Abstract
Replication of human immunodeficiency virus (HIV) involves obligatory sequential processes. Following viral entry and reverse transcription, the newly synthesized viral DNA integrates into the host chromatin. Integration is mandatory for viral production, yet HIV infection of CD4 T cells in vivo results in high levels of nonintegrated DNA. The biological potential of nonintegrated HIV DNA is unclear; however, prior work has demonstrated a limited transcription of the nef gene by nonintegrated HIV in infected quiescent T-cell populations. In a kinetic analysis of HIV infection of metabolically active transformed and primary CD4 T cells, we find an unexpected transient expression of both early and late message by nonintegrated HIV DNA. However, only the early multiply spliced transcript was measurably translated. This restriction of protein expression was due in part to inadequate Rev function, since expression of Rev in trans resulted in the expression of the late structural gene gag by nonintegrated HIV DNA.
Retroviruses, such as human immunodeficiency virus (HIV), undergo reverse transcription to form a double-stranded DNA, which is then integrated into the host chromatin. The integrated provirus transcribes new genomic RNAs and mRNAs for virion production (32). This ordered sequence, centered on integration, is mandatory for HIV replication, and yet the natural process of HIV infection yields nonintegrated DNA (22, 26); indeed, during the asymptomatic phase in HIV-infected individuals, nonintegrated HIV DNA levels can reach 99% of total viral DNA (8). The biological contribution of nonintegrated DNA is unclear, but previous reports have shown that nonintegrated viral DNA has limited transcriptional activity (1, 7, 9, 30, 33). For example, infection by integrase-defective HIV can induce β-galactosidase expression from an HIV long terminal repeat (LTR) reporter gene in a HeLa indicator line (1, 9, 33). Presumably, the HIV tat gene is expressed in the absence of functional integrase activity. Additionally, work from this and another laboratory (28, 34) has provided evidence that HIV infection of resting CD4 T-cell populations does not lead to detectable integration or replication but does result in the transcription of the nef gene. However, the rate of completion of steps prior to transcription, such as reverse transcription, is diminished in quiescent cells (14, 28), and detection of nef transcripts requires several days of incubation (28, 34). It seems possible that the restricted transcription seen following infection by integrase mutants or in quiescent cells is an aberration that exists only where integration is highly impeded. Indeed, infection of cells by integrase mutants results in amplified levels of circular HIV DNA (9, 33), and such forms of DNA have been demonstrated to possess transcriptional activity in transfection assays (6). The question arises whether this transcription from HIV DNA is a function of the inhibited integration activity or is a normal process of HIV infection that has yet to be defined.
In this report, we demonstrate that transcription from nonintegrated DNA is a normal, early step in HIV replication and that nonintegrated DNA has full capacity to synthesize all classes of viral transcripts, both the early, multiply spliced and the late, singly spliced and nonspliced transcripts, prior to integration.
MATERIALS AND METHODS
Viruses and cells.
Wild-type HIV-1NL4-3, the integrase mutant HIV-1IN/D116N (9), and primary CD4 T cells were prepared as described previously (34). Equal p24 levels of HIV-1NL4-3 and HIV-1IN/D116N were used in infection studies (500 ng of p24 per 106 cells; multiplicity of infection, <0.05). Cells were incubated with virus for 2 to 3 h, then pelleted and resuspended in fresh culture medium. Viral production was defined by p24 secretion (HIV type 1 [HIV-1] p24 antigen assay; Beckman Coulter). Induction of Rev in ΔKS Jurkat cells was carried out as previously described (5). Nef-expressing Jurkat cells, made by retroviral transduction, have also been described previously (25).
DNA detection.
Nuclear DNA was purified from infected cells following lysis (for 5 min at 4°C with a solution containing 50 mM KCl, 10 mM HEPES-NaOH [pH 7.6], 5 mM MgCl2, and 0.05% NP-40). Nuclei were then pelleted in a microcentrifuge, washed once with the lysis buffer, and then resuspended with the nuclear lysis solution from the Wizard Genomic DNA purification kit (Promega). PCR amplification of 1-LTR circles was carried out by using primers LTR-nef2 (5′-TGGGTTTTCCAGTCACACCTCAG-3′) and LTR-gag (5′-GATTAACTGCGAATCGTTCTAGC-3′) in a solution containing 50 μl of 10× PCR buffer, 125 μM deoxynucleoside triphosphates, 1.5 mM Mg2+, 50 pmol of each primer, and 1 U of SuperTaq Plus DNA polymerase (Ambion), with 35 cycles of 20 s at 94°C and 90 s at 68°C. Quantitative PCR analyses of late reverse transcription (RT) products and 2-LTR circles were achieved with an ABI Prism 7700 sequence detection system as described previously (4), with the following modifications for detection of the late RT product: the forward primer was 5′ LTR-U5 (5′-AGATCCCTCAGACCCTTTTAGTCA-3′), the reverse primer was 3′ gag (5′-TTCGCTTTCAAGTCCCTGTTC-3′), and the probe was FAM-U5/gag [5′-(FAM)-TGTGGAAAATCTCTAGCAGTGGCGCC-(TAMRA)-3′]. Each incubation contained 300 nM each primer and 200 nM probe. The standard used for both late RT and 2-LTR circle quantification was constructed by using a plasmid containing a complete 2-LTR region (pLTR-2C, cloned by amplification of infected cells with 5′-TGGGTTTTCCAGTCACACCTCAG-3′ and 5′-GATTAACTGCGAATCGTTCTAGC-3′). Measurements were determined in duplicate, ranging from 10 to 106 copies, mixed with DNA from uninfected cells.
