Abstract
Point mutations that resulted in a substitution of the conserved 3′-penultimate cytidine in genomic RNA or the RNA negative strand of the self-amplifying replicon of the Flavivirus Kunjin virus completely blocked in vivo replication. Similarly, substitutions of the conserved 3′-terminal uridine in the RNA negative or positive strand completely blocked replication or caused much-reduced replication, respectively. The same preference for cytidine in the 3′-terminal dinucleotide was noted in reports of the in vitro activity of the RNA-dependent RNA polymerase (RdRp) for the other genera of Flaviviridae that also employ a double-stranded RNA (dsRNA) template to initiate asymmetric semiconservative RNA positive-strand synthesis. The Kunjin virus replicon results were interpreted in the context of a proposed model for initiation of RNA synthesis based on the solved crystal structure of the RdRp of φ6 bacteriophage, which also replicates efficiently using a dsRNA template with conserved 3′-penultimate cytidines and a 3′-terminal pyrimidine. A previously untested substitution of the conserved pentanucleotide at the top of the 3′-terminal stem-loop of all Flavivirus species also blocked detectable in vivo replication of the Kunjin virus replicon RNA.
The Flaviviridae comprise three genera of positive-strand RNA viruses (Flavivirus, Pestivirus, and Hepacivirus). Their infectious genomes have a single open reading frame and no 3′-poly(A) tail (17). The 5′- and 3′-penultimate and/or -terminal nucleotides of genomic RNA are conserved within each genus, and they are each adjacent to one or more conserved terminal stem-loops: e.g., for Hepacivirus RNA (4, 18, 37), Flavivirus RNA (8, 36, 39, 41, 43, 48), and Pestivirus RNA (3, 13, 53). Other common features are the similarities in gene order and function of the major viral enzymes in NS3 (protease and helicase) and in NS5/NS5B (RNA-dependent RNA polymerase [RdRp]) (17, 30).
The Flavivirus RNA replication strategy has been established with Kunjin virus (KUN) (11, 12, 24, 49-51). The genomic or RNA positive strand is translated, a replication complex is formed cotranslationally on the 3′ untranslated region (UTR) of the template RNA, and it transcribes an RNA negative strand that remains base paired with the RNA positive strand as a replicative form (RF). We found no evidence for the presence of a free RNA negative strand in KUN replicon-transfected cells (22). We have defined the consensus composition of the KUN replication complex (RC) as NS1, NS3, NS5, and hydrophobic nonstructural proteins NS2A and NS4A plus the RF (24, 34, 50, 51). The RF functions as a recycling template (shown by the kinetics of pulse-chase labeling) for asymmetric and semiconservative replication of progeny RNA positive strands by the viral replicase, and no subgenomic RNA is synthesized (11). Purified 32P-labeled KUN RF was converted to a replicative intermediate (RI) when presented as a substrate to a lysate of KUN-infected cells (2). The Pestivirus Bovine viral diarrhea virus (BVDV) (15, 47) and Hepacivirus Hepatitis C virus (HCV) (1) appear to use a similar strategy.
It has recently become apparent that a cytidine residue is present as the penultimate and/or terminal base at the 3′ end of many non-polyadenylated viral RNA positive strands. In Flavivirus species, the conserved 5′- and 3′-terminal nucleotides end as 5′AGU/A-G/A/UCUOH3′, and the RNA positive strand terminates as A/UCUOH3′ (7, 8, 21, 36, 41, 43, 48). Their roles in Flavivirus replication have not been explored, in contrast to the numerous in vitro studies using recombinant HCV NS5B and a variety of single-stranded RNA templates (e.g., references 20, 27, 37, 40, and 54). We decided to investigate the role in Flavivirus replication of the 3′-terminal dinucleotide by an in vivo approach because relevant experiments with recombinant NS5/NS5B assays in vitro were shown to be relatively inefficient and primer dependent but not template specific; furthermore RNA synthesis mostly occurred by “copyback” on the single-stranded template rather than by production of a de novo RNA transcript (44). Other examples of similar RdRp assays are those done with HCV recombinant NS5B (described above), recombinant Flavivirus NS5 of Dengue 1 virus (45) and KUN (16), and recombinant NS5B of the Pestivirus BVDV (19). In contrast, experiments with infectious Flavivirus RNA would retain the native forms of the templates for synthesis of both polarities of RNA, and all cellular components of the RC would be available. Therefore, we elected to use the self-amplifying KUN replicon or subgenomic RNA (22) (KUN RNA GenBank accession no. D00246, L24511, and L24512). The KUN replicon appears to replicate by the same strategy as full-length genomic RNA (23-25, 33). In this report, we show that the penultimate C at the 3′ terminus of KUN RNA positive and negative strands was essential for KUN RNA replication. In addition, mutations that changed the 3′-terminal nucleotide (uridine) of either strand blocked or restricted replication.
