Abstract
Leishmania RNA virus (LRV) is a double-stranded RNA virus that infects some strains of the protozoan parasite Leishmania. As with other totiviruses, LRV presumably expresses its polymerase by a ribosomal frameshift, resulting in a capsid-polymerase fusion protein. We have demonstrated previously that an LRV capsid-polymerase polyprotein is specifically cleaved by a Leishmania-encoded cysteine protease. This study reports the purification of this protease through a strategy involving anion-exchange chromatography and affinity chromatography. By using a Sepharose-immobilized lectin, concanavalin A, we isolated a fraction enriched with LRV polyprotein-specific protease activity. Analysis of the active fraction by sodium dodecyl sulfate-polyacrylamide gel electrophoreses and silver staining revealed a 50-kDa protein that, upon characterization by high-pressure liquid chromatography electrospray tandem mass spectrometry (electrospray ionization/MS/MS), was identified as a cysteine protease of trypanosomes. A partial amino acid sequence derived from the MS/MS data was compared with a protein database using BLAST software, revealing homology with several cysteine proteases of Leishmania and other trypanosomes. The protease exhibited remarkable temperature stability, while inhibitor studies characterized the protease as a trypsin-like cysteine protease—a novel finding for Leishmania. To elucidate substrate preferences, a panel of deletion mutations and single-amino-acid mutations were engineered into a Gag-Pol fusion construct that was subsequently transcribed and translated in vitro and then used in cleavage assays. The data suggest that there are a number of cleavage sites located within a 153-amino-acid region spanning both the carboxy-terminal capsid region and the amino-terminal polymerase domain, with LRV capsid exhibiting the greatest susceptibility to proteolysis.
Leishmania RNA virus (LRV) is a double-stranded RNA (dsRNA) virus that persistently infects some strains of the parasitic protozoan Leishmania, the causative agent of leishmaniasis. The LRV genus consists of viruses that infect both New World and Old World Leishmania protozoan parasites. To date, the New World viruses number greater than 12 confirmed species (7), while only one Old World virus has been documented. Nucleotide sequence analysis and PCR fingerprinting of both New World and Old World viruses support a virus-parasite coevolution with infection probably occurring prior to the divergence of Leishmania into different lineages (24).
The New World LRV genome possesses a dsRNA segment of approximately 5.3 kb in length. Four open reading frames (ORFs) are encoded on the positive-sense strand, with ORF 1 and ORF X encoding small coding sequences that share no homology with any known protein sequence. The small ORFs correspond to the 5′ untranslated region, which is 449 nucleotides in length in LRV 1-4 and shows greater than 90% identity between LRV1-1 and LRV1-4 at the nucleotide level (19). ORF 2 encodes the 82-kDa viral capsid protein, while ORF3 encodes the RNA-dependent RNA polymerase consensus motifs. The polymerase domain overlaps the capsid domain by 71 nucleotides and is presumably expressed by a translational frameshift (11). However, a capsid-polymerase fusion protein has yet to be identified in vivo, suggesting the possibility of proteolytic processing of the fusion protein.
We have previously reported the specific cleavage of a synthetic LRV capsid-polymerase fusion protein by a host-encoded protease (17). This observation provides support for a hypothesis that a capsid-polymerase fusion protein is produced in vivo but undergoes posttranslational processing to yield the 95- and 82-kDa antigens detected in cell lysates. The role of the proteolytic processing in the virus life cycle has yet to be deciphered but has been postulated to play a role in virus maturation or viral genome copy number regulation.
To further understanding of the virus-parasite interaction, we purified, identified, and characterized the Leishmania-encoded protease responsible for LRV processing. The protease shares homology with several cysteine proteases of Leishmania and other trypanosomes, one of which is an L. mexicana homolog that was previously identified as a virulence factor (15). The protease is a stable protein that retains activity under a wide range of pH and temperature conditions and has been further characterized as a trypsin-like cysteine protease. Finally, we describe the complex substrate requirements for cleavage.
MATERIALS AND METHODS
Parasite culture conditions.
The parasite used in this study was an L. guyanensis stock MHOM/BR/75/M4147 (M4147). The promastigote stage of the parasite was maintained at 25°C in supplemented M199 medium (Invitrogen). The supplemented M199 medium contained 5% fetal bovine serum (Gibco-BRL), 100 U of penicillin per ml, 100 μg of streptomycin per ml, 40 mM HEPES (pH 7.4), 0.1 mM hypoxanthine, 0.0005% hemin, and 5% sterile human urine. Parasites were cultured in a volume of 200 ml in T175 tissue culture flasks.
Harvesting and lysis of promastigotes.
Parasites were grown to late log phase and harvested by centrifugation in a 250-ml volume at 4°C at 3,000 × g for 15 min. The supernatant was poured off, and the pellets were resuspended in 25 ml of phosphate-buffered saline (PBS), combined into a single centrifuge bottle, and then centrifuged once again at 4°C at 3,000 × g for 10 min. Promastigotes (wet weight, 9 g) were lysed by three freeze-thaw cycles at −20 and 37°C in 8 ml of lysis buffer (50 mM Tris [pH 8.0], 0.3 M sucrose, 1× protease inhibitor cocktail [Roche]). The lysate was clarified by centrifugation at 10,000 × g for 20 min. The supernatant was filtered through a 0.45-μm-pore-size syringe filter. The filtered lysate was stored at 4°C for chromatography.
Anion-exchange chromatography.
Two HiTrap Q (Amersham Pharmacia Biotech) columns were equilibrated in start buffer (50 mM Tris [pH 8.0]) according to the manufacturer's suggestions. Filtered lysate was passed through the coupled anion-exchange columns at a flow rate of 2.5 ml/min. The flow through was passed two additional times over the columns to ensure maximum binding. The columns were washed with 5 column volumes (CV) of start buffer. Pilot experiments using a 1-ml HiTrap Q column and stepped salt elution gradients had shown that specific cleavage activity was associated with fractions eluted with 230 mM NaCl (data not shown). Therefore, the columns were washed twice with two buffers containing start buffer with 150 mM NaCl and 200 mM NaCl, respectively, until the optical density at 260 nm was nil to eliminate irrelevant proteins. Finally, the column was eluted with 5 CV of 50 mM Tris [pH 8.0] containing 230 mM NaCl. Fractions were assayed with the protease cleavage assay.