Assay for integration.
Nuclear DNA was purified, and aliquots equivalent to 1/10 of the DNA were subjected to amplification by Alu-LTR PCR, using the Alu primer (5′-TCCCAGCTACTGGGGAGGCTGAGG-3′) and HIV-1 LTR primer L1 (5′-AGGCAAGCTTTATTGAGGCTTAAGC-3′) as described previously (34). Following Alu-LTR PCR, a second round of PCR was carried out by using an aliquot equivalent to 1/500 of the PCR product and the LTR-specific primer pair L2 (5′-digoxigenin-CTGTGGATCTACCACACACAAGGCTAC-3′) and L3 (5′-GCTGCTTATATGTAGCATCTGAGGGC-3′). The PCR was performed in a solution containing 50 μl of Promega PCR buffer, 150 μM deoxynucleoside triphosphates, 1.5 mM Mg2+, 50 pmol of each primer, and 2 U of Taq DNA polymerase (Promega), with 35 cycles of 20 s at 94°C, 30 s at 65°C, and 40 s at 72°C. Following gel electrophoresis, blots were incubated with a horseradish peroxidase-labeled sheep antibody for digoxigenin (1:1,000 dilution) (Boehringer Mannheim). The light signal was captured on a cooled charge-coupled device (CCD) camera (Alpha Innotech) by using SuperSignal chemiluminescence substrate (Pierce).
Quantitative RT-PCR.
Total cellular poly(A)+ mRNA was purified with a MicroPoly(A)Pure mRNA isolation kit (Ambion) and then treated with DNase I (DNA-free kit; Ambion). Reverse transcription was accomplished by using the RETROscript First-Strand Synthesis kit (Ambion) with random decamers as the first-strand primers. Following cDNA synthesis, PCR was carried out as described previously (34). Briefly, primers F2 (5′-TAATCGGCCGAACAGGGACTTGAAAGCGAAAG-3′) and B4 (5′-CCATCGATTGCGTCCCAGAAGTTCCACAATCC-3′) were used to amplify viral transcripts, particularly doubly spliced transcripts such as tat, rev, and nef. For relative quantification, cellular β-actin transcripts were coamplified. To distinguish individual transcripts, the PCR products were hybridized with the digoxigenin-labeled probe P1 (5′-GCTGACTTCCTGGATGCTTCCAGG-3′), P2 (5′-CCTGCCATAGGAGATGCCTAAGGC-3′), P3 (5′-GAGCTCTTCGTCGCTGTCTCCGCT-3′), or P4 (5′-GTGCTAAGGATCCGTTCACTAATCG-3′). For analysis of singly spliced viral transcripts such as the env transcript, primers F2 and B3 (5′-CCCATCTCCACAAGTGCTGATACTTC-3′) were used, whereas for the vif transcript, F2 and B2 (5′-CTAGGTCAGGGTCTACTTGTGTGC-3′) were used. For analysis of full-length viral transcripts, the primer pair F2 and B1 (5′-digoxigenin-TCTGAAGGGATGGTTGTAGCTGTCC-3′) was used.
Immunodetection of viral proteins.
Viral proteins were detected by resolving proteins on a sodium dodecyl sulfate (SDS)-4 to 20% polyacrylamide gel and electroblotting onto a 0.2-μm-pore-size nitrocellulose membrane. A 1:1,000 dilution of a human anti-HIV antiserum was incubated with the membrane, followed by a peroxidase-conjugated goat anti-human secondary antiserum (dilution, 1:2,000; Kirkegaard & Perry Laboratories). The Nef protein was detected by using a 1:1,000 dilution of a sheep anti-Nef antiserum (21), followed by a peroxidase-conjugated rabbit anti-sheep secondary antibody (dilution, 1:20,000; Kirkegaard & Perry Laboratories). Inclusion of recombinant Nef protein (a generous gift from Paul Wingfield, National Institute of Arthritis and Musculoskeletal and Skin Diseases) served as a control. The Rev protein was detected by using a 1:500 dilution of an anti-Rev rabbit antiserum (kindly provided by Barbara Felber, National Institute of Allergy and Infectious Diseases) followed by a peroxidase-conjugated goat anti-rabbit secondary antibody (dilution, 1:1,000; Kirkegaard & Perry Laboratories). The immunoreactive product was detected by chemiluminescence as described elsewhere (34).
RESULTS
To examine the early events in viral infection, we performed kinetic analysis of the sequential processes of retroviral infection: reverse transcription, integration, and transcription from the provirus in cells fully supportive of HIV replication. As a negative control for integration, we used an HIV integrase-defective virus, the D116N mutant (9). In these studies CEM-50 T cells were infected with equivalent levels of wild-type HIV or the D116N virus. The CEM line is readily infected, and unlike primary cell preparations, all cells within the population have a uniform propensity for HIV infection. Additionally, by eliminating the variation seen with cells from different donors, quantitative comparisons were possible over multiple experiments.
Kinetic measurement of viral reverse transcription, integration, and transcription by quantitative PCR.
