Abstract
Hepatitis C virus (HCV) core protein plays an important role in the formation of the viral nucleocapsid and a regulatory protein involved in hepatocarcinogenesis. In this study, we have identified proteasome activator PA28γ (11S regulator γ) as an HCV core binding protein by using yeast two-hybrid system. This interaction was demonstrated not only in cell culture but also in the livers of HCV core transgenic mice. These findings are extended to human HCV infection by the observation of this interaction in liver specimens from a patient with chronic HCV infection. Neither the interaction of HCV core protein with other PA28 subtypes nor that of PA28γ with other Flavivirus core proteins was detected. Deletion of the PA28γ-binding region from the HCV core protein or knockout of the PA28γ gene led to the export of the HCV core protein from the nucleus to the cytoplasm. Overexpression of PA28γ enhanced the proteolysis of the HCV core protein. Thus, the nuclear retention and stability of the HCV core protein is regulated via a PA28γ-dependent pathway through which HCV pathogenesis may be exerted.
Hepatitis C virus (HCV) is the causative agent in most cases of acute and chronic non-A, non-B hepatitis (16, 51). Over 50% of patients with acute infection evolve into a chronic carrier state (26), and persistent infection frequently results in chronic hepatitis. Chronic HCV infection may lead to the development of cirrhosis and eventually hepatocellular carcinoma (21, 51). HCV belongs to the Flaviviridae family, a family that also includes Japanese encephalitis virus (JEV) and Dengue fever virus (DEN), and possesses a viral genome consisting of a single positive-strand RNA of approximately 9.6 kb and encoding approximately 3,000 amino acids in a single polypeptide (9, 58). HCV proteins are produced as a single polypeptide that is posttranslationally cleaved by host cellular peptidases and viral proteases to yield at least 10 viral proteins (7, 10, 12, 54).
A comparison of the genome structure of HCV with other flaviviruses, as well as the observation of a specific interaction of viral sense RNA with HCV core protein in cells (53, 68), suggests that the HCV core protein forms the nucleocapsid with viral genome RNA. An HCV core protein consisting of the N-terminal 191 amino acids is generated by protein cleavage by host signal peptidase(s) (37, 52). The HCV core protein is further processed into a mature core protein lacking its C-terminal hydrophobic region by either an unknown host protease (52, 65) or by a signal peptide peptidase (36). The matured core protein is retained on the endoplasmic reticulum (ER) either by an interaction with immature core protein on the ER membrane (29) or via E1 envelope protein (32). The C-terminal hydrophobic region between amino acids 174 and 191 is essential for HCV core protein anchoring on the ER membrane and for the signal sequence of E1 protein to translocate into the ER lumen. Core proteins truncated at the C termini are mainly localized in the nucleus and, to lesser extent, in the cytoplasm (8, 55). Further processing of the HCV core protein yields a 16-kDa product whose C terminus is near amino acid 151; this protein translocates into the nucleus (30, 31, 55).
We have reported that hepatic steatosis and hepatocellular carcinoma are induced in transgenic mice expressing the HCV core protein, suggesting that the HCV core protein has an oncogenic activity in liver. These data further suggest that the cellular components responsible for HCV-induced carcinogenesis exist not only in humans but also in mice (39). Thus, the identification of core-binding partners in mammalian cells could potentially clarify the molecular mechanism(s) of HCV-induced hepatocarcinogenesis. Several cytoplasmic and nuclear proteins have been reported to bind the HCV core protein to both induce carcinogenesis and facilitate virion formation. A report has suggested that the HCV core protein may sequester LZIP, a putative tumor suppressor, in the cytoplasm, with a resulting enhancement of carcinogenesis of NIH 3T3 cells (18). The HCV core protein interacts with the C-terminal region of p53 and enhances its transcriptional activity through augmentation of p53 DNA binding affinity (46). A putative cellular RNA helicase, primarily localized in the nucleus and to a lesser extent in the cytoplasm, interacts with the N-terminal 40 amino acids of the HCV core protein and is colocalized with the HCV core protein in both cellular locations (33, 67). It was recently reported that the HCV core protein directly binds and activates STAT3 by phosphorylation through a JAK-independent pathway; cells overexpressing both HCV core protein and STAT3 exhibited anchorage-independent growth and tumorigenesis (66). These reports suggest that the HCV core protein functions in both the nucleus and cytoplasm.
In this report, we identify proteasome activator PA28γ (11S regulator) as an HCV core binding protein by the yeast two-hybrid system. It is well known that PA28γ enhances the latent proteasome activity of the 20S proteasome and is predominantly localized in the nucleus (48, 62). PA28γ is conserved across the animal kingdom from invertebrates to vertebrates (34, 47), although the biological significance of PA28γ is largely unknown. Here, we demonstrate through several lines of evidence that PA28γ specifically interacts with the HCV core protein and remains in the nucleus, consequently regulating its stability.
MATERIALS AND METHODS
Plasmids.
Human PA28γ cDNA was isolated from a human fetal brain library by the advanced yeast two-hybrid technique (MATCHMAKER two-hybrid system 3; Clontech, Palo Alto, Calif.) with amino acids 1 to 173 of the HCV core protein as bait. The gene encoding HCV core protein was amplified from HCV strain J1 (genotype 1b) (2) and cloned into the pGBKT7 vector (pGBKT7HCVCore173). The cDNA of PA28γ was amplified by PCR with Pfu turbo DNA polymerase (Stratagene, La Jolla, Calif.) and cloned into pEFFlagpGPKpuro (17), pEGFP-C3, and pDsRed2N1; the sequence was verified by DNA sequencing. The gene encoding PA28γ, with amino acids 82 to 90 deleted, was amplified by splicing the overlapping extension (13, 15) and cloned into pDsRed2N1 (Clontech). Other mutant constructs of the HCV core protein were introduced into pCAG-GS (45) and pEGFP-C3 (Fig. 1). The genes encoding the core proteins of DEN (amino acids 1 to 100) and JEV (amino acids 1 to 105), both lacking the C-terminal hydrophobic regions, were amplified by PCR and cloned into pEGFP-C3. F protein was shown to be synthesized by ribosomal frameshift of the core protein-coding sequence (64). The gene encoding F protein of the −2/+1 frame attached to a Flag tag at N terminus was generated by deletion of one adenine in codon 10 and then introduced into the pEFFlagpGKpuro vector. The gene encoding human Bad or human FKBP attached to a hemagglutinin (HA) tag were isolated from human fetal libraries and introduced into pIRESbleo (Clontech). Mouse anti-Flag (M2), mouse anti-HA (HA.11), and mouse anti-cytochrome c oxygenase subunit IV (20E) antibodies were purchased from Sigma (St. Louis, Mo.), Babco (Richmond, Calif.), and Molecular Probes (Eugene, Oreg.), respectively. Horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit antisera were purchased from ICN Pharmaceuticals (Aurora, Ohio). Rabbit antisera against synthetic peptides corresponding to amino acids 70 to 85 of PA28γ, 5 to 19 of PA28α, or 15 to 31 of PA28β were purchased from AFFINITI (Exeter, United Kingdom).
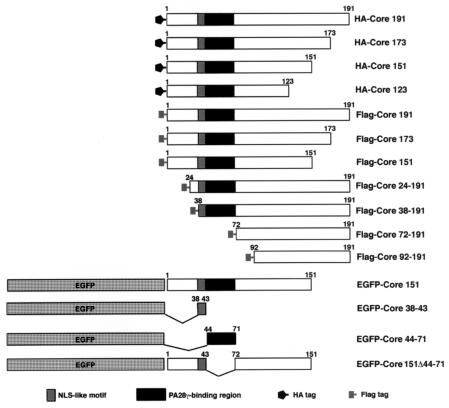
FIG. 1.
Expression constructs of HCV core protein used in this study. The genes encoding HA-tagged HCV mutants or Flag-tagged HCV mutants were cloned into pCAG-GS vectors (45) while the genes encoding EGFP-fused proteins were introduced into pEGFP-C3. Other vectors are described in the text or in figure legends.
Yeast two-hybrid assay and library screening.