Concanavalin A chromatography.
The active fraction obtained from anion-exchange chromatography was dialyzed overnight using 10-kDa cutoff dialysis tubing (Pierce) against buffer 1 (25 mM Tris-HCl [pH 7.4]). The dialyzed fraction was adjusted to a final concentration of 150 mM NaCl, 5 mM CaCl2, and 5 mM MnCl2 and then passed over an equilibrated concanavalin A (ConA) column as previously described with some modifications (9). Briefly, 4 ml of ConA-agarose (Sigma) was packed into an Econocolumn (Bio-Rad), washed extensively, and equilibrated with buffer 2 (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM MnCl2, 1 mM MgCl2). The fraction was passed over the column at a flow rate of 0.5 ml/min at room temperature. The flowthrough was reapplied to the column two additional times to ensure maximum binding. Next, the column was washed with buffer 2 until the optical density at 280 nm reached a baseline reading. Prior to elution, the column was equilibrated with 1 CV of buffer 3 (buffer 2 containing 0.5 M α-methyl mannoside) and was then incubated at 37°C for 1 h. Finally, the column was eluted with 5 CV of prewarmed (37°C) buffer 3. Fractions were assayed using an in vitro protease cleavage assay.
In vitro transcription and translation.
In vitro transcription and translation were performed using the TNT-coupled reticulocyte lysate system (Promega) according to the manufacturer's recommendations. Briefly, 2 μg of plasmid DNA, encoding the LRV capsid-polymerase polyprotein (17), was incubated at 37°C for 90 min in a TNT reaction mix containing T7 polymerase and [35S]methionine (Dupont-NEN). Translation products were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE) (10). The gel was dried and then exposed to Biomax-AR film (Kodak) overnight.
Protease cleavage assay.
In vitro-translated products (5 μl) were incubated in a cleavage assay containing 50 mM Tris (pH 7.5), 100 mM NaCl, 2 mM dithiothreitol, and 4 μl of protease fraction in a 15-μl final volume. Reactions were incubated for 40 min at 30°C and terminated by incubating at 100°C for 4 min with 7.5 μl of 3× SDS sample buffer. Reaction products were fractionated by SDS-PAGE as previously described (10).
Immunoprecipitation.
Cleavage reaction products were characterized by immunoprecipitation using LRV 1-4 capsid-specific and LRV 1-4 polymerase-specific polyclonal antisera. Briefly, 15 μl of in vitro-transcribed LRV capsid-polymerase substrate was incubated with cleavage assay buffer and 3 μl of ConA-eluted fraction (40 ng of total protein) in a final volume of 45 μl for 40 min at 30°C. To each reaction, the appropriate polyclonal antiserum (5 μl) was added with 50 μl of TPBS (i.e., 0.05% Tween 20 and 1× PBS), and the reaction was incubated with gentle shaking at 4°C for 1 h. Protein A Sepharose (Sigma) was then added, and the reaction was incubated for 1 additional h at 4°C. The immune complex was pelleted by low-speed centrifugation (1,500 × g for 5 min) and washed six times with 200 μl of TPBS. Finally, 35 μl of 3× SDS sample buffer was added, followed by a 5-min incubation at 100°C. The supernatant was resolved by SDS-PAGE and then visualized by autoradiography.
Substrate gel electrophoresis.
Substrate gel electrophoresis was performed as previously described with slight modifications (15, 18). Briefly, samples were mixed with 3× SDS sample buffer lacking β-mercaptoethanol. Samples were loaded on a SDS-12% polyacrylamide gel copolymerized with 0.2% gelatin and were then electrophoresed at constant voltage (120 V) for 1.5 h at 4°C. The gel was then washed with 200 ml of 2.5% (vol/vol) Triton X-100 for 1 h at room temperature. Next, the gel was transferred to a container containing activity buffer (100 mM sodium acetate [pH 5.5] and 1 mM dithiothreitol) and incubated overnight at 37°C. Finally, the gel was stained with Coomassie blue stain for 1 h, followed by a 3-h destaining procedure as previously described (10).
Mass spectrometry.
Identification of stained bands after SDS-PAGE was accomplished by mass spectrometry. Slices of the gel containing protein bands of interest were digested with trypsin according to standard protocols based on the initial work of Shevchenko et al. (20). Briefly, protein bands of interest were excised from the gel and destained prior to digestion. The in-gel digestion was performed with 5 to 10 ng of modified trypsin (Promega) per ml in 40 mM ammonium bicarbonate and incubated overnight at 37°C. The resulting digests were analyzed by matrix-assisted laser desorption ionization-time of flight (mass spectrometry) [MALDI-TOF (MS)] using an Applied Biosystems Voyager-DE STR (Framingham, Mass.) operated in reflector mode using delayed extraction. Samples were applied to the target and embedded in a UV-absorbing matrix consisting of a saturated solution of 2,5-dihydroxybenzoic acid (Sigma) in 50% acetonitrile-0.1% trifluoroacetic acid. The peptide mass maps produced by MALDI-TOF (MS) were searched against the published databases by means of the MS-Fit module in Protein Prospector (http://prospector.ucsf.edu/ucsfhtml4.0/msfit.htm) to provide information about the identity of the protein(s) in each band. Further characterization of samples required capillary high-pressure liquid chromatography-electrospray tandem mass spectrometry (HPLC-ESI/MS/MS) on a Thermo Finnigan LCQ ion trap mass spectrometer in conjunction with a Michrom BioResources MAGIC 2002 micro-HPLC and a microspray interface. Capillary online HPLC separation of tryptic peptides was conducted using the following conditions: column, New Objectives PicoFrit, 75-mm inside diameter, packed to 10 cm with Vydac 218MSB5; mobile phase A, 0.5% acetic acid-0.005% trifluoroacetic acid in water; mobile phase B, 90% acetonitrile-0.5% acetic acid-0.005% trifluoroacetic acid in water; gradient, 2% B to 72% B in 30 min; flow rate, 0.4 ml/min. Data-dependent acquisition was employed to generate a survey scan followed by four collision-induced dissociation MS/MS spectra. The uninterpreted MS/MS data were analyzed by SEQUEST, a component of the LCQ software.