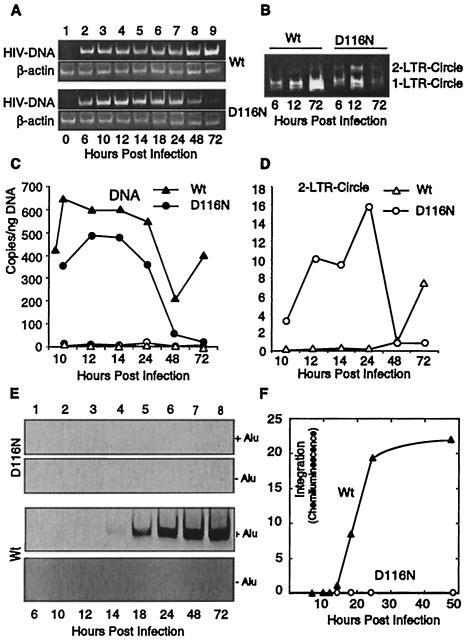
Following infection by either wild-type or integrase mutant D116N HIV, viral DNA synthesis (late RT product) was detected at the earliest time point (6 h postinfection) (Fig. 1A), and DNA expression from the D116N mutant was transient. Transient formation of 1-LTR and 2-LTR circular DNAs was also seen following infection by the D116N virus (Fig. 1B). These experiments were then repeated using the more quantitative real-time PCR. DNA levels for the D116N mutant approximated those of the wild-type virus, although DNA expression by the mutant virus approached background by 2 to 3 days (Fig. 1C). We simultaneously measured the generation of 2-LTR circles by quantitative PCR (Fig. 1D). We found that by 12 h postinfection, the level of 2-LTR DNA circles generated by the integrase mutant virus was an order of magnitude greater than that generated by wild-type virus infection. This is consistent with the previous findings of enhanced circle formation following infection by integrase-defective HIV (9, 33). 1-LTR circles, which are not quantifiable by this analytical PCR method, were generated by both wild-type and D116N infection (Fig. 1B). All forms of DNA observed following D116N mutant infection were transient.
FIG. 1.
Viral reverse transcription and integration in wild-type- and D116N mutant-infected cells. CEM-50 cells (106) were infected with equivalent levels of wild-type (Wt) HIV-1NL4-3 or an integrase mutant, HIV-1IN/D116N (D116N). Cells were harvested at different times postinfection, and nuclear DNA was purified. (A) Total cellular DNA was amplified by PCR as previously described (34). The sample immediately following the addition of virus was defined as the time zero sample. (B) The 1-LTR circle was also measured by PCR amplification as described in Materials and Methods. Both the 1-LTR circle and the 2-LTR circle were amplified by the primer pair as indicated. (C) Real-time PCR. The same DNA used in panels A and B was subjected to real-time PCR quantification using primers targeting the late RT product (filled triangles and circles) and 2-LTR circles (open triangles and circles). (D) Expansion of the 2-LTR circle data from panel C. (E) Viral DNA integration in cells infected with wild-type or D116N mutant HIV. Total cellular DNAs from D116N mutant- or wild-type-infected cells, harvested at different times postinfection (lanes 1 to 8), were subjected to Alu-PCR amplification (+Alu) as described in Materials and Methods. To ensure that the amplification was specific to integrated viral DNA, amplification was also carried out in the absence of the Alu primer (−Alu). The PCR products were analyzed by gel electrophoresis, transferred to a nylon membrane, and hybridized with a digoxigenin-labeled probe. (F) Plot of amplification signals as captured on a cooled CCD camera. Each experiment was reproduced three or more times.
HIV DNA integration was then measured by the Alu assay. As expected, the D116N integrase mutant displayed no activity (Fig. 1E). The earliest time point for detection of integration by wild-type virus was 14 h postinfection. Following this initial measurable integration, the signal increased linearly with time, up to 24 h. Extrapolation of this line showed zero integration events at a point between 13 and 14 h (Fig. 1F). These findings are consistent with the results of prior analyses by real time PCR (4) and by Southern blotting (36), where integration was not demonstrable prior to 12 h postinfection.
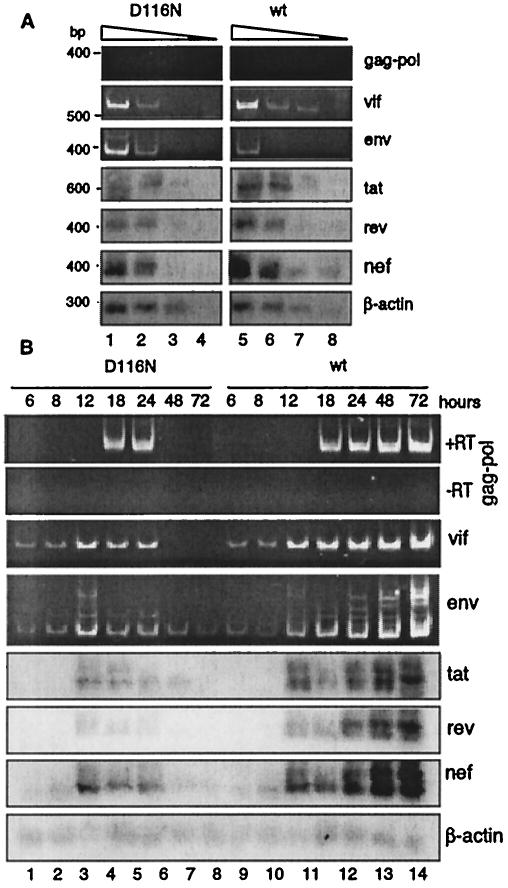
We then compared the transcriptional activities of wild-type and integrase mutant HIV in these T cells. We first examined serial dilutions of viral mRNA from infected cells to ensure that the PCR analysis was within the range of concentration dependency (Fig. 2A) and then examined the kinetics of viral transcript generation (Fig. 2B). In contrast to the previously demonstrated predominant expression of nef in resting CD4 T cells (28, 34), the nonintegrating DNA from the D116N mutant HIV unexpectedly generated all forms of transcripts. Two other features became apparent. First, the transcripts generated by the nonintegrating HIV were transiently expressed in the actively proliferating cells, with nef transcripts peaking at 12 h postinfection; secondly, the early kinetics of transcript formation appeared similar for the D116N mutant and wild-type HIV.