The bait vector, pGBKT7HCVCore173, described above, was introduced into Saccharomyces cerevisiae strain AH109. The yeast containing pGBKT7HCVCore173 were grown in yeast extract-peptone-dextrose medium and transfected with library plasmids based on pACT2 (Clontech), a vector encoding ampicillin resistance. Clones (2 × 106) from a human fetal brain library were screened (Clontech). The yeast clones encoding pGBKT7-53 and pGADT7-T (Clontech) were used as a positive control while yeast containing pGBKT7 and pGADT7 were considered the negative control. Yeast colonies grown on dropout plates lacking tryptophan, leucine, histidine, and adenine were inoculated on two new dropout plates lacking leucine and tryptophan. One of the two plates was subjected to β-galactosidase assay according to the method of Duttweiller (11), and the other plate was kept at 4°C as a master plate. One of 85 dropout-plate-positive clones led to dark blue staining on a β-galactosidase assay plate to the same extent as the positive control and was called C1-24. The total DNA was recovered from C1-24 and introduced into Escherichia coli strain JM109. The plasmid from C1-24 was recovered from the clones independently grown on the Luria-Bertani plate containing 10 μg of ampicillin/ml. The sequences of the insert were determined by using the Big Dye terminator reaction mixture and an ABI Prism 310 genetic analyzer (Applied Biosystems Japan, Tokyo, Japan). All 3 clones contained the same insert encoding human PA28γ.
Transfection, immunoprecipitation, and immunoblotting.
Plasmid vectors were transfected into HeLa or 293T cells by liposome-mediated transfection. Immunoprecipitation and liposome-mediated transfection were reported previously (38). Human and mouse liver samples were washed twice with cold phosphate-buffered saline (PBS) and then homogenized in 10 volumes of 20 mM Tris-HCl (pH 7.4) containing 135 mM NaCl, 1% Triton X-100, 10% glycerol, supplemented with 0.5 μg of Pefabloc (Pentapharm, Munich, Germany)/ml, 1 mM phenylmethylsulfonyl fluoride, 1 μg of soybean trypsin inhibitor/ml, 50 mM NaF, and 5 mM Na3VO4 (Tris lysis buffer). Mouse monoclonal antibodies to the HCV core protein (clones 11-4, 11-10, and 11-14) (5) were used for immunoprecipitation and immunoblotting (38).
Laser scanning confocal microscopy.
Transfectants were grown on glass slides at 37°C overnight, washed twice with PBS, and fixed with 4% paraformaldehyde for 15 min at room temperature. Enhanced green fluorescent protein (EGFP) fusion protein-expressing cells were examined directly. For samples requiring immunostaining, cells were washed twice with PBS after fixation, permeabilized for 15 min at room temperature with PBS containing 0.5% Triton X-100, and incubated in PBS containing 1% bovine serum albumin (PBS-BSA) in order to block nonspecific binding. Cells were then incubated at room temperature for 30 min in PBS-BSA containing 1 μg of rabbit anti-PA28γ antiserum (AFFINITI)/ml and mouse anti-HA antibody. Cells were washed three times with PBS-BSA and incubated at room temperature for 30 min in PBS-BSA containing 0.5 μg of Alexa Fluor 488-conjugated anti-rabbit immunoglobulin G (IgG) and Alexa Fluor 594-conjugated anti-mouse IgG antisera (Molecular Probes)/ml. After being washed three times with PBS-BSA, all samples were visualized with a Bio-Rad (Tokyo, Japan) confocal laser-scanning microscope.
Mouse embryonic fibroblasts isolated from PA28γ knockout mice.
Embryonic fibroblasts from PA28γ knockout mice were prepared as previously described (42). Cells were cultured at 37°C (5% CO2) in Dulbecco's modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin, streptomycin, sodium pyruvate, and nonessential amino acids. Plasmid vectors were transfected into cells after three passages, and the intracellular localization of proteins was examined.
Time-lapse microscopy.
HeLa cells or mouse embryonic fibroblasts were seeded on 35-mm-diameter culture dishes, grown overnight to 70% confluence, and transfected with 2.5 μg of plasmid DNA with Lipofectamine 2000 per the manufacturer's instructions (Invitrogen Corp., Carlsbad, Calif.). For immunofluorescent microscopy, cells were incubated at 37°C overnight after transfection, trypsinized, reseeded, and cultivated for 20 h. For time-lapse microscopy, cells were seeded on 35-mm-diameter dishes, grown for 24 h after transfection, and incubated in Dulbecco's modified Eagle’s medium containing 10% fetal calf serum at 37°C. Cells were viewed at 37°C (5% CO2) with an Olympus (Tokyo, Japan) IX71 and Cool SNAP HQ charge-coupled device camera (Roper Scientific JAPAN, Tokyo, Japan). Digital images were analyzed with Metamorph software (Universal Imaging, Downingtown, Pa.).
Effect of MG132 on stability of the HCV core protein.
To determine the effect of proteasome inhibitors on the degradation of HCV core proteins in the presence of overexpressed PA28γ, 2 × 105 293T cells were transfected with expression plasmids encoding the HCV core protein and PA28γ by lipofection on 35-mm-diameter plates. The proteasome inhibitor MG132 (Sigma) or solvent, dimethyl sulfoxide, was added as a 10−3 volume of medium into wells at 8 h posttransfection. Cells were harvested at 24 h posttransfection and lysed in lysis buffer as described above. Proteins were detected by Western blotting with mouse monoclonal anti-Flag, anti-HA, goat polyclonal anti-actin (Santa Cruz Biotechnology, Santa Cruz, Calif.) or rabbit polyclonal anti-PA28γ.
RESULTS
Isolation of PA28γ cDNA from human libraries.
To determine the protein(s) that interact with HCV core protein in mammalian cells, we choose to employ a yeast two-hybrid system with the HCV core protein as bait. Human fetal brain and liver libraries were used for this screening because it is not known whether the target protein is specifically expressed in the liver. Many light blue colonies emerged on dropout plates, but these were eliminated from further screening so that proteins exhibiting strong binding could be examined more fully. Several clones exhibited dark blue color on a dropout plate containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside to an extent similar to that of the positive control containing p53 and large T antigen. No gene has been included which has previously been reported as a core-binding protein in the dark blue colonies, and we selected the darkest one. The total DNA was extracted from this clone and introduced into E. coli strain JM109 with the goal of recovering the pACT2 plasmid encoding the candidate core-binding protein. The nucleotide sequence of the DNA insert was determined from three independent colonies. The sequence isolated from the positive clone included the 5′ and 3′ noncoding regions as well as the full coding region of proteasome activator PA28γ; all sequences were in frame. There are two splicing variants of PA28γ in human tissue (3, 23). The isolated cDNA of PA28γ encoded the major isoform that is comprised of 254 amino acids; this isoform demonstrates 100% identity with mouse PA28γ based on amino acid sequence. The isolated pACT2 plasmid containing PA28γ cDNA was introduced into yeast strain AH109 together with either an empty bait plasmid, pGBKT7, or a plasmid encoding the HCV core protein, pGBKT7HCVCore173, in order to confirm that the isolated plasmid encodes an HCV core-binding protein. The yeast clone containing pACT2-PA28γ and pGBKT7HCVCore173 grew on a dropout plate deficient in leucine, tryptophan, histidine, and adenine, but the yeast clone containing pACT2-PA28γ and pGBKT7 did not (data not shown). These data suggest that PA28γ binds to the HCV core protein in yeast. The cDNAs of HCV core protein and its mutants were introduced into several mammalian expression vectors as shown in Fig. 1.
Interaction of the HCV core protein with PA28γ in mammalian cells, livers of HCV core transgenic mice, and a patient with chronic hepatitis C.
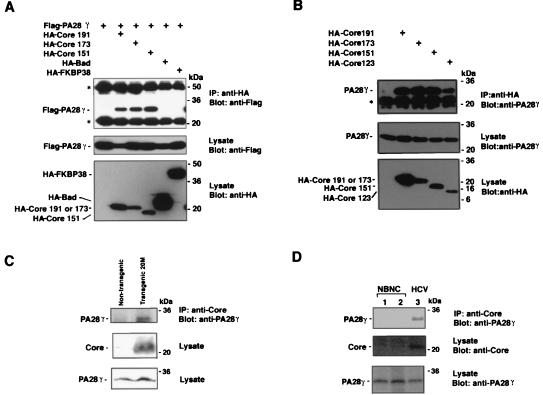
Because it is generally known that many false-positive clones are identified by using the yeast two-hybrid system, protein-protein interaction and coincidence of intracellular localization between bait and prey proteins should be examined in mammalian cells. When Flag-tagged PA28γ (Flag-PA28γ) was coexpressed in 293T cells with HA-Core191, HA-Core173, HA-Core151, HA-Bad, or HA-FKBP, Flag-PA28γ was coprecipitated with HA-Core191, HA-Core173, and HA-Core151 but not with HA-Bad and HA-FKBP by mouse anti-HA antibody. The interaction of Flag-PA28γ with HA-Bad and HA-FKBP was not observed even though these constructs were expressed at a higher level than the HA-Core proteins (Fig. 2A). To eliminate the possibility of an artificial interaction of the HCV core protein with PA28γ due to overexpression, the association of HCV core proteins with endogenous PA28γ was examined. Endogenous PA28γ was coprecipitated with HCV core proteins in HA-Core-expressing 293T cells but not in nontransfected cell lysates (Fig. 2B).