Protease inhibitor study.
ConA-eluted protease (total protein, 40 ng) was incubated with 1 μl of the prescribed protease inhibitor in a protease cleavage assay. After the 40-min incubation, the reactions were terminated and analyzed by SDS-PAGE. The following protease inhibitors (Roche) were examined for their ability to block polyprotein proteolysis: antipain dihydrochloride, aprotinin, E-64, EDTA, leupeptin, Pefabloc SC, phosphoramidon, and trypsin inhibitor. Inhibitors were prepared according to the manufacturer's recommendations.
Generation of recombinant LRV deletion and mutation constructs.
LRV mutants were constructed using the previously described pBSK-INFRAME plasmid (17) by site-directed PCR. Charged amino acid residues near the amino terminus of the polymerase domain were changed to alanines, and the individual PCR fragments were cloned into a pGem-T Easy vector (Promega). Mutated fragments were shuttled out of the subclones by excision with MluI and NcoI and then ligated into an MluI/NcoI-cut BSK-INFRAME plasmid. Deletions were made by using primers that had either an MluI site or BglII site incorporated into them. See Fig. 6 for a summary of the deletion and mutation constructs. The ORF 3 construct was generated by PCR using an XhoI primer corresponding to the amino terminus of the predicted reading frame encoding the LRV polymerase domain and a reverse XbaI primer corresponding to the 3′ end of the LRV genome. The restriction enzymes were used to shuttle the fragment into pBluescript vector (Stratagene). Generation of the ORF 2 construct has been described previously (17).
FIG. 6.
Schematic representation of deletion mutant constructs and single-amino-acid-mutant constructs. Mutants were assembled either by PCR-directed mutagenesis or splicing of overlapping ends as described in Materials and Methods. The results of the cleavage assay are noted to the right.
RESULTS
Purification of a cysteine-like protease.
Nine grams of the New World parasite L. braziliensis guyanensis promastigotes were harvested and lysed as detailed in Materials and Methods. After clarification and filtration, the extract was passed through a HiTrap Q Sepharose column as described above. A microaliquot (200 μl) of each fraction eluted from the NaCl gradient was concentrated 10-fold in a 10-kDa cutoff microconcentrator (Millipore) and then assayed in the protease cleavage assay. The fraction eluting at 250 mM NaCl readily cleaved LRV capsid-polymerase substrate. Further gradient analysis revealed that optimal elution occurred at 230 mM NaCl (data not shown).
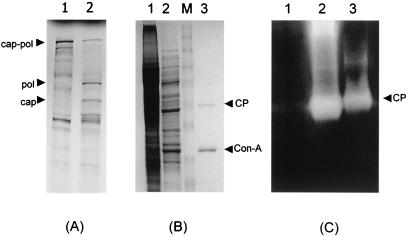
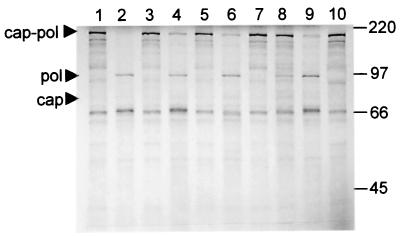
We have previously demonstrated that in vitro cleavage of the LRV capsid-polymerase can be blocked using cysteine protease (CysP)-specific inhibitors (17). Although there is no literature describing CysP of L. braziliensis, there is substantial data regarding CysP of L. mexicana, a New World parasite, and related kinetoplasts such as the trypanosomes. Based on the characteristics of the capsid-polymerase-specific protease—broad pH tolerance, high susceptibility to leupeptin, and high endopeptidase activity—we hypothesized that the protease could potentially be either a cathepsin B or cathepsin L protease. Since cathepsin L proteases have been shown to possess glycosylation sites (1), ConA was used as a potential affinity chromatography step. The active fraction from anion-exchange chromatography was applied to the column at room temperature as described in Materials and Methods. The eluted fraction was concentrated 10-fold and then incubated with LRV Gag-Pol substrate. The fraction retained cleavage activity (Fig. 1A), while SDS-PAGE analysis showed that the ConA-eluted fraction contained only two proteins of approximately 50 and 30 kDa (Fig. 1B). Substrate gel electrophoresis demonstrated that the ConA-eluted fraction was devoid of contaminating proteases compared to the S15 fraction (Fig. 1C).
FIG. 1.
Cleavage activity and protein profile of the highly purified CysP. (A) In vitro cleavage products were resolved on an SDS-10% polyacrylamide gel and visualized by autoradiography. Note the cleavage products in the positive control (lane 2) migrating at 82 and 95 kDa (capsid and polymerase, respectively). (B) Protein profile of active fractions from each purification step. Promastigote lysate, anion-exchange fraction, and ConA-eluted fraction were resolved on an SDS-10% polyacrylamide gel (lanes 1, 2, and 3). Note the 49-kDa CYSP and 30-kDa ConA contaminant (arrows). Lane M, prestained molecular mass marker (Invitrogen). CP, cysteine protease. (C) Substrate gel electrophoresis was performed as described in Materials and Methods. Lanes 1 to 3 represent the size marker, S15 fraction, and highly purified ConA-eluted CysP, respectively. The arrow indicates the location of the 50-kDa CYSP. CP, cysteine protease.