FIG. 2.
Early HIV transcription is similar for CEM cells infected by wild-type (wt) or nonintegrating D116N mutant HIV. (A) Comparison of viral transcription at 12 h postinfection. mRNAs from 104 infected cells were harvested. For each sample, mRNA was purified, treated with DNase I, serially diluted (1:5), and reverse transcribed using random decamers. One-fourth of the cDNA was subjected to PCR amplification using the primer pair F2-B4 as described in Materials and Methods. To ensure that comparisons were quantitative, samples were normalized by the content of the coamplified cellular β-actin transcript. One-fifth of the product was applied to a 2% agarose gel, transferred to a nylon membrane, and hybridized with digoxigenin-labeled probes specific for tat, tat-rev (rev panel), tat-rev-nef (nef panel), or human β-actin. The same cDNA was further amplified by the primer pair F2-B3, F2-B2, or F2-B1 to detect singly spliced env and vif viral transcripts or unspliced gag-pol transcripts. Amplified products were resolved on a 6% polyacrylamide gel and visualized by ethidium bromide staining. (B) Time course analysis of viral transcription in wild-type and D116N mutant infection. Samples taken at each time point following infection were analyzed as for panel A and normalized by the β-actin transcript. To ensure that the amplified unspliced transcripts were not derived from viral DNA, mRNA was also directly subjected to amplification using F2-B1 without reverse transcription (panel −RT).
Lack of viral replication by integrase-defective virus.
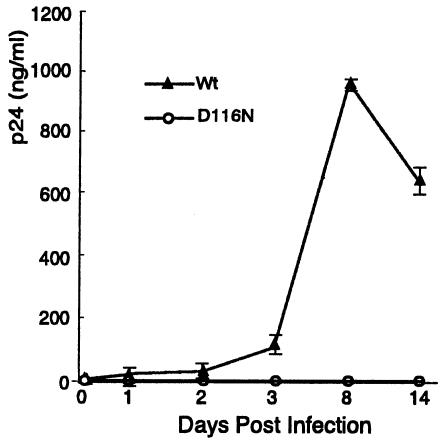
These findings raised a concern that the CEM-50 cells might support viral replication of the integrase mutant. Certain transformed T-cell lines, such as the human T-cell lymphotropic virus type 1 (HTLV-1)-transformed MT-4 cell line, can complement integrase-defective HIV and permit limited viral replication (18, 29). Although all classes of viral transcripts were generated following infection by the integrase-defective virus (Fig. 2), the CEM line did not support the replication of D116N mutant HIV (Fig. 3). This finding is consistent with previous demonstrations that CEM cells are not permissive for replication of HIV possessing the integrase mutation (9, 18, 33).
FIG. 3.
Integrase-defective D116N HIV does not replicate in CEM cells. Cells were infected with either wild-type (Wt) or D116N mutant HIV at equal viral loads. Viral replication was monitored by measuring extracellular p24 levels. Data are means of triplicate determinations. Error bars, standard deviations.
Diketo acid integrase inhibitor does not affect early transcription.
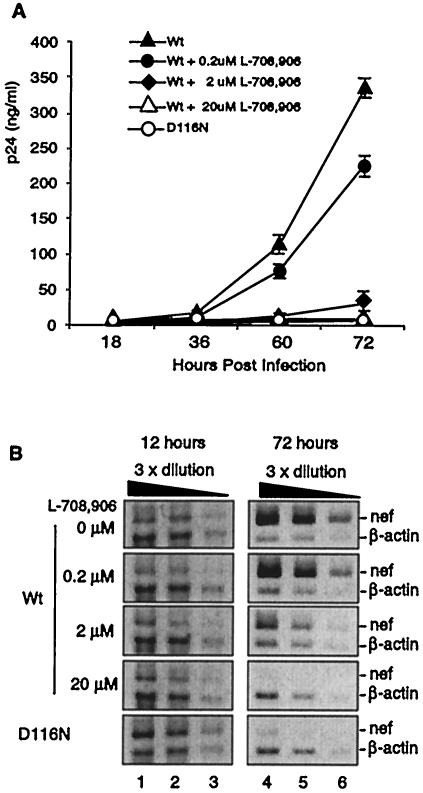
Independently of the inability of integrase-negative virus to replicate, the similar kinetics of early transcription following infection of cells by either wild-type HIV or the nonintegrating D116N mutant suggest that the early transcripts could be generated by nonintegrated DNA after infection by either virus. The peaking of transcript levels at 12 h postinfection is in contrast to the earliest detectable integration at approximately 14 h postinfection. The reduction of transcript synthesis after 24 h with the D116N mutant and the rapidly increasing levels with wild-type virus are presumably mediated by the absence or presence of the stable integration of HIV DNA, respectively. However, to further exclude the possibility that this early transcriptional activity by wild-type virus was mediated by an integration event undetectable by the Alu assay, we made use of the diketo acid integrase inhibitor L-708,906. This compound inhibits strand transfer by HIV-1 integrase without affecting other known functions (13). Inhibition of viral replication followed an expected dose-response curve (13) (Fig. 4A), where 20 μM L-708,906 prevented measurable replication of wild-type virus. We then compared increasing doses of the inhibitor to levels of Nef transcript generated, as measured at 12 and 72 h postinfection (Fig. 4B). Prevention of integration by increasing concentrations of inhibitor eliminated the Nef transcript at 72 h postinfection, consistent with the inhibition of formation of a stable integrated provirus. However, the inhibitor, even at the highest concentration, did not alter Nef transcription at the 12-h time point, providing evidence that the early RNA synthesis from wild-type virus is from nonintegrated DNA. Thus, there appears to be no dependence on integration for this early transcriptional activity immediately following infection.