FIG. 2.
Interaction of HCV core protein with PA28γ. (A) Human embryonic kidney 293T cells were transfected with the expression plasmids encoding HCV core proteins and/or PA28γ. A Flag epitope tag was added to PA28γ at its amino terminus while an HA epitope tag was added to the HCV core protein and control proteins (Bad and FKBP38). PA28γ was able to be coimmunoprecipitated with the HCV core protein by anti-HA antibody and was visualized by Western blotting with anti-Flag antibody. HA-Core 151 was expressed by using two times amount of plasmid DNA compared to HA-Core 191 and 173 because coexpression with PA28γ decreased the amount of HA-Core 151. (B) 293T cells were transfected with the expression plasmids encoding HA-tagged HCV core proteins. Endogenous PA28γ was coimmunoprecipitated with HCV core proteins and was detected by immunoblotting with anti-PA28γ antiserum. Asterisks indicate IgG bands. (C) Liver homogenates of HCV core transgenic mice and nontransgenic mice were immunoprecipitated (IP) with anti-core antibody; endogenous PA28γ was coprecipitated with HCV core protein and was detected by immunoblotting with anti-PA28γ antibody. (D) Liver homogenates of non-B and non-C hepatitis patients (lanes 1 and 2) and a patient with chronic hepatitis C (lane 3) were immunoprecipitated with anti-core antibody. Endogenous PA28γ was coimmunoprecipitated with HCV core protein and was detected by immunoblotting with rabbit anti-PA28γ antiserum.
Hepatic steatosis and hepatocellular carcinoma have been shown to be induced in transgenic mice expressing the HCV core protein; in this system, expression levels of the HCV core protein in mouse livers were similar to those in patients with chronic hepatitis C (25, 39). The amino acid sequence of human PA28γ is identical to that of mouse PA28γ (22, 23, 44). Liver tissue of HCV core transgenic and nontransgenic mice were homogenized in lysis buffer. Endogenous PA28γ was coprecipitated with HCV core protein by anti-HCV core antibody in liver lysates of HCV core transgenic mice but not in those of nontransgenic mice (Fig. 2C), indicating that the HCV core protein specifically interacts with PA28γ in the liver of core transgenic mice. To further confirm the specific interaction of the HCV core protein with endogenous PA28γ, this interaction was examined in liver specimens from a patient with chronic hepatitis C infection (Fig. 2D). Endogenous PA28γ was also coprecipitated with HCV core protein in liver lysates from this patient (Fig. 2D, lane 3), but not in patients with non-B and non-C hepatitis (Fig. 2D, lanes 1 and 2), by anti-HCV core antibody. These results indicate that the HCV core protein specifically binds to PA28γ not only in mammalian cell lines but also in liver tissue.
Intracellular localization of the HCV core protein with PA28α, β, and γ.
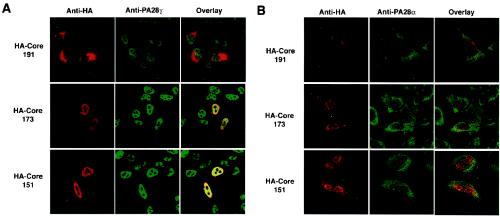
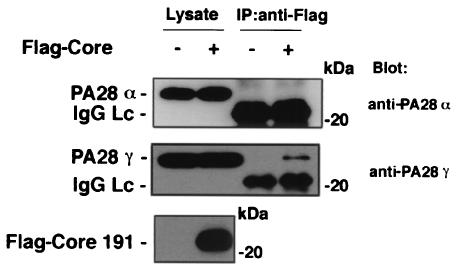
The nonessential, flexible loop region of PA28, termed the homologue-specific insert region, lies between the N terminus of the protein and the proteasome activation domain. This region does not show any homology with other PA28 isotypes (49). The nuclear localization signal (NLS) of PA28γ encompasses amino acids 82 to 90 and lies within the homologous specific insert (amino acids 72 to 102); there is no NLS in PA28α and β. PA28γ is primarily localized to the nucleus in mammalian cells through its NLS motif, but PA28α and β are predominantly found in the cytoplasm (6). Figure 3 shows the intracellular localization of the HCV core protein and endogenous PA28γ and PA28α. HA-Core191 was predominantly detected in the cytoplasm and to a lesser extent in the nucleus or perinuclear region in HeLa cells. Conversely, HA-Core173 and HA-Core151 were primarily found in the nucleus with less cytoplasmic staining. Endogenous PA28γ was visualized by indirect immunostaining with polyclonal rabbit anti-PA28γ antiserum and was predominantly detected in the nucleus of HeLa cells irrespective of the expression of HCV core proteins. HA-Core191 was partially colocalized with PA28γ in the nucleus. In contrast to these findings, a large proportion of HA-Core151 or 173 was found to be colocalized with PA28γ in the nucleus. PA28α and β share 41.3 and 33.6% homology to PA28γ, respectively. A heteroheptamer of PA28α and β binds to the 20S proteasome in the cytoplasm to activate the peptidase activity of this proteasome (1). Endogenous PA28α was predominantly detected in the cytoplasm and, to a lesser extent, in the nucleus. When HA-Core191 was expressed in HeLa cells, it was mainly localized to the cytoplasm, but it did not colocalize with PA28α. When HA-Core151 and 173 were expressed in HeLa cells, endogenous PA28α was not translocated from the cytoplasm to the nucleus, and no colocalization with HCV core proteins was observed. Similar results were also obtained in 293T cells (data not shown). Endogenous PA28α was not able to be coimmunoprecipitated with Flag-HCV Core191 in 293T cells. Endogenous PA28γ, however, was clearly coprecipitated with the core protein (Fig. 4). Endogenous PA28β was not colocalized with HCV core proteins in HeLa cells by indirect immunostaining (data not shown). These data indicate that the HCV core protein interacts with PA28γ but not with PA28α and β.
FIG. 3.
Intracellular localization of HCV core protein with PA28α and γ. HeLa cells were transfected with plasmids encoding HA-Core proteins and then fixed with paraformaldehyde. The HCV core protein was visualized by indirect immunostaining with anti-HA antibody. Endogenous PA28γ and α were visualized by indirect immunostaining with rabbit anti-PA28γ (A) and anti-PA28α (B) antisera, respectively. All samples were observed with a confocal microscope.
FIG. 4.
HCV core protein does not bind to PA28α. After overnight cultivation, 293T cells were transfected with the expression plasmid encoding Flag-HCV Core191. Immunoprecipitation (IP) was performed as described in Materials and Methods. Endogenous PA28α and PA28γ coprecipitated with HCV core protein were stained with anti-PA28α and anti-PA28γ antisera, respectively. Lc, light chain.
Intracellular localization of Flaviviridae core proteins with PA28γ.
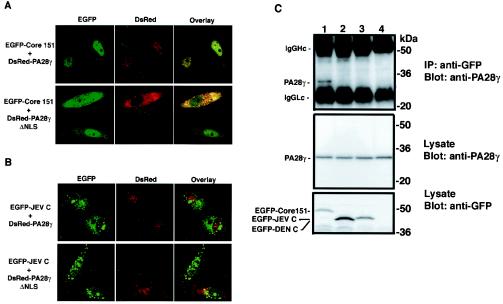
The interaction of the HCV core protein with PA28γ was demonstrated by coimmunoprecipitation, and the colocalization of these proteins was examined by immunostaining. It was still unknown, however, whether the HCV core protein interacts with PA28γ under living cell conditions. Since the nuclear localization of PA28γ is dependent on a c-Myc-like NLS, deletion of the NLS in PA28γ should shift its localization into the cytoplasm. When PA28γ was fused to the C terminus of the red fluorescence protein (DsRed) (DsRed-PA28γ) and coexpressed with EGFP-Core151 in HeLa cells (Fig. 1), EGFP-Core151 colocalized with DsRed-PA28γ in the nucleus (Fig. 5A, upper panels). In the presence of DsRed-PA28γ lacking the NLS (DsRed-PA28γΔNLS), however, EGFP-Core151 was predominantly detected in the cytoplasm and was colocalized with DsRed-PA28γΔNLS (Fig. 5A, lower panels). The detection of EGFP-Core151 in the nucleus of cells overexpressing DsRed-PA28γΔNLS was probably due to the interaction of the core protein with endogenous PA28γ in the nucleus. The cytoplasmic localization of EGFP-Core151 was also detected with DsRed-PA28γΔNLS in 293T cells (data not shown). These data indicate that the HCV core protein binds to PA28γ in living cells.