Characterization of cleavage products by immunoprecipitation.
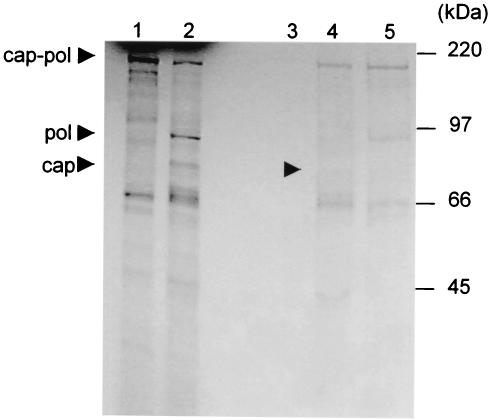
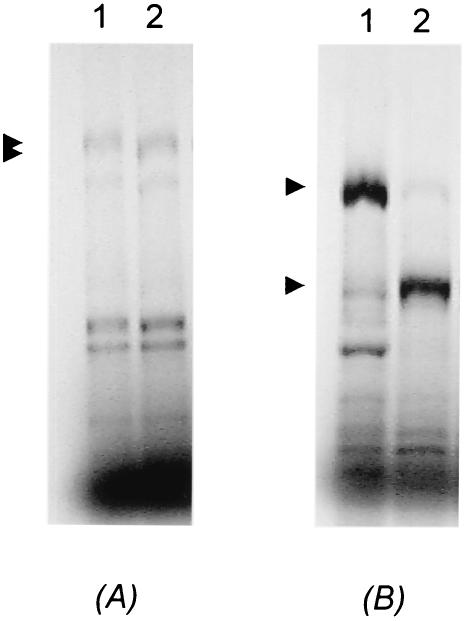
Time course experiments with ConA-eluted protease have correlated hydrolysis of the Gag-Pol full-length fusion protein with the appearance of smaller products of approximately 95 and 82 kDa (data not shown). To confirm the identity of the products, the cleavage reaction was characterized by immunoprecipitation with LRV 1-4 capsid- and LRV 1-4 polymerase-specific polyclonal antisera. As shown in Fig. 2, both capsid and polymerase antisera recognized the fusion protein. The 95- and 82-kDa proteins were recognized by polymerase and capsid antisera, respectively.
FIG. 2.
Characterization of the Gag-Pol cleavage products by immunoprecipitation with capsid- and polymerase-specific antisera. In vitro cleavage reactions were immunoprecipitated with preimmune antiserum, capsid-specific antiserum, and polymerase-specific antiserum (lanes 3 to 5, respectively). The immune complexes were resolved on an SDS-10% polyacrylamide gel and visualized by autoradiography. Note the cleavage products in the positive control (lane 2) migrating at 82 and 95 kDa (capsid and polymerase, respectively). Lane 1 is a buffer-only control.
Protein identification by mass spectroscopy.
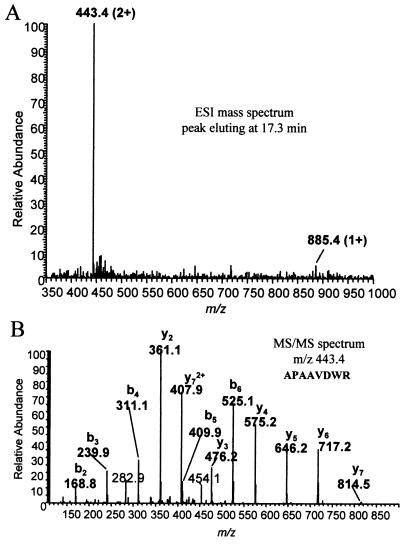
Analysis of the ConA-eluted fraction by SDS-PAGE and Coomassie blue staining revealed at least two proteins in the active fraction, approximately 50 and 30 kDa. To identify the proteins in this fraction, the bands were excised, trypsinized, and analyzed by MALDI-TOF (MS) as detailed in Materials and Methods. The peptide mass maps generated were compared with published databases, which identified the 30-kDa band as ConA (data not shown). MS data obtained from the 50-kDa band produced well-isolated peaks; however, no database hits were identified (data not shown). Further characterization of the 50-kDa band by HPLC-ESI/MS/MS (Fig. 3A) revealed that the polypeptide had sequence similarity with a CysP protein of trypanosomes. A partial amino acid sequence was derived from the MS/MS (Fig. 3B) data and then searched against a protein database using BLAST software. BLAST results showed that there was a short, nearly exact match for several CysP of Leishmania and other trypanosomes as shown in Table 1. A major characteristic of these CysP homologs is the ability to hydrolyze gelatin (14). Indeed, substrate gel electrophoresis using the active ConA fraction further confirmed that the 50-kDa protein was the CysP responsible for the cleavage activity (Fig. 1C).
FIG. 3.
Mass spectroscopy and amino acid determination. The highly purified CysP fraction was resolved by SDS-PAGE, and then the bands were excised and digested with trypsin. The resulting digests were analyzed by HPLC-ESI/MS/MS on a Thermo Finnigan LCQ ion trap mass spectrometer in conjunction with a Michrom BioResources MAGIC 2002 micro-HPLC and a microspray interface MALDI-TOF (MS). (A) A graph of the electro-spray mass spectroscopy data eluting at 17.3 min; (B) the tandem mass spectroscopy data of m/z 443.4. Also illustrated on the graph is the deduced amino acid sequence for the CysP.
TABLE 1.
MS/MS amino acid comparison
| Amino acid start | Sequencea | Organism |
|---|---|---|
| 121 | EVGGAPAAVDWRARG | T. rangeli |
| 119 | EVVGAPAAVDWRARG | T. cruzi |
| 122 | TTGRAPAAVDWREKG | T. brucei |
| 121 | ADLSAVDAVDWREKG | L. mexicana |
Boldface type indicates homology with the MS/MS data sequence, APAAVDWR.
Effects of temperature and pH on cleavage activity.