FIG. 4.
Early viral transcription is resistant to the integrase inhibitor L-708,906. (A) Effect of the integrase inhibitor on viral replication. CEM cells were treated with various amounts of L-708,906 1 h before infection with wild-type (Wt) or D116N mutant HIV. Virus replication was monitored by measuring p24 production in three independent determinations. Values are means and standard deviations. (B) Viral mRNA from the infected cells, harvested at 12 or 72 h, was analyzed by quantitative RT-PCR as in Fig. 2.
Restricted translation of HIV transcripts by integrase-defective virus.
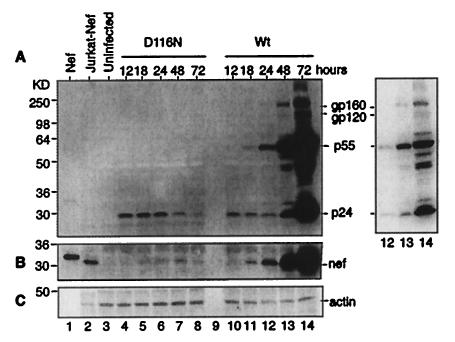
Given the unexpected expression of all transcripts by the integrase mutant in active cells, we then looked for expression of viral proteins. A Western blot analysis of an SDS gel of infected cells was developed with human anti-HIV antisera, as shown in Fig. 5A. Gag product p24 was detected at the earliest time points for both the wild-type and the D116 mutant virus; however, p55gag, the precursor to p24, became visible in wild-type virus infection only after 1 day and gained prominence at 48 h postinfection. Since both viruses displayed the early p24, but in the absence of precursor, we assume that the early presence of this protein was residual from the virions added to the cell suspension (as evidence, p24 could be detected immediately after infection [data not shown]). The lack of structural protein synthesis in the D116N mutant virus infection is consistent with the nonreplicative capacity of this integrase mutant. A separate Western blot analysis demonstrated the expression of Nef protein at 24 to 48 h postinfection for both viruses, and for the D116N mutant virus, Nef protein expression peaked at 48 h (Fig. 5B). The transient nature of protein expression in the absence of integration is presumably due to the loss of DNA and transcript, and the expression of Nef protein is consistent with the earlier finding of Nef protein production in quiescent T cells in the absence of detectable integration (34). These Western blot analyses define the restricted generation of viral products in the absence of integration. The presence of Nef implies that Tat and Rev may also be expressed, but at low concentrations these latter proteins are poorly detected by Western blot analysis.
FIG. 5.
Viral protein synthesis is limited to early products in the absence of integration. CEM cells were infected as in Fig. 1 and were harvested at the indicated time points. A total of 5 × 105 cells were loaded onto an SDS-14% polyacrylamide gel, resolved, and electroblotted onto a nitrocellulose membrane for Western blot analysis (lanes 4 to 14). (A) A human anti-HIV antiserum was incubated with the membrane, followed by a goat anti-human secondary antiserum conjugated with peroxidase. The right panel is a shorter exposure of lanes 12 to 14. (B) Following signal detection, the blot was stripped and reblotted with a sheep anti-Nef antiserum, followed by a rabbit anti-sheep secondary antibody conjugated to peroxidase. (C) As a control, the same blot was also reblotted with a monoclonal antibody against human actin and a goat anti-mouse secondary antibody conjugated to peroxidase. To serve as Nef controls, 0.5 ng of purified HIV-1 Nef protein (lane 1), 5 × 105 Jurkat cells expressing the nef gene of HIV-1NL4-3 (lane 2), and 5 × 105 uninfected CEM cells (lane 3) were applied to the gel. Wt, wild type.
Rev function is required for p55 synthesis.
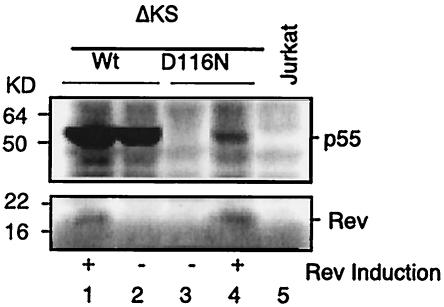
The finding that protein translation was limited to multiply spliced message suggested an inadequate Rev function. Rev is required for nuclear export and translation of the partially spliced and unspliced message (10, 11, 16). Although the Rev transcript from nonintegrated DNA was detected (Fig. 2), the level of transcript was low and its expression was transient. To test this hypothesis, we infected cells that contained an inducible Rev gene with the D116N integrase mutant virus. The presence of this in trans, induced Rev protein permitted the translation of Gag message (unspliced) to precursor p55 protein (Fig. 6), but at a reduced level relative to that for the wild-type virus. Expression of the structural p55 protein was optimal when Rev was induced at the time of viral infection, but viral replication remained undetectable (data not shown), presumably a result of diminished and transiently expressed Rev. Thus, inadequate Rev activity, as generated in D116N mutant virus infection, will lead to restricted expression of multiply spliced (Rev-independent) HIV gene products. We also conclude that the unspliced transcripts, as synthesized by nonintegrated HIV DNA, were functional.