FIG. 5.
Interaction of Flaviviridae core proteins with PA28γ. HeLa cells were transfected with the expression plasmids encoding DsRed-PA28γ (upper panels) or DsRed-PA28γΔNLS (lower panels) together with EGFP-Core151 (A) or EGFP-JEV C (B). All samples were observed with a confocal microscope. (C) The cells were transfected with the expression plasmid encoding EGFP-Core151 (lane 1), EGFP-DEN C (lane 2), or EGFP-JEV C (lane 3) and then harvested at 36 h posttransfection. EGFP fusion proteins were precipitated with anti-GFP antibody. Endogenous PA28γ was coimmunoprecipitated with anti-GFP antibody and then was visualized by immunoblotting with anti-PA28γ antiserum. The untransfected cells were used as a negative control (lane 4). IP, immunoprecipitation; Hc, heavy chain; Lc, light chain.
DEN and JEV are both members of the Flaviviridae family, which also includes HCV (35, 50). The HCV core protein shares 22 and 30% homology with the DEN and JEV core proteins within the N-terminal 50 amino acids, respectively. Also similar to HCV, the core proteins of DEN and JEV are basic. The EGFP-fused JEV core protein lacking the C-terminal hydrophobic region (EGFP-JEV C) can be visualized in both the cytoplasm and nucleus (Fig. 5B, upper panels). The intracellular localization of EGFP-JEV C was quite distinct from that of DsRed-PA28γ, and coexpression with DsRed-PA28γΔNLS did not affect the subcellular localization of the protein (Fig. 5B, lower panels). Similar results were obtained by coexpression of the EGFP-fused DEN core protein lacking the C-terminal hydrophobic region (EGFP-DEN C). EGFP-DEN C was not colocalized with DsRed-PA28γ and was not affected by expression of DsRed-PA28γΔNLS (data not shown). Endogenous PA28γ was coprecipitated with EGFP-Core151 by anti-GFP antibody but not with EGFP-DEN C or EGFP-JEV C (Fig. 5C). These data suggest that PA28γ specifically interacts with the HCV core protein but not with DEN and JEV core proteins in living cells.
Mapping of the PA28γ-binding region of the HCV core protein.
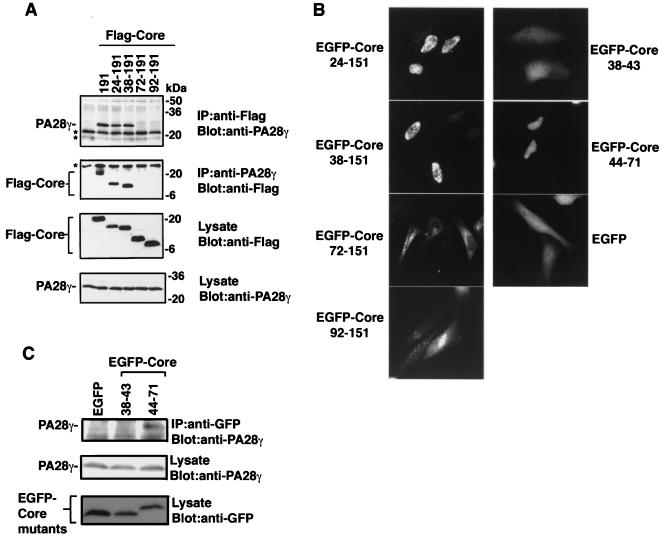
To determine the region of the HCV core protein responsible for PA28γ binding, the interactions of PA28γ with deletion mutants of the HCV core protein were examined. When Flag-Core mutants (Fig. 1) were expressed in 293T cells, endogenous PA28γ was coimmunoprecipitated with Flag-Core191, Flag-Core24-191, and Flag-Core38-191 by anti-Flag antibody but not with Flag-Core72-191 and Flag-Core92-191; the levels of protein expression were the same for all constructs (Fig. 6A). Conversely, Flag-Core191, Flag-Core24-191, and Flag-Core38-191, but not Flag-Core72-191 and Flag-Core92-191, were coprecipitated with endogenous PA28γ by anti-PA28γ antibody. These results indicate that the N-terminal 37 amino acids of the HCV core protein are not involved in the interaction with PA28γ. Because HA-Core151 was shown to interact with PA28γ (Fig. 2A) and localized to the nucleus (Fig. 3A), we examined the effect of deletion the N-terminal amino acids on the localization of Core-151 in living cells by using EGFP-Core151 (Fig. 5A). EGFP-Core24-151 and EGFP-Core38-151 were localized entirely within the nucleus, and EGFP-Core72-151 and EGFP-Core92-151 were predominantly localized in the cytoplasm (Fig. 6B). These results give rise to the question of whether amino acids 38 to 71 of the HCV core protein might be involved in the interaction with PA28γ and in the nuclear localization of the HCV core protein. To determine the precise region of the HCV core protein responsible for binding with PA28γ, we constructed additional mutant core proteins, EGFP-Core38-43 and EGFP-Core44-71 (Fig. 1). EGFP-Core44-71 was primarily localized to the nucleus, but EGFP-Core38-43 displayed a diffuse cellular staining similar to that of EGFP alone (Fig. 6B). EGFP-Core44-71, but not EGFP-Core38-43, was coprecipitated with endogenous PA28γ by rabbit anti-GFP antiserum in 293T cells (Fig. 6C). These results suggest that a cluster of amino acids from 44 to 71 in the HCV core protein is responsible for both its interaction with PA28γ and its nuclear localization.
FIG. 6.
Mapping of the PA28γ-binding region in HCV core protein. (A) 293T cells were transfected with plasmids encoding Flag-tagged HCV core deletion mutants. Endogenous PA28γ was coimmunoprecipitated with an anti-Flag antibody and was visualized by immunoblotting with anti-PA28γ antiserum. (B) Intracellular localization of EGFP-Core deletion mutants in 293T cells. All samples were observed at 24 h posttransfection. (C) EGFP-tagged HCV core deletion mutants were expressed in 293T cells. Endogenous PA28γ was coimmunoprecipitated with anti-GFP antiserum and then was visualized by immunoblotting with anti-PA28γ antiserum. IP, immunoprecipitation.
Deletion of the PA28γ-binding region or knockout of PA28γ leads to export of the HCV core protein from nucleus to cytoplasm.
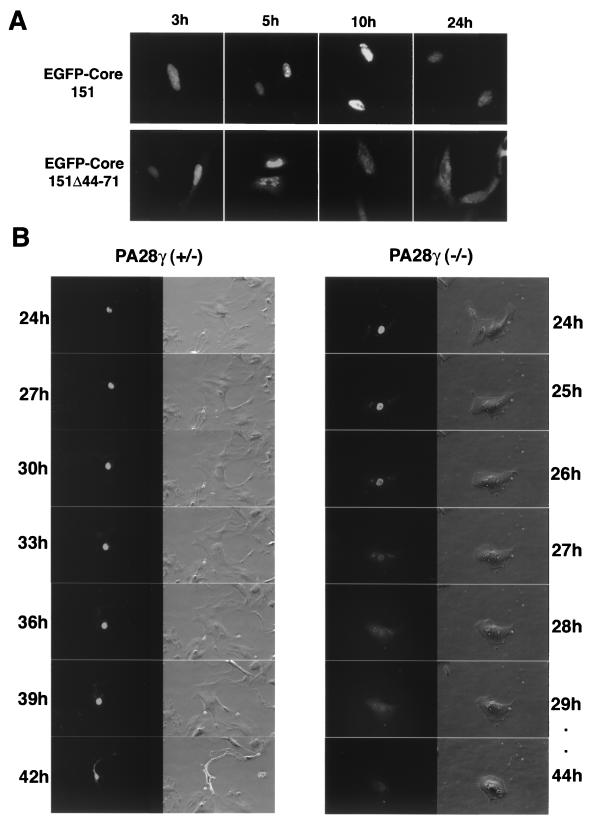
To determine whether the PA28γ-binding region identified in HCV core protein amino acids 44 to 71 functioned as anNLS, the localization of a deletion mutant lacking amino acids 44 to 71 was determined (Fig. 7A). EGFP-Core151 was detected in the nucleus of HeLa cells and retained there until at least 48 h posttransfection. Conversely, EGFP-Core151Δ44-71 (Fig. 1) was detected in the nucleus at 3 h posttransfection and gradually translocated into the cytoplasm. Most of the EGFP-Core151Δ44-71 was detected in the cytoplasm at 24 h posttransfection. These results indicate that HCV core protein amino acids 44 to 71 have a function in both PA28γ binding and nuclear retention. To further confirm this observation, we examined embryonic fibroblasts derived from PA28γ knockout mice (2) (Fig. 7B). When EGFP-Core151 was expressed in PA28γ+/−or PA28γ−/−mouse embryonic fibroblasts, EGFP-Core151 was localized to the nucleus at 24 h posttransfection, irrespective of PA28γ expression. EGFP-Core151 was retained in the nucleus of PA28γ+/−mouse embryonic fibroblasts until 42 h posttransfection, when cell death was induced (Fig. 7B, left panels). In PA28γ−/−fibroblasts, however, EGFP-Core151 was exported to the cytoplasm at 27 h posttransfection and no cell damage was observed until 44 h posttransfection (Fig. 7B, right panels). These data clearly indicate that an interaction with PA28γ is essential for the nuclear retention of the HCV core protein.
