R. Longnecker, and T. S. Jardetzky. 2002. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol. Cell 9:375-385. [DOI] [PubMed] [Google Scholar]
- 31.Nicola, A. V., A. M. McEvoy, and S. E. Straus. 2003. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J. Virol. 77:5324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pertel, P., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 33.Pertel, P. E. 2002. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol. 76:4390-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakisaka, T., T. Taniguchi, H. Nakanishi, K. Takahashi, M. Miyahara, W. Ikeda, S. Yokoyama, Y. F. Peng, K. Yamanishi, and Y. Takai. 2001. Requirement of interaction of nectin-1 alpha/HveC with afadin for efficient cell-cell spread of herpes simplex virus type 1. J. Virol. 75:4734-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shukla, D., M. Dal Canto, C. L. Rowe, and P. G. Spear. 2000. Striking similarity of murine nectin-1α to human nectin-1α (HveC) in sequence and activity as a gD receptor for alphaherpesvirus entry. J. Virol. 74:11773-11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13-22. [DOI] [PubMed] [Google Scholar]
- 37.Shukla, D., and P. G. Spear. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Investig. 108:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 39.Struyf, F., W. M. Martinez, and P. G. Spear. 2002. Mutations in the N-terminal domains of nectin-1 and nectin-2 reveal differences in requirements for entry of various alphaherpesviruses and for nectin-nectin interactions. J. Virol. 76:12940-12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takai, Y., and H. Nakanishi. 2003. Nectin and afadin: novel organizers of intercellular junctions. J. Cell Sci. 116:17-27. [DOI] [PubMed] [Google Scholar]
- 41.Tugizov, S. M., J. W. Berline, and J. M. Palefsky. 2003. Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat. Med. 9:307-314. [DOI] [PubMed] [Google Scholar]
- 42.Wang, F. Z., S. M. Akula, N. P. Pramod, L. Zeng, and B. Chandran. 2001. Human herpesvirus 8 envelope glycoprotein K8.1A interaction with the target cells involves heparan sulfate. J. Virol. 75:7517-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, F. Z., S. M. Akula, N. Sharma-Walia, L. Zeng, and B. Chandran. 2003. Human herpesvirus 8 envelope glycoprotein B mediates cell adhesion via its RGD sequence. J. Virol. 77:3131-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, X., and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J. Virol. 72:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, X., W. J. Kenyon, Q. Li, J. Mullberg, and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wittels, M., and P. G. Spear. 1991. Penetration of cells by herpes simplex virus does not require a low pH-dependent endocytic pathway. Virus Res. 18:271-290. [DOI] [PubMed] [Google Scholar]
- 47.Yoon, M., and P. G. Spear. 2002. Disruption of adherens junctions liberates nectin-1 to serve as receptor for herpes simplex virus entry. J. Virol. 76:7203-7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon, M., A. Zago, D. Shukla, and P. G. Spear. 2003. Mutations in the N termini of herpes simplex virus type 1 and 2 gDs alter functional interactions with the entry/fusion receptors HVEM, nectin-2, and 3-O-sulfated heparan sulfate but not with nectin-1. J. Virol. 77:9221-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zago, A., and P. G. Spear. 2003. Differences in the N termini of herpes simplex virus type 1 and 2 gDs that influence functional interactions with the human entry receptor nectin-2 and an entry receptor expressed in Chinese hamster ovary cells. J. Virol. 77:9695-9699. [DOI] [PMC free article] [PubMed] [Google Scholar]