FIG. 6.
In the absence of integration, induced Rev expression permits late structural protein synthesis. ΔKS Jurkat cells, which possess a Rev-inducible system based on the tetracycline operon, were infected with wild-type (Wt) virus (lanes 1 and 2) or the D116N mutant (lanes 3 and 4) in the absence (lanes 2 and 3) or presence (lanes 1 and 4) of Rev induction by the addition of tetracycline at the time of infection. Uninfected Jurkat cells were used as a control (lane 5). For Western blot analysis, 5 × 105 cells were loaded onto an SDS-4 to 20% polyacrylamide gel. The blot was probed with a human anti-HIV serum (top panel), as described in the legend to Fig. 5A, and with a rabbit anti-Rev serum reacted with a goat anti-rabbit secondary antibody (bottom panel). p55gag synthesis was detected by Western blotting in D116N mutant infection with Rev induction (lane 4).
Early transcriptional activity is also seen in HIV infection of primary CD4 T cells.
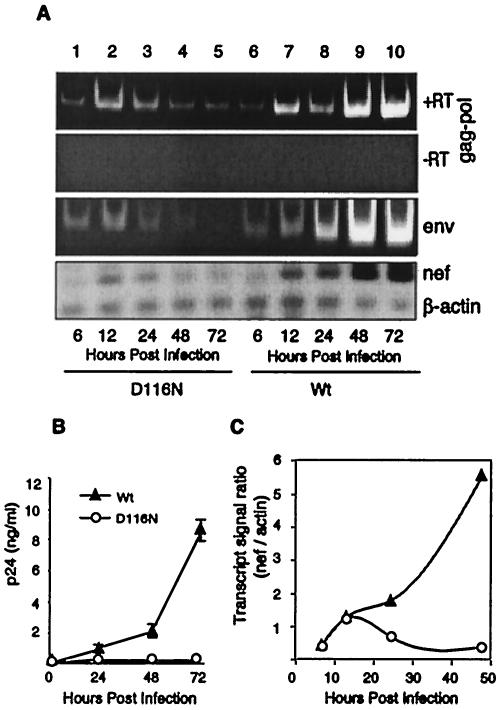
We then examined early transcriptional activity following wild-type and D116N mutant HIV infection of activated primary CD4 T cells. Consistent with the results from metabolically active CEM cells, all classes of transcripts were synthesized, and the transcriptional activities for the earliest time points were identical for the D116N mutant and wild-type viral infections (Fig. 7A). The expression of transcripts from the D116N mutant was transient, with a peak of multiply spliced Nef transcript at 12 h postinfection. As in the CEM cells, viral replication in primary T cells was absent following HIV D116N mutant infection (Fig. 7B), in agreement with previous findings (7, 33).
FIG. 7.
Early HIV transcription from nonintegrated DNA in primary human T cells. Primary human CD4 T cells from peripheral blood of healthy donors were purified, stimulated with anti-CD3/CD28-conjugated beads at a ratio of 5 beads per cell, and then infected with wild-type (Wt) or D116N mutant virus as described in the legend to Fig. 1. mRNA molecules from infected cells were subjected to RT-PCR analyses as described in the legend to Fig. 2. (A) Multiply spliced nef, singly spliced env, and unspliced transcripts. (B) The presence of multiple transcripts in infected cells did not lead to replication of the D116N mutant in stimulated primary T cells, as measured by extracellular p24 production. Data are means for triplicate determinations plus standard deviations. (C) For comparison of transcriptional activity in cells infected with wild-type versus D116N mutant HIV, the ratio of chemiluminescent signals between nef and β-actin was plotted for each time point. These results were reproduced using primary CD4 T cells from multiple donors (data not shown).
To permit a comparison of HIV transcriptional activities in wild-type versus D116N mutant infection, we plotted the ratio of the transcript signal for Nef to that for an internal control, β-actin. As demonstrated in Fig. 7C, relative to β-actin transcript levels, Nef levels were similar for wild-type and D116N mutant infections at 12 h postinfection, suggesting that the wild-type virus undergoes nonintegration transcriptional activity similar to that of the nonintegrating D116N mutant virus. The ratio also permitted a comparison of Nef transcript synthesis at different times postinfection, with the 12-h preintegration level representing approximately 20% of the level achieved by 48 h postinfection for the wild-type virus.
DISCUSSION
The findings presented here demonstrate that reverse-transcribed, nonintegrated HIV DNA functions as a template and that this transcription is a normal early process in HIV infection of T cells. Furthermore, in active T cells, the preintegration DNA has the full capacity to synthesize all classes of viral transcripts. This is in contrast to the previous demonstration of limited transcription in resting CD4 T cells (28, 34). This limitation is likely due to the low metabolic state seen in quiescent T cells, which suppresses infection processes (3, 14, 28, 30, 35).