FIG. 7.
Intracellular localization of HCV core protein. (A) EGFP-Core151 and EGFP-Core151Δ44-71 were expressed in HeLa cells. The intracellular localization of EGFP-Core151 (upper panels) or EGFP-Core151Δ44-71 (lower panels) was observed at 3, 5, 10, and 24 h posttransfection. (B) Intracellular localization of EGFP-Core 151 in embryonic fibroblasts prepared from a PA28γ+/− mouse (left) or from a PA28γ−/− mouse (right) was examined from 24 to 42 h posttransfection by time-lapse microscopy.
Degradation of HCV core protein via PA28γ-dependent pathway.
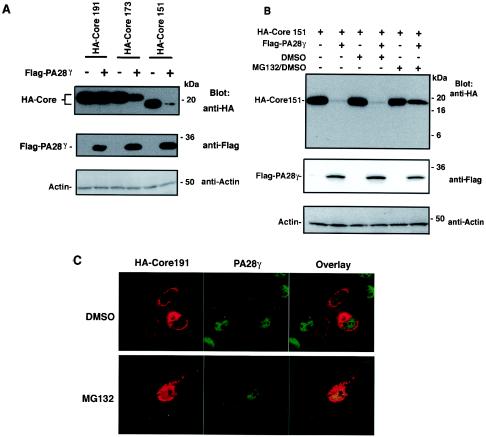
It was previously reported that HCV core proteins truncated at the C termini (HCV Core151 and 173), although normally rapidly degraded, were able to be detected after the addition of a proteasome inhibitor (57). To determine the effect of PA28γ expression on the stability of HCV core protein, HA-Core191, HA-Core173, or HA-Core151 was coexpressed with Flag-PA28γ in 293T cells. The amounts of HA-Core173 and HA-Core151 were decreased by overexpression of Flag-PA28γ, but expression levels of HA-Core191 were unchanged (Fig. 8A). Degradation of HA-Core151 by PA28γ overexpression was eliminated by the addition of the proteasome inhibitor MG132 (Fig. 8B), thus suggesting that nucleus-localized HCV core protein undergoes degradation by the proteasome in a PA28γ-dependent manner. To confirm the nuclear localization and degradation of the processed HCV core proteins derived from HA-Core191, MG132 was added to HeLa cells transfected with the plasmid encoding HA-Core191 (Fig. 8C). Treatment with MG132 enhanced the expression of HCV core protein colocalized with endogenous PA28γ in the nucleus of HeLa cells expressing HA-Core191. F protein was generated by the −2/+1 ribosomal frameshift in the gene encoding HCV core protein (64). The expected molecular mass of the F protein of the J1 strain is about 14 kDa. Endogenous PA28γ was coprecipitated by anti-Flag antibody with Flag-Core151 but not with Flag-F protein (Fig. 9). These results suggest that the HCV core protein is processed by the cleavage of the C-terminal hydrophobic region and that the truncated core protein or the mature protein is translocated into the nucleus and degraded in a PA28γ-dependent manner.
FIG. 8.
The proteasome inhibitor MG132 blocks degradation of the HCV core protein. (A) 293T cells were transfected with the expression plasmids encoding HA-Core 191, 173, or 151 together with an empty plasmid or a plasmid encoding Flag-PA28γ. Cell lysates were analyzed by immunoblotting with anti-HA, anti-Flag, or anti-actin antibodies. (B) 293T cells were transfected with an expression plasmid encoding HA-Core 151 with either an empty plasmid or a plasmid encoding Flag-PA28γ. Cells were treated with either MG132 in dimethyl sulfoxide (DMSO) or dimethyl sulfoxide alone as a control where indicated. (C) HeLa cells transfected with plasmid encoding HA-Core191 were treated with 30 μM MG132 at 10 h posttransfection and then fixed with paraformaldehyde at 24 h posttransfection. Endogenous PA28γ and HA-Core were stained with rabbit anti-PA28γ antiserum and mouse anti-HA antibody, respectively. All samples were observed with a confocal microscope. +, present; −, absent.
FIG. 9.
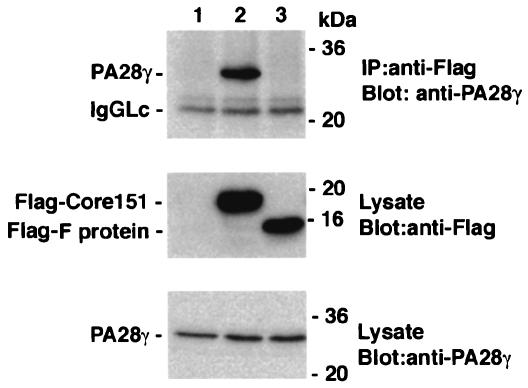
The F protein of the −2/+1 frame does not bind to PA28γ. The plasmid encoding Flag-Core151 (lane 2) or Flag-F (lane 3) protein was transfected into 293T cells and harvested at 36 h posttransfection. Endogenous PA28γ was coimmunoprecipitated with anti-Flag antibody and then was visualized by immunoblotting with anti-PA28γ antiserum. The cells transfected with the empty plasmid were used as a negative control (lane 1). IP, immunoprecipitation; Lc, light chain.
DISCUSSION
The mechanism of hepatocellular carcinoma development in patients with chronic hepatitis C remains unclear. It has been demonstrated that expression of the HCV core protein alone is sufficient for the induction of hepatic steatosis and hepatocellular carcinoma in transgenic mice (28, 39, 41). These findings suggest that the HCV core protein plays a pivotal role in the development of hepatocellular carcinoma. In this study, we isolated PA28γ from a human fetal brain library as a host protein that specifically binds to the HCV core protein. We further suggest that HCV core protein interaction with PA28γ correlates with the retention of HCV core protein in the nucleus and regulates the stability of the HCV core protein in a proteasome-dependent manner. There are two isoforms of PA28γ in humans, a major form and a splicing variant that contains an additional 13 amino acids in the second helix domain. The second isoform is detected only in the human fetal brain and is not found in other human tissues or other mammals (3, 23). In this screen, we did not obtain the splicing variant of PA28γ from the human fetal brain library; it is, therefore, still unknown whether the human-specific isoform of PA28γ binds to the HCV core protein.
The C-terminal hydrophobic region of the HCV core protein is processed by host proteases such as signal peptidase and/or intramembrane proteases. The processed, mature HCV core protein transferred into lipid droplets when a full length of core protein was expressed by an alphavirus expression system (14, 27, 36). However, the mature core protein remained in the ER when the full length of core protein was expressed by transfection in this study (Fig. 3). This discrepancy might be due to the difference in expression systems, cell lines, and genotypes of the HCV clone.