Structure and conformation of the KUN replicon RNA.
The replicon RNA has had all of the structural genes (C, prM, and E) deleted except for the first 20 codons of the core (or C) gene, which are required for replication, but otherwise it is identical to KUN genomic RNA (22). In addition to the conserved Flavivirus 5′- and 3′-terminal trinucleotides, there are conserved complementary sequences within the KUN core coding region and 3′ UTR (5′ nucleotides 137 to 144 and 3′ nucleotides −97 to −104, respectively) that are able to base pair and cause cyclization of the viral RNA (26). We also showed that mutations in these cyclization sequences (CS) blocked the RNA synthesis in vivo unless they were compensatory mutations that could base pair in the same way as the original CS (26).
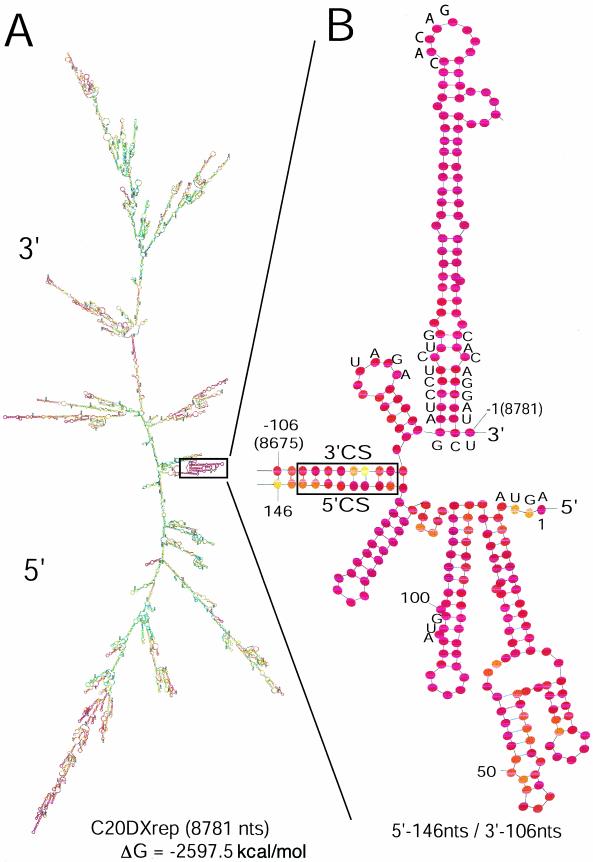
A computer-generated secondary structure of KUN replicon RNA (7.5 kb) in a characteristic minimum free energy configuration is shown in Fig. 1; it represents the longest secondary structure presented for a Flavivirus RNA. The low pairing numbers (P-num values) (38) of the bases in the area of interest shown in Fig. 1B suggest that the secondary structure as drawn is actually in the preferred configuration and is of high biological significance. The salient features in the 5′- and 3′-terminal sequences are the base pairing of CS, the presence of 5′- and 3′-terminal stem-loops each within the last 100 nucleotides and their spatial proximity to each other, retention of the Flavivirus conserved non-base-paired pentanucleotide 5′CACAG3′ at the top of the 3′-terminal stem-loop (48), and the non-base-paired terminal nucleotide. Notably, the 3′-stem-loop retains the base of the stem required to form a conserved pseudoknot (43).
FIG.1.
Computer-predicted model showing the topology of the RNA of the KUN replicon C20DXrep, prepared by using the mfold program and presented as the global MinE structure with bases colored according to their P-num values (38). (A) Model of the complete genome. (B) Enlargement of the inset region in panel A that represents only the 5′-terminal 146 nucleotides (nts) (numbering from 5′ is 1 to 146) and the 3′-terminal 106 nucleotides (numbering −1 to −106 from the terminus). The stem-loop structures in the enlarged inset region are slightly rearranged so the structures of the 5′- and 3′-terminal regions are more clearly depicted. CS identifies the base-paired cyclization sequence (see text). Bases indicated as A, C, G, or U are shown for the terminal nucleotides, the AUG translation initiation codon (at positions 97 to 99), the base-paired region in the lower portion of the 3′-terminal stem-loop, the Flavivirus-conserved pentanucleotide at the top of this loop, and the proposed pseudoknot sequences. Low P-num values (<3%) are represented by red bases, while increasing P-num values are represented by the colors orange, yellow, green (medium), and blue through black (high).