Our findings are reminiscent of some early demonstrations of viral replication from integration-defective retroviruses, such as spleen necrosis virus (23), visna virus (12), and HIV (7, 18, 29). Various approaches have added stipulations to these initial findings (15, 18). For example, the replication of integrase-defective HIV in the transformed T-cell line MT-4 may be due to complementation by viral gene products of the transforming (and integrated) HTLV-1 provirus (18). Complementation between HIV and HTLV-1 infections has been amply demonstrated (17, 24, 27). In our system, when primary human CD4 T cells or CEM cells were used, we observed an absolute requirement of integration for viral replication. This is supported by the finding that late viral structural proteins were not detected in cells infected with integrase-defective virus (Fig. 5A).
The transcriptional activity from integrase-defective HIV and from nonintegrated wild-type HIV appears to be dependent on intact retroviral processes that normally lead up to integration. For example, inhibition of reverse transcription (34), or mutation of the nuclear localization signal region of the viral integrase (2), greatly diminishes measurable transcriptional or Tat-dependent reporter activity. This suggests that, along with the Rev dependency for gene expression (Fig. 6), this transcriptional activity occurs under the conditions of normal retroviral infection and is dependent on essential retroviral processes.
The nature of the transcribing DNA remains unknown, but there is evidence that all forms, linear DNA and 1-LTR and 2-LTR circles, can transcribe in transfection assays (6). Previous studies with integrase-defective HIV have demonstrated the enhanced expression of 2-LTR circles (as seen here in Fig. 1D), coincidental with the induction of a Tat-dependent reporter (9, 33). Additionally, inhibition of wild-type HIV integration by a diketo acid integrase inhibitor, as used here, resulted in an apparent increase in DNA circles (13). In our experiments, when both the viral DNA and transcripts were quantitatively compared, neither the mutant nor the inhibitor dramatically affected early transcriptional levels. We conclude that the level of transcription from nonintegrated DNA does not appear to be defined by the concentration of circular 2-LTR DNA. Since a quantitative difference could not be established for 1-LTR or linear DNA in our experimental approaches, both remain candidates for transcriptional activity. Furthermore, although transcriptional activity (Fig. 2B) precedes measurable integration (Fig. 1F), we do not conclude that these are sequential events for the same DNA molecule.
It is not known how much of the nonintegrated HIV DNA found in HIV-infected individuals has transcriptional capacity. Approximately 99% of total HIV DNA in CD4 T cells seen in patients during the asymptomatic phase of HIV infection exists as a nonintegrated, linear form (8), but this DNA is replication incompetent and appears to have undergone host cell modifications. Measurements of the level of HIV DNA integration in short-term cellular systems have also been reported, with some experiment-dependent variation. With Alu PCR (4), linker-primer PCR (31), and an Alu real-time PCR assay at an early time point (19), integration levels were 5 to 10% of total reverse-transcribed DNA, whereas a subtractive method suggested that integration levels could exceed 50% of total DNA (36). Nonetheless, the infection process of HIV will involve the generation of nonintegrating DNA, perhaps with greater prominence in in vivo infection.
This early transcriptional activity would provide a means for HIV gene expression, particularly Nef and Tat expression, in the absence of integration and productive viral infection. Both of these proteins are known to enhance T-cell activity (20, 25), as well as viral replication following infection of resting T cells (34). Additionally, the capacity of a nonintegrating HIV to transiently generate viral products suggests that nonintegrating lentiviral vectors could serve to express low levels of protein for therapeutic or vaccine purposes, without the permanency of an integrated retrovirus or disruption of normal cellular genes.
Acknowledgments
We thank Thomas Trischmann for providing elutriated lymphocytes, Gene Major for use of his BL-3 facility, Terrence Burke for providing the integrase inhibitor L-708,906, Alan Engelman and Malcolm Martin for viral DNA, Celsa Spina and Barbara Felber for antisera, V. Kalyanaraman for the CEM-50 cells, Paul Wingfield for recombinant Nef protein, and the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, for human anti-HIV Ig (NABI and NHLBI) and DKS Jurkat cells (Joseph Sodroski). We also thank Klaus Strebel, Chris Buck, and Gary Nabel for comments and criticism concerning the manuscript.