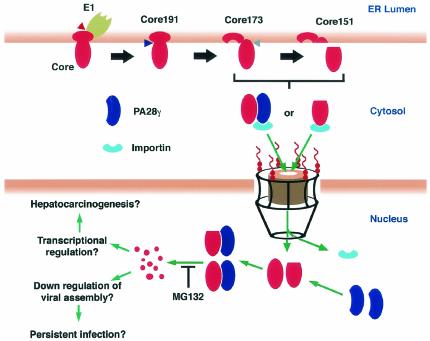
When fused to EGFP, the PA28γ-binding region of the HCV core protein (EGFP-Core44-71) migrated into the nucleus, indicating that this region may function as an NLS. Deletion of the PA28γ-binding region from the HCV core protein (EGFP-Core151Δ44-71) or depletion of PA28γ from cells, however, did not eliminate nuclear transport of the HCV core protein, suggesting the presence of an alternative mechanism for the nuclear transport of the HCV core protein other than its interaction with PA28γ. Within the C-terminally truncated HCV core protein there exist three putative NLSs consisting of a cluster of basic amino acids (8, 55). β-Galactosidase-fused C-terminal truncated core protein lacking one of these clusters (β-gal-Core123Δ38-43) was localized primarily in the cytoplasm rather than the nucleus in COS cells (55); an EGFP-fused mutant, EGFP-Core151Δ38-43, however, was localized in the nucleus in the HeLa and 293T cell lines (data not shown). These results suggest that there are at least two possible mechanisms, PA28γ dependent and PA28γ independent, leading to nuclear transport of the HCV core protein. EGFP-Core151Δ38-43 and EGFP-Core151Δ44-71 are translocated into the nucleus by the PA28γ-dependent and -independent pathways, respectively. Both pathways may be mediated through importin or importin-like molecules because PA28γ has a c-Myc-like NLS in its homolog-specific region. Furthermore, the interaction with PA28γ was shown by time-lapse microscopy to play an important role in the retention of the HCV core protein in the nucleus. HCV core proteins lacking the PA28γ-binding region, EGFP-Core151Δ44-71 and EGFP-Core151, were exported from the nucleus to the cytoplasm in HeLa cells and embryonic fibroblasts derived from PA28γ knockout mice, respectively. The nuclear exporting signal was found in the C-terminal half of the HCV core protein and plays a role in the export of the HCV core protein from the nucleus to the cytoplasm (R. Suzuki, S. Sakamoto, T. Tsutsumi, A. Rikimaru, T. Shimoike, S. Machida, Y. Matsuura, T. Miyamura, and T. Suzuki, unpublished data). The putative PA28γ-dependent and -independent translocation of the HCV core protein from the cytoplasm to the nucleus, as well as the possible functions and fates of the HCV core protein in the nucleus, are illustrated in Fig. 10.
FIG. 10.
Putative PA28γ-dependent and -independent translocation of the HCV core protein from the cytoplasm into the nucleus and possible function and fate. A precursor HCV core protein (Core191) is processed from a polyprotein in the ER by signal peptidase (red triangle), and then signal peptide peptidase (blue triangle) cleaves the C-terminal hydrophobic region, resulting in generation of mature core protein (Core173). The C-terminal region of the mature core protein is further processed by an unknown host protease (gray triangle) (Core151). Core173 and Core151 migrate into the nucleus through the nuclear pore complex, presumably by importin or an importin-PA28γcomplex. HCV core proteins interacting with PA28γ in the nucleus are degraded by the proteasome. This proteolysis is inhibited by the addition of the proteasome inhibitor MG132. Nuclear localization and degradation of the HCV core protein may induce down regulation of viral assembly, which may contribute to the maintenance of persistent infection with HCV. Furthermore,the resulting HCV core polypeptides might have some role as transcriptional regulators of host genomes, which are involved in hepatocarcinogenesis.
Although many host proteins have been reported to interact with the HCV core protein in relation to carcinogenesis (18, 33, 46, 66, 67), this is the first report demonstrating the interaction of the HCV core protein with an endogenously expressed host protein. In the livers of HCV core transgenic mice, the HCV core protein was primarily detected in the cytoplasm but some protein was found in the nucleus, albeit to a lesser extent (40). PA28γ was shown to coimmunoprecipitate with HCV core proteins irrespective of their intracellular localization (Fig. 2 and 3), suggesting that the core proteins bind to PA28γ after cell disruption. HCV core proteins truncated at the C terminus (HCV Core151 and 173) migrated into the nucleus and were degraded by ubiquitin-mediated proteolysis (57). In this study, overexpression of PA28γ led to the degradation of the HCV core protein; this degradation was able to be partially blocked by the proteasome inhibitor MG132. Additionally, HCV core protein was detected in the nucleus of a HeLa cell expressing the full-length HCV core protein in the presence of MG132 (Fig. 8). These results suggest that the HCV core protein migrates into the nucleus and is then promptly degraded by the nuclear proteasome.
The F protein generated by ribosomal frameshift in the gene encoding the core protein was mainly localized in the cytoplasm and degraded by the proteasome (63). Although the expected mass of 14 kDa of the F protein from strain J1 was not detected in HeLa cells expressing HA-Core151 even in the presence of MG132 (Fig. 8B), we examined the interaction of the protein of −2/+1 frame of the gene encoding the HCV core protein with PA28γ. Lack of interaction of endogenous PA28γ with the F protein (Fig. 9) suggests that PA28γ specifically interacts with the HCV core protein but not with the F protein.
Hepatitis B virus X factor (HBx) alone induces hepatocellular carcinoma in mice (20, 24), suggesting that HBx plays an important role in hepatocellular carcinoma. HBx bound to PSMA7 and PSMC1, subunits of PA700 and the 20S proteasome, respectively, leads to the enhancement of the transcription activities of AP-1 and VP-16 (69). Like HBx, the HCV core protein is processed by the proteasome in a PA28γ-dependent manner. An HCV core protein with the same molecular mass as HCV Core151 was detected in cells in the presence of MG132 (57). The proteasome is well known to regulate many transcription factors such as NF-κB, p53, and c-Myc, etc. (4). For example, NF-κB and its inhibitor IκB are degraded by the proteasome, resulting in translocation of active NF-κB into the nucleus (19). Upon processing, the active form of NF-κB acquires transcription activity that regulates many biological functions such as cell proliferation (43). The HCV core protein is known as a regulatory factor that modulates some signaling pathways as well as affecting expression levels of a variety of proteins under the control of different promoters (reviewed in reference 56). The short-lived, C-terminally truncated HCV core protein may acquire an as yet undetermined biological function in the nucleus. Additionally, peptides derived from the HCV core protein that has been processed by the PA28γ-activated proteasome may play some role in the transcriptional regulation that is involved in hepatocellular carcinogenesis.
The PA28γ homopolymer is able to associate with the 20S proteasome (60) and strongly activates the peptidase activity of the latent proteasome (48). The PA28α/β heteropolymer forms a hybrid proteasome with the 20S proteasome and PA700; this complex efficiently enhances antigen processing in an ATP-dependent manner (59). The PA28γ homopolymer, PA700, and the 20S proteasome may also form a hybrid proteasome that may be responsible for the proteolysis of the HCV core protein in the nucleus. PA28γ knockout mice demonstrate no abnormality other than growth retardation; this suggests that PA28γ is either dispensable for host physiological function or that suitable compensation mechanisms exist within the organism (42). Translocation and degradation of the HCV core protein by the PA28γ-activated proteasome in the nucleus may also contribute to the establishment and maintenance of persistent infection of HCV through the down regulation of viral assembly.
Although the biological significance of PA28γ is not well understood, in this study we have demonstrated new mechanisms by which PA28γ translocates and retains the HCV core protein in the nucleus; PA28γ is also involved in the proteolysis of the HCV core protein. Another nuclear proteasome activator, PA200, was recently purified from bovine testis and was demonstrated to enhance the peptidase activity but not the protease activity of the 20S proteasome (61). This report suggests that PA200 may be the functional homologue of PA28 in the nucleus. PA200 is predominantly localized to the nucleus and demonstrates homology to yeast and worm proteins that are implicated in the repair of DNA double-strand breaks. Thus, nuclear proteasome activity may be associated with DNA repair. Therefore, it may be possible that the interaction of PA28γ with the HCV core protein results in a perturbation of DNA repair activity through the nuclear proteasome, and these changes may subsequently induce hepatocellular carcinoma in humans and mice.
In conclusion, we have demonstrated that PA28γ specifically interacts with the HCV core protein in cell culture as well as in the livers of both HCV core transgenic mice and a patient with chronic hepatitis C. This interaction correlates to the nuclear retention and degradation of C-terminally truncated HCV core proteins. Understanding the precise function of PA28γ may give us new insight into virus-cell interactions and lead to a greater understanding of the pathogenicity of HCV infection. Establishment of HCV core transgenic mice deficient in PA28γ gene expression will allow the direct assessment of the involvement of PA28γ in the development of hepatocellular carcinoma induced by HCV core protein; these experiments are under way.