Mutations in the 3′-terminal dinucleotide of the KUN replicon RNA affecting replication.
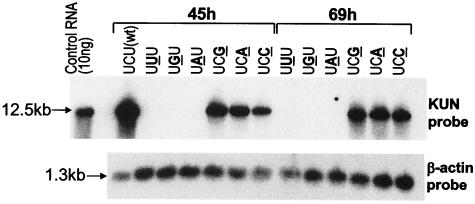
The cDNA of KUN replicon RNA was mutated in the conserved dinucleotide by PCR-directed mutagenesis of KUN replicon cDNA repPACβ-gal, which is able to express a β-galactosidase reporter gene (31) with the appropriate primers (data not shown). RNA was transcribed by SP6 polymerase and electroporated into BHK cells as previously described (26). Mutation of the penultimate C—changing it to U, G, or A in the replicon RNA—completely eliminated the capacity for replication of KUN replicon RNA transcribed from the cDNA. Thus, no KUN RNA replication was detected by Northern blot analysis of RNA from cells at 45 or 69 h after transfection of the mutated RNA (Fig. 2) and by staining cells with 5-bromo-4-chloro-3-uridylyl-β-d-galactosidase (X-Gal) (data not shown). Mutation of the terminal nucleotide U to produce replicon RNA ending in G, A, or C caused a significant inhibition (30 to 60%; Table 1) of RNA synthesis assayed at 45 h postinfection, and by 69 h, some inhibition was still present (Fig. 2), with the signals in Northern blots only 70% of that detected for wild-type RNA at 45 h. These results are summarized in Tables 1 and 2 and show that the penultimate C and the 3′-terminal pyrimidine (U) are absolutely essential or required for efficient replication, respectively.
FIG. 2.
Analysis of RNA copied from 3′-mutated KUN replicon RNA by Northern blotting. Approximately 10 μg of total cell RNA was extracted from BHK cells at 45 and 69 h after transfection with 5 μg of KUN replicon RNAs mutated in the penultimate cytidine or the 3′-terminal uridine and subjected to Northern blot hybridization with 32P-labeled cDNA probes detecting replicon RNA and β-actin mRNA as previously described (31). The mutated nucleotides are shown in boldface and underlined. Densitometry analysis of the blots is shown in Table 1.
TABLE 1.
Effect on replication of mutations in the 5′- and 3′-terminal dinucleotides of KUN replicon RNA
| RNA | Sequencea
|
% RNA replication ofb:
|
||
|---|---|---|---|---|
| 5′-RNA positive strand-3′ | RNA negative strand-3′ | 45 h | 69 h | |
| Wild type | 5′AGU-UCUOH3′ | ACUOH3′ | 100 | NA |
| 3′-terminal mutations | 5′AGU-UuUOH3′ | 0 | 0 | |
| -UgUOH3′ | 0 | 0 | ||
| -UaUOH3′ | 0 | 0 | ||
| 5′AGU-UCgOH3′ | 70 | 70 | ||
| -UCaOH3′ | 60 | 70 | ||
| -UCcOH3′ | 40 | 70 | ||
| 5′-terminal mutations | 5′uGU | ACaOH3′ | 0 | |
| 5′gGUc | ACcOH3′ | 0 | ||
| 5′AaU | AuUOH3′ | 0 | ||
| 5′AuUc | AaUOH3′ | 0 | ||
Terminal dinucleotides are shown in boldface, and the mutated nucleotides are shown in lowercase.
RNA replication by the replicons with 3′-terminal mutations was quantified as a percentage of that of wild-type replicon RNA at 45 h by densitometry of the autoradiograms used in the Northern blot analysis shown in Fig. 2. NA, no assay done for wild-type RNA.
Using the more sensitive RNase protection assay, RNA negative strands copied from these 5′-mutated RNAs transcribed by cellular RNA polymerase from transfected plasmid DNAs were detected (Fig. 3B).
TABLE 2.