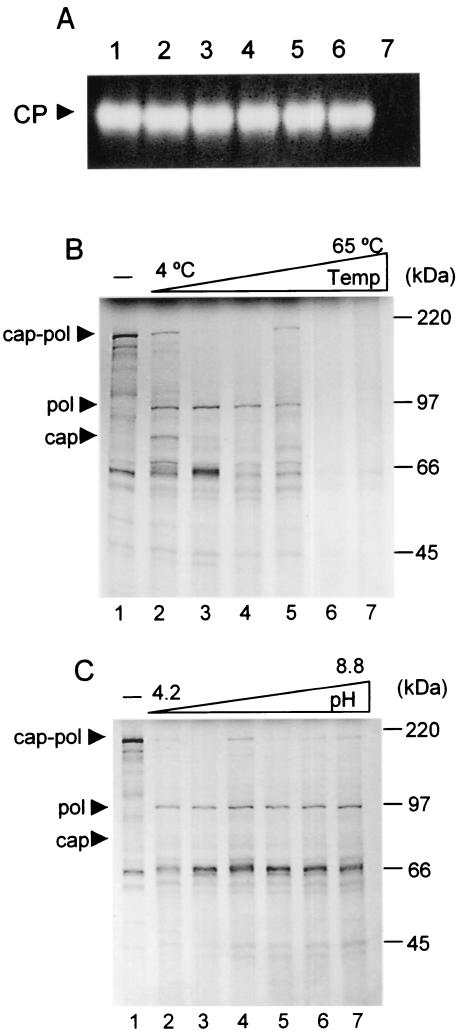
The purified protease demonstrated remarkable temperature stability as assayed by substrate gel electrophoresis. Aliquots (1.5 μl) of ConA-purified protease were incubated for 2 min in a 10-μl reaction containing 50 mM Tris (pH 6.8) at the temperatures indicated in Fig. 4. The samples were then assayed by substrate gel electrophoresis as described. The protease was surprisingly stable, retaining the ability to readily hydrolyze gelatin after treatment temperatures ranging from 4 to 65°C (Fig. 4A).
FIG. 4.
Temperature and pH optimization of purified CysP. (A) The purified CysP is heat stable. Highly purified enzyme was incubated at 4, 25, 37, 45, 55, and 65°C (lanes 1 to 6). Substrate gel electrophoresis was performed as described in Materials and Methods. Gelatinase activity was retained even after incubation at 65°C. The buffer-only control shows no gelatinase activity (lane 7). (B) In vitro cleavage assays were incubated at 4, 25, 37, 45, 55, and 65°C (lanes 2 to 7, respectively). Above 55°C, the substrate is hydrolyzed, though specificity is lost as evidenced by the absence of cleavage products. (C) pH optimization experiments. In vitro cleavage assays were incubated in buffers at pH 4.5, 5.5, 6.2, 7.2, 8.0, and 8.8 (lanes 2 to 7). The protease showed no preference for pH. Note that lane 1 contains the negative control at pH 5.5.
The broad range of temperature tolerance is echoed in temperature optimization studies using LRV gag-pol polyprotein as substrate. Figure 4B illustrates the broad range at which the protease remains functional. In the experiment, cleavage assays were incubated for a full 40 min at the indicated temperatures, with cleavage apparent at all the temperatures assayed. The optimal temperature ranged from 25 to 37°C, while at temperatures above 55°C, although the substrate is hydrolyzed, specificity is lost, as evidenced by the absence of cleavage products. It is interesting that the protease remains active even on ice, although the efficiency of cleavage is reduced by approximately 50%.
The stability and versatility of the protease also resonate in pH optimization experiments. In the study, microaliquots were incubated in an in vitro cleavage assay LRV Gag-Pol substrate in buffers ranging in pH from 4.5 to 8.8 as described. The samples were resolved by SDS-PAGE. The protease showed no preference for pH as illustrated in Fig. 4C. The protease was similarly efficient at digesting capsid-polymerase substrate at pH values ranging from 4.5 to 8.8.
Protease inhibitor studies.
To better characterize the nature of the CysP specificity, in vitro protease inhibitor studies were performed as detailed in Materials and Methods. Protease inhibitor studies reveal that the protease exhibits a dual susceptibility to both cysteine protease inhibitors and trypsin protease inhibitors. Cleavage was unaffected by serine or metalloprotease inhibitors such as aprotinin, EDTA, and phosphoramidon (Fig. 5, lanes 4, 6, and 9). However, cleavage activity was blocked by both CysP inhibitors (Fig. 5, lanes 3, 5, and 7) and trypsin inhibitors (Fig. 5, lanes 8 and 10). The CysP inhibitor cystatin also inhibits proteolysis (data not shown).
FIG. 5.
Protease inhibitor study. The following protease inhibitors (Roche) were examined for their ability to block polyprotein proteolysis (lanes 3 to 10, respectively): antipain dihydrochloride, aprotinin, E-64, EDTA, leupeptin, Pefabloc SC, phosphoramidon, and trypsin inhibitor. Cleavage was blocked by both CysP inhibitors (lanes 3, 5, and 7) and trypsin inhibitors (lanes 8 and 10). Lanes 1 and 2 represent the buffer-only control and the protease control, respectively.
Protease substrate preferences.