Acknowledgments
We thank T. Shioda for advice on confocal microscopy, S. Ogawa for assistance in sequencing and the preparation of experimental reagents, and T. Abe, K. Okamoto, and Y. Mori for critical reading of the manuscript.
This work was supported in part by grants-in-aid from the Ministry of Health, Labor, and Welfare, the program for Promotion of Fundamental Studies in Health Sciences of the Organization for Drug ADR Relief, R&D Promotion, and Product Review of Japan to Y.M. and T.M. and the Ministry of Education, Science, and Culture of Japan to Y.M.
REFERENCES
- 1.Ahn, J. Y., N. Tanahashi, K. Akiyama, H. Hisamatsu, C. Noda, K. Tanaka, C. H. Chung, N. Shibmara, P. J. Willy, J. D. Mott, C. A. Slaughtere, and G. N. DeMartino. 1995. Primary structures of two homologous subunits of PA28, a gamma-interferon-inducible protein activator of the 20S proteasome. FEBS Lett. 366:37-42. [DOI] [PubMed] [Google Scholar]
- 2.Aizaki, H., Y. Aoki, T. Harada, K. Ishii, T. Suzuki, S. Nagamori, G. Toda, Y. Matsuura, and T. Miyamura. 1998. Full-length complementary DNA of hepatitis C virus genome from an infectious blood sample. Hepatology 27:621-627. [DOI] [PubMed] [Google Scholar]
- 3.Albertsen, H. M., S. A. Smith, S. Mazoyer, E. Fujimoto, J. Stevens, B. Williams, P. Rodriguez, C. S. Cropp, P. Slijepcevic, M. Carlson, M. Robertson, P. Bradley, E. Lawrence, T. Harrington, Z. Mei Sheng, R. Hoopes, N. Sternberg, A. Brothman, R. Callahan, B. A. J. Ponder, and R. White. 1994. A physical map and candidate genes in the BRCA1 region on chromosome 17q12-21. Nat. Genet. 7:472-479. [DOI] [PubMed] [Google Scholar]
- 4.Almond, J. B., and G. M. Cohen. 2002. The proteasome: a novel target for cancer chemotherapy. Leukemia 16:433-443. [DOI] [PubMed] [Google Scholar]
- 5.Aoyagi, K., C. Ohue, K. Iida, T. Kimura, E. Tanaka, K. Kiyosawa, and S. Yagi. 1999. Development of a simple and highly sensitive enzyme immunoassay for hepatitis C virus core antigen. J. Clin. Microbiol. 37:1802-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks, P., G. Fuertes, R. Z. Murray, S. Bose, E. Knecht, M. C. Rechsteiner, K. B. Hendil, K. Tanaka, J. Dyson, and J. Rivett. 2000. Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem. J. 346:155-161. [PMC free article] [PubMed] [Google Scholar]
- 7.Bukh, J., R. H. Purcell, and R. H. Miller. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. USA 91:8239-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, S. C., J. H. Yen, H. Y. Kang, M. H. Jang, and M. F. Chang. 1994. Nuclear localization signals in the core protein of hepatitis C virus. Biochem. Biophys. Res. Commun. 205:1284-1290. [DOI] [PubMed] [Google Scholar]
- 9.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 10.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, A. J. Weiner, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duttweiller, H. M. 1996. A highly sensitive and non-lethal beta-galactosidase plate assay for yeast. Trends Genet. 12:340-341. [DOI] [PubMed] [Google Scholar]
- 12.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 14.Hope, R. G., and J. McLauchlan. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81:1913-1925. [DOI] [PubMed] [Google Scholar]
- 15.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 16.Houghton, M., A. Weiner, J. Han, G. Kuo, and Q. L. Choo. 1991. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology 14:381-388. [PubMed] [Google Scholar]
- 17.Huang, D. C., S. Cory, and A. Strasser. 1997. Bcl-2, Bcl-XL and adenovirus protein E1B19kD are functionally equivalent in their ability to inhibit cell death. Oncogene 14:405-414. [DOI] [PubMed] [Google Scholar]
- 18.Jin, D. Y., H. L. Wang, Y. Zhou, A. C. Chun, K. V. Kibler, Y. D. Hou, H. Kung, and K. T. Jeang. 2000. Hepatitis C virus core protein-induced loss of LZIP function correlates with cellular transformation. EMBO J. 19:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 20.Kim, C. M., K. Koike, I. Saito, T. Miyamura, and G. Jay. 1991. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature 351:317-320. [DOI] [PubMed] [Google Scholar]
- 21.Kiyosawa, K., T. Sodeyama, E. Tanaka, Y. Gibo, K. Yoshizawa, Y. Nakano, S. Furuta, Y. Akahane, K. Nishioka, and R. H. Purcell. 1990. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology 12:671-675. [DOI] [PubMed] [Google Scholar]
- 22.Knowlton, J. R., S. C. Johnston, F. G. Whitby, C. Realini, Z. Zhang, M. Rechsteiner, and C. P. Hill. 1997. Structure of the proteasome activator REGalpha (PA28alpha). Nature 390:639-643. [DOI] [PubMed] [Google Scholar]
- 23.Kohda, K., T. Ishibashi, N. Shimbara, K. Tanaka, Y. Matsuda, and M. Kasahara. 1998. Characterization of the mouse PA28 activator complex gene family: complete organizations of the three member genes and a physical map of the approximately 150-kb region containing the alpha- and beta-subunit genes. J. Immunol. 160:4923-4935. [PubMed] [Google Scholar]
- 24.Koike, K., K. Moriya, S. Iino, H. Yotsuyanagi, Y. Endo, T. Miyamura, and K. Kurokawa. 1994. High-level expression of hepatitis B virus HBx gene and hepatocarcinogenesis in transgenic mice. Hepatology 19:810-819. [PubMed] [Google Scholar]
- 25.Koike, K., T. Tsutsumi, H. Fujie, Y. Shintani, and M. Kyoji. 2002. Molecular mechanism of viral hepatocarcinogenesis. Oncology 62(Suppl. 1):29-37. [DOI] [PubMed] [Google Scholar]
- 26.Kuo, G., Q. L. Choo, H. J. Alter, G. L. Gitnick, A. G. Redeker, R. H. Purcell, T. Miyamura, J. L. Dienstag, M. J. Alter, C. E. Stevens, G. E. Tegtmeier, F. Bonino, M. Colombo, W. S. Lee, C. Kuo, K. Berger, F. R. Shuster, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 244:362-364. [DOI] [PubMed] [Google Scholar]
- 27.Lemberg, M. K., and B. Martoglio. 2002. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell 10:735-744. [DOI] [PubMed] [Google Scholar]
- 28.Lerat, H., M. Honda, M. R. Beard, K. Loesch, J. Sun, Y. Yang, M. Okuda, R. Gosert, S. Y. Xiao, S. A. Weinman, and S. M. Lemon. 2002. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology 122:352-365. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Q., C. Tackney, R. A. Bhat, A. M. Prince, and P. Zhang. 1997. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J. Virol. 71:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo, S. Y., F. Masiarz, S. B. Hwang, M. M. Lai, and J. H. Ou. 1995. Differential subcellular localization of hepatitis C virus core gene products. Virology 213:455-461. [DOI] [PubMed] [Google Scholar]
- 31.Lo, S. Y., M. Selby, M. Tong, and J. H. Ou. 1994. Comparative studies of the core gene products of two different hepatitis C virus isolates: two alternative forms determined by a single amino acid substitution. Virology 199:124-131. [DOI] [PubMed] [Google Scholar]
- 32.Lo, S. Y., M. J. Selby, and J. H. Ou. 1996. Interaction between hepatitis C virus core protein and E1 envelope protein. J. Virol. 70:5177-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mamiya, N., and H. J. Worman. 1999. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J. Biol. Chem. 274:15751-15756. [DOI] [PubMed] [Google Scholar]
- 34.Masson, P., O. Andersson, U. M. Petersen, and P. Young. 2001. Identification and characterization of a Drosophila nuclear proteasome regulator. A homolog of human 11 S REGgamma (PA28gamma). J. Biol. Chem. 276:1383-1390. [DOI] [PubMed] [Google Scholar]
- 35.Matsuura, Y., T. Harada, M. Makimura, M. Sato, H. Aizaki, T. Suzuki, and T. Miyamura. 1994. Characterization of HCV structural proteins expressed in various animal cells. Intervirology 37:114-118. [DOI] [PubMed] [Google Scholar]