Effect on replication of mutations in the 3′ pentanucleotide loop of KUN replicon RNA
| Type of sequence | Sequencea | Assay result at 69 h
|
|
|---|---|---|---|
| % Immuno- fluorescenceb | % X-Gal staining | ||
| Wild type | 5′CACAG 3′ | 100 | 100 |
| Mutated | 5′u c uAG 3′ | 0 | 0 |
The conserved pentanucleotide loop is located at the top of the terminal 3′ stem-loop (48). Mutated nucleotides are in lowercase.
KUN anti-NS3 antibodies were used in immunofluorescence assays.
Mutations in the 5′ terminus of KUN replicon RNA affecting replication.
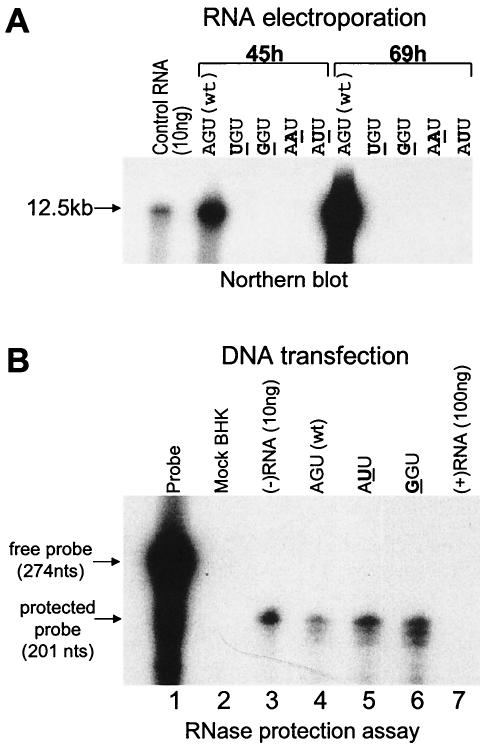
Because the template for Flavivirus RNA positive-strand synthesis is the RNA negative strand of the RF, the replicase complex must initiate synthesis by copying from the 3′ end of the RNA negative strand that terminates A/UCUOH. Note that cytidine is again the 3′-penultimate nucleotide with a 3′-terminal uridine as in the RNA positive strand. If the viral replicase engaged the RNA negative strand as a template in the RF for initiating RNA positive-strand synthesis in a similar manner to engagement of the free RNA positive strand as a template for RNA negative-strand synthesis, mutations causing loss of the 3′-penultimate C would be expected to block RNA positive-strand synthesis, preventing amplification, as observed in the previous section. Thus mutations from G to A or U in the second nucleotide at the 5′ end of the RNA positive strand should change the 3′ end of the RNA negative strand from -UCAOH to -UAAOH or UUAOH, respectively. When these changes were introduced into the KUN replicon repPACβ-gal by PCR-directed mutagenesis of cDNA with the appropriate primers (31), they completely blocked replication of the electroporated transcribed mutant RNA, as judged by negative results in Northern blots (Fig. 3A) and by a complete lack of in situ staining of transfected cells with X-Gal at 45 and 69 h (data not shown) in accord with the expectation based on loss of the 3′-penultimate C in the RNA negative strand. Note that the cDNA probe used is capable of detecting both RNA positive and negative strands and that X-Gal staining is a very sensitive detector of expression of genomic RNA, normally the most abundant RNA product.
FIG. 3.
Analysis of RNA copied from 5′-mutated replicon RNA. (A) Northern blot analysis with KUN replicon-specific 32P-labeled cDNA probe of ∼10 μg of total cell RNA extracted from BHK cells at 45 and 69 h after transfection with replicon RNAs containing native (wild type [wt]) and mutated 5′-terminal ribonucleotides. (B) Detection of KUN replicon RNA negative-strand synthesis in BHK cells at 48 h after transfection with plasmid DNAs containing KUN replicon cDNA with native and mutated 5′-terminal nucleotides. The probe was a 32P-labeled 274-nucleotide (nts) positive-sense RNA from the NS4A region transcribed in vitro, and the RNase protection assay was performed with an RPA-III kit, essentially as described by the manufacturer (Ambion, Inc., Austin, Tex.).