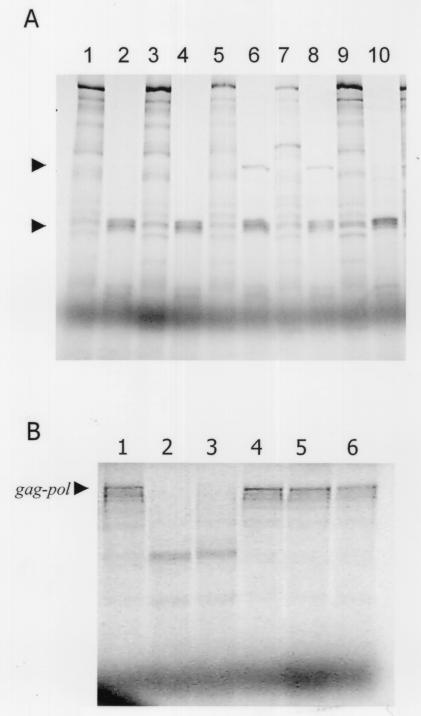
To define the substrate requirements for cleavage, a series of deletions and single-amino-acid mutations were constructed in a LRV gag-pol fusion construct as described in Materials and Methods. The single-amino-acid mutations were directed toward residues that were identified as cleavage sites for other trypsin-like cysteine proteases and also directed toward charged amino acids. None of the single-amino-acid mutations involving charged residues in the overlap region resulted in aberrant cleavage (Fig. 6). Expression of the ORF3 construct resulted in a 97-kDa protein of the predicted size for the polymerase enzyme (Fig. 6). Incubation of the product with purified CysP yielded a processed protein truncated by approximately 1 kDa (Fig. 7A). Expression of ORF2 construct in an in vitro transcription and translation assay generated an 82-kDa protein that, when incubated with the purified CysP, resulted in a processed protein degraded by approximately 20 kDa (Fig. 7B). Deletions ranging from 15 to 40 amino acids within the polymerase domain failed to block cleavage (Fig. 8A, lanes 1 to 4). Only a single large deletion (del gag-pol) spanning both the capsid and polymerase domains prevented Gag-Pol processing (Fig. 8B, lanes 4 to 6).
FIG. 7.
Protease cleavage assay using polymerase protein and capsid protein as substrate. (A) A truncated polymerase protein is observed in a protease cleavage assay with polymerase protein as substrate (lane 2). Arrows represent the location of polymerase protein and cleavage product, respectively. Lane 1 is the buffer-only negative control. (B) A truncated capsid protein is evident in a cleavage assay with capsid protein as substrate (lane 2). Arrows represent the location of polymerase protein and cleavage product, respectively. The buffer-only control is in lane 1.
FIG. 8.
(A) Cleavage of single-amino-acid mutants and deletion mutants. Cleavage assays were performed with del 15 (726 to 741) without (lane 1) or with (lane 2) CysP, del 30 (726 to 756) without (lane 3) or with (lane 4) CysP, K731 mutant without or with CysP (lanes 5 and 6, respectively), F766 mutant without or with CysP (lanes 7 and 8), and E7 (746) mutant without or with CysP (lanes 9 and 10). The arrows represent the 95-kDa polymerase cleavage product and the 66-kDa species of the capsid cleavage product. (B) Cleavage is abolished by a deletion spanning both the capsid and polymerase domains. Wild-type Gag-Pol and Gag-Pol del (644 to 797) were expressed in an in vitro transcription and translation assay, and the products were incubated in cleavage assay. Lanes 1 and 4 are wild-type and deletion Gag-Pol buffer-only control. Lanes 2 and 3 are wild-type Gag-Pol with CysP added at (40 and 120 ng, correspondingly), while lanes 5 and 6 are Gag-Pol del (644 to 797) with CysP added (at the concentrations indicated above).
DISCUSSION
Synthesis of a viral polymerase by ribosomal frameshifting is a common strategy for some members of the family Totiviridae (5, 22). In these systems, a capsid-polymerase fusion protein can be detected in vivo by immunoblotting. LRV is believed to use the same approach for expressing its polymerase, but presumably due to rapid processing of the capsid-polymerase polyprotein, a Gag-Pol type protein has not been identified. In this study, we purified the cellular protease responsible for proteolysis of the viral fusion protein and have identified it as a homolog of an L. mexicana CysP involved in parasite virulence. This is the first report of a protozoan protease that is important for parasite virulence also playing a role in a viral protein processing. In addition, biochemical characterization has allowed us to report for the first time a trypsin-like protease of Leishmania.
A major obstacle in purifying a cellular protease from Leishmania is that the parasite encodes many proteases with a variety of proteolytic specificities and overlapping specificities. As demonstrated by substrate gel electrophoresis, crude parasite lysates contain multiple proteases with gelatinase activity (Fig. 1C, lane 2). From previous experiments, we determined that cysteine protease inhibitors (17) block LRV Gag-Pol cleavage. Based on observed characteristics of the capsid-polymerase-specific protease, we hypothesized that the protease could be either a cathepsin B or cathepsin L protease. Since cathepsin L proteases have been shown to possess glycosylation sites (1), we decided to use the lectin ConA to capture the protease. A protease-enriched fraction was isolated that contained specific LRV Gag-Pol activity (Fig. 1A). Using substrate gel electrophoresis, a technique that can detect picogram quantities of proteases, we demonstrated that the fraction is free of contaminating proteases (Fig. 1C, lane 3).
Mass spectrometry analysis enabled us to identify the 30-kDa band as ConA, an artifact from the final purification step, while the 50-kDa protein was identified as a CysP with homology to other proteases of trypanosomes as well as Leishmania (Fig. 3 and Table 1). Proteases play vital roles in the physiology and virulence of parasitic protozoa. CysP in particular has been found to be relevant to several aspects of host-parasite interactions. CysPs of Trichomonas vaginalis and Entamoeba histolytica may function in immune evasion by degrading host immunoglobulin G, potentially limiting the effectiveness of the host humoral response (16, 21). Cruzipain, the major CysP of Trypanosoma cruzi, has been implicated in the escape of T. cruzi from the immune response. The CysP has been shown to completely degrade the Fc fragment of immunoglobulin G, thus leaving the Fab fragment intact on its surface (2). This event may serve to mask the parasite and protect it from destruction by complement. CysPs also function as virulence factors in Leishmania by facilitating survival in macrophages. Disruption of a CysP-encoding gene decreases infectivity to macrophages by 80% (15). In fact, the L. mexicana CPB gene array protease linked to increased macrophage survival is a homolog of the CysP protease that we identified in the present study (Table 1, L. mexicana sequence). That regulation of virus copy number is linked to an essential gene for parasite survival ensures the maintenance of a persistent infection.