- 36.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moradpour, D., T. Wakita, K. Tokushige, R. I. Carlson, K. Krawczynski, and J. R. Wands. 1996. Characterization of three novel monoclonal antibodies against hepatitis C virus core protein. J. Med. Virol. 48:234-241. [DOI] [PubMed] [Google Scholar]
- 38.Moriishi, K., D. C. Huang, S. Cory, and J. M. Adams. 1999. Bcl-2 family members do not inhibit apoptosis by binding the caspase activator Apaf-1. Proc. Natl. Acad. Sci. USA 96:9683-9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moriya, K., H. Fujie, Y. Shintani, H. Yotsuyanagi, T. Tsutsumi, K. Ishibashi, Y. Matsuura, S. Kimura, T. Miyamura, and K. Koike. 1998. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 4:1065-1067. [DOI] [PubMed] [Google Scholar]
- 40.Moriya, K., H. Fujie, H. Yotsuyanagi, Y. Shintani, T. Tsutsumi, Y. Matsuura, T. Miyamura, S. Kimura, and K. Koike. 1997. Subcellular localization of hepatitis C virus structural proteins in the liver of transgenic mice. Jpn. J. Med. Sci. Biol. 50:169-177. [DOI] [PubMed] [Google Scholar]
- 41.Moriya, K., H. Yotsuyanagi, Y. Shintani, H. Fujie, K. Ishibashi, Y. Matsuura, T. Miyamura, and K. Koike. 1997. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J. Gen. Virol. 78:1527-1531. [DOI] [PubMed] [Google Scholar]
- 42.Murata, S., H. Kawahara, S. Tohma, K. Yamamoto, M. Kasahara, Y. Nabeshima, K. Tanaka, and T. Chiba. 1999. Growth retardation in mice lacking the proteasome activator PA28gamma. J. Biol. Chem. 274:38211-38215. [DOI] [PubMed] [Google Scholar]
- 43.Murray, R. Z., and C. Norbury. 2000. Proteasome inhibitors as anti-cancer agents. Anticancer Drugs 11:407-417. [DOI] [PubMed] [Google Scholar]
- 44.Nikaido, T., K. Shimada, M. Shibata, M. Hata, M. Sakamoto, Y. Takasaki, C. Sato, T. Takahashi, and Y. Nishida. 1990. Cloning and nucleotide sequence of cDNA for Ki antigen, a highly conserved nuclear protein detected with sera from patients with systemic lupus erythematosus. Clin. Exp. Immunol. 79:209-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 46.Otsuka, M., N. Kato, K. Lan, H. Yoshida, J. Kato, T. Goto, Y. Shiratori, and M. Omata. 2000. Hepatitis C virus core protein enhances p53 function through augmentation of DNA binding affinity and transcriptional ability. J. Biol. Chem. 275:34122-34130. [DOI] [PubMed] [Google Scholar]
- 47.Paesen, G. C., and P. A. Nuttall. 1996. A tick homologue of the human Ki nuclear autoantigen. Biochim. Biophys. Acta 1309:9-13. [DOI] [PubMed] [Google Scholar]
- 48.Realini, C., C. C. Jensen, Z. Zhang, S. C. Johnston, J. R. Knowlton, C. P. Hill, and M. Rechsteiner. 1997. Characterization of recombinant REGalpha, REGbeta, and REGgamma proteasome activators. J. Biol. Chem. 272:25483-25492. [DOI] [PubMed] [Google Scholar]
- 49.Rechsteiner, M., C. Realini, and V. Ustrell. 2000. The proteasome activator 11 S REG (PA28) and class I antigen presentation. Biochem. J. 345:1-15. [PMC free article] [PubMed] [Google Scholar]
- 50.Roehrig, J. T., L. A. Staudinger, A. R. Hunt, J. H. Mathews, and C. D. Blair. 2001. Antibody prophylaxis and therapy for flavivirus encephalitis infections. Ann. N. Y. Acad. Sci. 951:286-297. [DOI] [PubMed] [Google Scholar]
- 51.Saito, I., T. Miyamura, A. Ohbayashi, H. Harada, T. Katayama, S. Kikuchi, Y. Watanabe, S. Koi, M. Onji, Y. Ohta, Q. Choo, M. Houghton, and G. Kuo. 1990. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 87:6547-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santolini, E., G. Migliaccio, and N. La Monica. 1994. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 68:3631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimoike, T., S. Mimori, H. Tani, Y. Matsuura, and T. Miyamura. 1999. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 73:9718-9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shoji, I., T. Suzuki, M. Sato, H. Aizaki, T. Chiba, Y. Matsuura, and T. Miyamura. 1999. Internal processing of hepatitis C virus NS3 protein. Virology 254:315-323. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki, R., Y. Matsuura, T. Suzuki, A. Ando, J. Chiba, S. Harada, I. Saito, and T. Miyamura. 1995. Nuclear localization of the truncated hepatitis C virus core protein with its hydrophobic C terminus deleted. J. Gen. Virol. 76:53-61. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki, R., T. Suzuki, K. Ishii, Y. Matsuura, and T. Miyamura. 1999. Processing and functions of hepatitis C virus proteins. Intervirology 42:145-152. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki, R., K. Tamura, J. Li, K. Ishii, Y. Matsuura, T. Miyamura, and T. Suzuki. 2001. Ubiquitin-mediated degradation of hepatitis C virus core protein is regulated by processing at its carboxyl terminus. Virology 280:301-309. [DOI] [PubMed] [Google Scholar]
- 58.Takamizawa, A., C. Mori, I. Fuke, S. Manabe, S. Murakami, J. Fujita, E. Onishi, T. Andoh, I. Yoshida, and H. Okayama. 1991. Structure and organization of the hepatitis C virus genome isolated from human carriers. J. Virol. 65:1105-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanahashi, N., Y. Murakami, Y. Minami, N. Shimbara, K. B. Hendil, and K. Tanaka. 2000. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J. Biol. Chem. 275:14336-14345. [DOI] [PubMed] [Google Scholar]
- 60.Tanahashi, N., K. Yokota, J. Y. Ahn, C. H. Chung, T. Fujiwara, E. Takahashi, G. N. DeMartino, C. A. Slaughter, T. Toyonaga, K. Yamamura, N. Shimbara, and K. Tanaka. 1997. Molecular properties of the proteasome activator PA28 family proteins and gamma-interferon regulation. Genes Cells 2:195-211. [DOI] [PubMed] [Google Scholar]
- 61.Ustrell, V., L. Hoffman, G. Pratt, and M. Rechsteiner. 2002. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 21:3516-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilk, S., W. E. Chen, and R. P. Magnusson. 2000. Properties of the nuclear proteasome activator PA28gamma (REGgamma). Arch. Biochem. Biophys. 383:265-271. [DOI] [PubMed] [Google Scholar]
- 63.Xu, Z., J. Choi, W. Lu, and J. H. Ou. 2003. Hepatitis C virus F protein is a short-lived protein associated with the endoplasmic reticulum. J. Virol. 77:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu, Z., J. Choi, T. S. Yen, W. Lu, A. Strohecker, S. Govindarajan, D. Chien, M. J. Selby, and J. Ou. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yasui, K., T. Wakita, K. Tsukiyama-Kohara, S. I. Funahashi, M. Ichikawa, T. Kajita, D. Moradpour, J. R. Wands, and M. Kohara. 1998. The native form and maturation process of hepatitis C virus core protein. J. Virol. 72:6048-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshida, T., T. Hanada, T. Tokuhisa, K. Kosai, M. Sata, M. Kohara, and A. Yoshimura. 2002. Activation of STAT3 by the hepatitis C virus core protein leads to cellular transformation. J. Exp. Med. 196:641-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.You, L. R., C. M. Chen, T. S. Yeh, T. Y. Tsai, R. T. Mai, C. H. Lin, and Y. H. Lee. 1999. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J. Virol. 73:2841-28453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, J., O. Yamada, H. Yoshida, T. Iwai, and H. Araki. 2002. Autogenous translational inhibition of core protein: implication for switch from translation to RNA replication in hepatitis C virus. Virology 293:141-150. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, Z., N. Torii, A. Furusaka, N. Malayaman, Z. Hu, and T. J. Liang. 2000. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J. Biol. Chem. 275:15157-15165. [DOI] [PubMed] [Google Scholar]