REFERENCES
- 1.Ansarilari, M. A., L. A. Donehower, and R. A. Gibbs. 1995. Analysis of human immunodeficiency virus type-1 integrase mutants. Virology 211:332-335. [DOI] [PubMed] [Google Scholar]
- 2.Bouyac-Bertoia, M., J. D. Dvorin, R. A. M. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 3.Bukrinsky, M. I., T. L. Stanwick, M. P. Dempsey, and M. Stevenson. 1991. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254:423-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, S. L., M. S. T. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 5.Cao, J., I. W. Park, A. Cooper, and J. Sodroski. 1996. Molecular determinants of acute single-cell lysis by human immunodeficiency virus type 1. J. Virol. 70:1340-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cara, A., A. Cereseto, F. Lori, and M. S. Reitz. 1996. HIV-1 protein expression from synthetic circles of DNA mimicking the extrachromosomal forms of viral DNA. J. Biol. Chem. 271:5393-5397. [DOI] [PubMed] [Google Scholar]
- 7.Cara, A., F. Guarnaccia, M. S. Reitz, Jr., R. C. Gallo, and F. Lori. 1995. Self-limiting, cell type-dependent replication of an integrase-defective human immunodeficiency virus type 1 in human primary macrophages but not lymphocytes. Virology 208:242-248. [DOI] [PubMed] [Google Scholar]
- 8.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 9.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinberg, M. B., R. F. Jarrett, A. Aldovini, R. C. Gallo, and F. Wongstaal. 1986. HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral-RNA. Cell 46:807-817. [DOI] [PubMed] [Google Scholar]
- 11.Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland, and G. N. Pavlakis. 1989. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 86:1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris, J. D., H. Blum, J. Scott, B. Traynor, P. Ventura, and A. Haase. 1984. Slow virus visna: reproduction in vitro of virus from extrachromosomal DNA. Proc. Natl. Acad. Sci. USA 81:7212-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 14.Korin, Y. D., and J. A. Zack. 1999. Nonproductive human immunodeficiency virus type 1 infection in nucleoside-treated G0 lymphocytes. J. Virol. 73:6526-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.List, J., and A. T. Haase. 1997. Integration of visna virus DNA occurs and may be necessary for productive infection. Virology 237:189-197. [DOI] [PubMed] [Google Scholar]
- 16.Malim, M. H., J. Hauber, R. Fenrick, and B. R. Cullen. 1988. Immunodeficiency virus Rev trans-activator modulates the expression of the viral regulatory genes. Nature 335:181-183. [DOI] [PubMed] [Google Scholar]
- 17.Moriuchi, H., M. Moriuchi, and A. S. Fauci. 1998. Factors secreted by human T lymphotropic virus type I (HTLV-I)-infected cells can enhance or inhibit replication of HIV-1 in HTLV-I-uninfected cells: implications for in vivo coinfection with HTLV-I and HIV-1. J. Exp. Med. 187:1689-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakajima, N., R. Lu, and A. Engelman. 2001. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: definition of permissive and nonpermissive T-cell lines. J. Virol. 75:7944-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Doherty, U., W. J. Swiggard, D. Jeyakumar, D. McGain, and M. H. Malim. 2002. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 76:10942-10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ott, M., S. Emiliani, C. VanLint, G. Herbein, J. Lovett, N. Chirmule, T. McCloskey, S. Pahwa, and E. Verdin. 1997. Immune hyperactivation of HIV-1-infected T cells mediated by Tat and the CD28 pathway. Science 275:1481-1485. [DOI] [PubMed] [Google Scholar]
- 21.Pandori, M. W., N. J. S. Fitch, H. M. Craig, D. D. Richman, C. A. Spina, and J. C. Guatelli. 1996. Producer-cell modification of human immunodeficiency virus type 1: Nef is a virion protein. J. Virol. 70:4283-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pang, S., Y. Koyanagi, S. Miles, C. Wiley, H. V. Vinters, and I. S. Y. Chen. 1990. High-levels of unintegrated HIV-1 DNA in brain-tissue of AIDS dementia patients. Nature 343:85-89. [DOI] [PubMed] [Google Scholar]
- 23.Panganiban, A. T., and H. M. Temin. 1983. The terminal nucleotides of retrovirus DNA are required for integration but not virus production. Nature 306:155-160. [DOI] [PubMed] [Google Scholar]
- 24.Rimsky, L., J. Hauber, M. Dukovich, M. H. Malim, A. Langlois, B. R. Cullen, and W. C. Greene. 1988. Functional replacement of the HIV-1 Rev protein by the HTLV-1 Rex protein. Nature 335:738-740. [DOI] [PubMed] [Google Scholar]
- 25.Schrager, J. A., and J. W. Marsh. 1999. HIV-1 Nef increases T cell activation in a stimulus-dependent manner. Proc. Natl. Acad. Sci. USA 96:8167-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw, G. M., B. H. Hahn, S. K. Arya, J. E. Groopman, R. C. Gallo, and F. Wongstaal. 1984. Molecular characterization of human T-cell leukemia (lymphotropic) virus type III in the acquired immune deficiency syndrome. Science 226:1165-1171. [DOI] [PubMed] [Google Scholar]
- 27.Siekevitz, M., S. F. Josephs, M. Dukovich, N. Peffer, F. Wongstaal, and W. C. Greene. 1987. Activation of the HIV-1 LTR by T-cell mitogens and the transactivator protein of HTLV-I. Science 238:1575-1578. [DOI] [PubMed] [Google Scholar]
- 28.Spina, C. A., J. C. Guatelli, and D. D. Richman. 1995. Establishment of a stable, inducible form of human immunodeficiency virus type 1 DNA in quiescent CD4 lymphocytes in vitro. J. Virol. 69:2977-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevenson, M., S. Haggerty, C. A. Lamonica, C. M. Meier, S. K. Welch, and A. J. Wasiak. 1990. Integration is not necessary for expression of human immunodeficiency virus type 1 protein products. J. Virol. 64:2421-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevenson, M., T. L. Stanwick, M. P. Dempsey, and C. A. Lamonica. 1990. HIV-1 replication is controlled at the level of T-cell activation and proviral integration. EMBO J. 9:1551-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vandegraaff, N., R. Kumar, C. J. Burrell, and P. Li. 2001. Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA. J. Virol. 75:11253-11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varmus, H. E., and P. O. Brown. 1989. Retroviruses, p. 53-108. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 33.Wiskerchen, M., and M. A. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, Y. T., and J. W. Marsh. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503-1506. [DOI] [PubMed] [Google Scholar]
- 35.Zack, J. A., A. M. Haislip, P. Krogstad, and I. S. Chen. 1992. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J. Virol. 66:1717-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]