LRV is an ancient virus that coevolved with its protozoan host (24). Since there is no evidence of horizontal transfer, the virus had to maintain a copy number compatible with host survival. It was hypothesized that the cleavage of LRV Gag-Pol protein is an important step in the virus's replication cycle, possibly functioning as either a virus maturation step or as a mechanism to maintain viral copy number. The fact that leishmaniasis is a zoonotic disease requires that the enzyme that processes viral proteins be robust. It must remain functional at low temperatures, like those encountered when in the sandfly vector, and at relatively high temperatures, like those in the human host. There may even be pH considerations when the parasite transforms into the intracellular amastigote within the macrophage. The characteristics of the purified CysP are consistent with an enzyme required for maintaining a viral infection at every stage of the disease. Enlisting a protease with such a dynamic range of pH and temperature tolerance has undoubtedly ensured the survival of the totivirus within the ancient protozoan.
Protease inhibitor studies have classified the protease as a trypsin-like cysteine protease (Fig. 5). There is no previous report of a trypsin-like cysteine protease expressed by any species of Leishmania (J. C. Mottram, personal communication). Although there are reports of a trypsin-like cysteine protease in T. brucei, it is a multisubunit protease that comprises the 20S proteosome (8, 12, 25). The protease is more reminiscent of the virally expressed 3C proteases of picornaviruses and caliciviruses. In these systems, the trypsin-like cysteine protease is required for processing of viral polyprotein into mature functional products. Blocking polyprotein processing adversely affects replication and virus maturation. Because of similarities to the calicivirus protease, initial attempts at substrate mapping focused on regions near the capsid-polymerase junction that resembled calicivirus cleavage sites. In each of these cases, cleavage was not abolished (Fig. 6), suggesting that the Leishmania CysP has unique cleavage sites.
Cleavage assays with the polymerase protein revealed the presence of a cleavage site near the amino terminus of the polymerase domain (Fig. 7A). Capsid proteolysis appeared to be more extensive with cleavage resulting in a 20-kDa truncation of the capsid protein (Fig. 7B). That the polymerase protein is more resistant to proteolysis than is the capsid protein is probably due to folding and the inaccessibility of cleavage sites. Deletion constructs targeting the amino terminus of the polymerase destabilized the protein. In cleavage assays with full-length Gag-Pol, the 95-kDa polymerase fragment was initially resistant to subsequent cleavage; however, 15 to 30 amino acid deletions at the amino terminus resulted in extensive proteolysis (Fig. 8A). Even a single-amino-acid mutation to a critical residue in the amino terminus can have this effect. For example, mutation of residues K and F results in a cleavage pattern similar to wild type, while mutation E7 results in complete degradation of the 95-kDa fragment. It is this instability of the altered Gag-Pol fusion that has undoubtedly added to the difficulty of defining a single protease cleavage site. In addition, throughout the study, the cleavage products appeared to vary between experiments. A possible explanation is that the experiments were performed at different times; although the highly purified eluent was stored at 4°C, the protease was still active, albeit at a decreased efficiency, thus resulting in degradation of the protease or activation of CysP precursor protein.
Initial cleavage events may unveil cryptic sites in the capsid domain, leading to extensive processing of the capsid protein. This is supported by the observation that a single 153-amino-acid deletion spanning both the capsid and polymerase junctions can block cleavage (Fig. 8B). Because the capsid protein can spontaneously assemble when full-length protein is expressed (3), it is unlikely that the extensive proteolysis is part of a maturation step to prime capsid assembly. Gag-Pol cleavage is more likely to be a mechanism to control viral genome copy number, with subsequent proteolysis functioning to eliminate potential harmful viral products. In a manner similar to the Yeast L-A virus, LRV probably requires dimerization of Gag-Pol fusion proteins for ultimate binding and packaging of its dsRNA genome (4-6, 23). By cleaving the fusion protein at the junction of the Gag-Pol domains, dimerization and packaging would be aborted, consequently affecting copy number. As we have previously reported, the capsid protein possesses site-specific endoribonuclease activity (13), and this endoribonuclease activity is dependent on fully assembled virus particles (Y.-T. Ro and J. L. Patterson, unpublished data). It is possible that LRV capsid also targets host RNA, since computer analysis has shown that similarity of the cleavage site to Leishmania rRNA (R. Carrion and J. L. Patterson, unpublished data). The extensive proteolysis of the capsid protein is likely to be a mechanism by which capsid assembly is averted, endoribonuclease activity is abolished, and the integrity of the host cell is maintained.
These studies provide a molecular characterization of the protease responsible for LRV Gag-Pol cleavage. The protease remains functional under a variety of temperature variations and is unaffected by various pH values, which may have contributed to its coevolution with its protozoan host. There appear to be numerous cleavage sites throughout the capsid and polymerase junction, many of which are hidden by protein conformation. Subsequent processing of the capsid domain may serve to eliminate viral proteins that may compromise parasite survival. Taken together, these data suggest that the role of the protease in Gag-Pol cleavage is to be one of regulating the persistent viral infection. More studies are required to definitively implicate Gag-Pol cleavage as a regulatory mechanism.
Acknowledgments
We thank Burton Beames and Steven Beverley for scientific advice. We also thank Patterson laboratory members Monica Avalos, Alex Hamill, Monica Ogg, and Ansyha Ticer for helpful discussions. We thank the UTHSCSA MS core. We thank April Hopstetter for assistance in preparing the manuscript.
R.C. received partial support through NIH training grant T32 AI07522. Y.-T.R. received support through a grant (R05-2000-000-00102-0) from the Basic Research Program of the Korea Science and Engineering Foundation.
REFERENCES
- 1.Barrett, A. J., and H. Kirschke. 1981. Cathepsin B, cathepsin H, and cathepsin L. Methods Enzymol. 80:535-561. [DOI] [PubMed] [Google Scholar]
- 2.Bontempi, E., and J. J. Cazzulo. 1990. Digestion of human immunoglobulin G by the major cysteine proteinase (cruzipain) from Trypanosoma cruzi. FEMS Microbiol. Lett. 58:337-341. [DOI] [PubMed] [Google Scholar]
- 3.Cadd, T. L., and J. L. Patterson. 1994. Synthesis of viruslike particles by expression of the putative capsid protein of Leishmania RNA virus in a recombinant baculovirus expression system. J. Virol. 68:358-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinman, J. D., and R. B. Wickner. 1992. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 66:3669-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujimura, T., J. C. Ribas, A. M. Makhov, and R. B. Wickner. 1992. Pol of gag-pol fusion protein required for encapsidation of viral RNA of yeast L-A virus. Nature 359:746-749. [DOI] [PubMed] [Google Scholar]
- 6.Fujimura, T., and R. B. Wickner. 1988. Gene overlap results in a viral protein having an RNA binding domain and a major coat protein domain. Cell 55:663-671. [DOI] [PubMed] [Google Scholar]
- 7.Guilbride, L., P. J. Myler, and K. Stuart. 1992. Distribution and sequence divergence of LRV1 viruses among different Leishmania species. Mol. Biochem. Parasitol. 54:101-104. [DOI] [PubMed] [Google Scholar]
- 8.Hua, S., W. Y. To, T. T. Nguyen, M. L. Wong, and C. C. Wang. 1996. Purification and characterization of proteasomes from Trypanosoma brucei. Mol. Biochem. Parasitol. 78:33-46. [DOI] [PubMed] [Google Scholar]
- 9.Labriola, C., M. Sousa, and J. J. Cazzulo. 1993. Purification of the major cysteine proteinase (cruzipain) from Trypanosoma cruzi by affinity chromatography. Biol. Res. 26:101-107. [PubMed] [Google Scholar]
- 10.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Lee, S. E., J. M. Suh, S. Scheffter, J. L. Patterson, and I. K. Chung. 1996. Identification of a ribosomal frameshift in Leishmania RNA virus 1-4. J. Biochem. (Tokyo) 120:22-25. [DOI] [PubMed] [Google Scholar]
- 12.Lomo, P. O., T. H. Coetzer, and J. D. Lonsdale-Eccles. 1997. Characterization of a multicatalytic proteinase complex (20S proteasome) from Trypanosoma brucei brucei. Immunopharmacology 36:285-293. [DOI] [PubMed] [Google Scholar]
- 13.MacBeth, K. J., Y. T. Ro, L. Gehrke, and J. L. Patterson. 1997. Cleavage site mapping and substrate-specificity of Leishmaniavirus 2-1 capsid endoribonuclease activity. J. Biochem. (Tokyo) 122:193-200. [DOI] [PubMed] [Google Scholar]
- 14.Mottram, J. C., M. J. Frame, D. R. Brooks, L. Tetley, J. E. Hutchison, A. E. Souza, and G. H. Coombs. 1997. The multiple cpb cysteine proteinase genes of Leishmania mexicana encode isoenzymes that differ in their stage regulation and substrate preferences. J. Biol. Chem. 272:14285-14293. [DOI] [PubMed] [Google Scholar]
- 15.Mottram, J. C., A. E. Souza, J. E. Hutchison, R. Carter, M. J. Frame, and G. H. Coombs. 1996. Evidence from disruption of the lmcpb gene array of Leishmania mexicana that cysteine proteinases are virulence factors. Proc. Natl. Acad. Sci. USA 93:6008-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Provenzano, D., and J. F. Alderete. 1995. Analysis of human immunoglobulin-degrading cysteine proteinases of Trichomonas vaginalis. Infect. Immun. 63:3388-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ro, Y. T., S. M. Scheffter, and J. L. Patterson. 1997. Specific in vitro cleavage of a Leishmania virus capsid-RNA-dependent RNA polymerase polyprotein by a host cysteine-like protease. J. Virol. 71:8983-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson, C. D., and G. H. Coombs. 1990. Characterization of three groups of cysteine proteinases in the amastigotes of Leishmania mexicana mexicana. Mol. Biochem. Parasitol. 42:269-276. [DOI] [PubMed] [Google Scholar]
- 19.Scheffter, S., G. Widmer, and J. L. Patterson. 1994. Complete sequence of Leishmania RNA virus 1-4 and identification of conserved sequences. Virology 199:479-483. [DOI] [PubMed] [Google Scholar]
- 20.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 21.Tran, V. Q., D. S. Herdman, B. E. Torian, and S. L. Reed. 1998. The neutral cysteine proteinase of Entamoeba histolytica degrades IgG and prevents its binding. J. Infect. Dis. 177:508-511. [DOI] [PubMed] [Google Scholar]
- 22.Wang, A. L., H. M. Yang, K. A. Shen, and C. C. Wang. 1993. Giardiavirus double-stranded RNA genome encodes a capsid polypeptide and a gag-pol-like fusion protein by a translation frameshift. Proc. Natl. Acad. Sci. USA 90:8595-8599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weeks, R. S., J. L. Patterson, K. Stuart, and G. Widmer. 1992. Transcribing and replicating particles in a double-stranded RNA virus from Leishmania. Mol. Biochem. Parasitol. 52:207-213. [DOI] [PubMed] [Google Scholar]
- 24.Widmer, G., and S. Dooley. 1995. Phylogenetic analysis of Leishmania RNA virus and Leishmania suggests ancient virus-parasite association. Nucleic Acids Res. 23:2300-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan, J., Q. Cheng, C. B. Li, and S. Aksoy. 2001. Molecular characterization of two serine proteases expressed in gut tissue of the African trypanosome vector, Glossina morsitans morsitans. Insect Mol. Biol. 10:47-56. [DOI] [PubMed] [Google Scholar]