Mutation of the 5′-terminal nucleotide of replicon RNA from A to U or G (changing U to A or C in the terminal nucleotide of the cRNA negative strand) also caused complete inhibition of detectable RNA synthesis by Northern blot analysis (Fig. 3A) and in situ X-Gal staining (results not shown). A single mutation of 5′AG to 5′GG in the genome of West Nile virus was reported to substantially delay establishment of infection until reversion had occurred in the recovered virus (52). The 5′-terminal substitutions for A in the KUN replicon produced different effects in the RF template. Thus, the ensuing change in the RNA negative strand from -ACUOH 3′ to -ACAOH 3′ caused loss of the normally conserved terminal uridine. The change to 5′GGU in the RNA positive strand introduced a second terminal GC base pair in the RF helix, hence possibly impairing access by the replicase to the penultimate cytidine of the RNA negative-strand template. If small amounts of mutant replicon RNA positive strands were produced following RNA negative-strand synthesis by copying of the electroporated 5′ mutant RNA, they would have been readily detected by X-Gal staining of their translated products. This clearly did not occur.
The scenarios presented above implied that RNA negative-strand synthesis could occur on the electroporated 5′-mutated replicon RNA, but these mutant RNA negative strands could not function as templates for synthesis of progeny replicon RNA. Thus, only a very small number of RNA negative strands could be synthesized (one per 5′-mutated replicon template), and these would not be detected in the Northern blot assays, as shown (Fig. 3A). Therefore, a different strategy was devised to allow synthesis and accumulation of multiple copies of replicon RNA per cell, with either wild-type (5′AGU) or mutated (5′AUU, 5′GGU) 5′-terminal sequences. This was achieved by constructing plasmids containing replicon cDNA inserts preceded by a cytomegalovirus promoter (46) so that the replicon RNA with the 5′ mutations could be transcribed in cells from the transfected plasmid DNA into multiple RNA positive strands. Each multiply transcribed RNA could be translated, forming an RC (24), and be copied into an RNA negative strand if the 5′ mutations blocked only RNA positive-strand synthesis in accord with the projected scenarios. This accumulation of RNA negative strands in each plasmid DNA-transfected cell would increase the sensitivity of detection. After incubation of these cells for 48 h, the total RNA was extracted for RNase protection assays to detect any accumulation of newly synthesized RNA negative strands (Fig. 3B). These were readily detected in cells transfected with plasmids transcribing both wild-type and 5′ mutant replicons, thus establishing that the block in amplification of the transfected 5′ mutant replicons must be due to their inability to be copied from their newly synthesized daughter strands as templates. Note that the positive-strand probe used for Fig. 3B could detect as little as 10 ng of RNA negative strand (lane 3), but no labeled band was detected in a sample containing a large excess (100 ng) of RNA positive strand (lane 7), thus demonstrating the specificity of the assay. The results (summarized in Table 1) thus show that both of the conserved bases in the 3′-terminal dinucleotide -CUOH of RNA positive and negative strands are required for replication of the KUN replicon. The significance of these observed effects is discussed later. Note in Fig. 3B that that there was less synthesis of RNA negative strands in cells transfected with the plasmid DNA for the wild-type replicon, presumably because of the normal undefined control mechanisms that limit late flavivirus RNA negative-strand synthesis in the presence of as much as a 1,000-fold excess of accumulated positive strands of replicon RNA (42), whereas the transcribed 5′ mutant replicons that were copied into RNA negative strands were not amplified as in the normal replicative cycle.
Mutations in the conserved pentanucleotide loop of the 3′-terminal stem-loop of KUN replicon RNA affecting replication.
A role in Flavivivirus replication of the conserved pentanucleotide loop 5′CACAG3′ (48) in the 3′-terminal stem-loop has not been previously reported. We have suggested that it might provide a “landing pad” for assembly of components of the RC during their translation to facilitate initiation and transcription into RNA negative strands (24, 51), similar to the in vitro binding to the 3′-terminal stem-loop that occurs with NS3 and NS5 specified by Japanese encephalitis virus (10). We therefore tested any requirement for the conserved pentanucleotide by introducing mutations in KUN replicon repPACβ-gal as described above, changing the conserved sequence to 5′UCUAG3′ (data not shown). Replicon RNA transcribed from the mutated cDNA was electroporated into BHK cells that were then incubated for 69 h. When the cells were assayed for evidence of replicon replication by immunofluorescence with anti-KUN NS3 antibodies (50) or by X-Gal staining, no positively stained cells were detected (Table 2). Hence, although the M-fold structure of the long stem remained, as shown in Fig. 1B, mutations in the conserved pentanucleotide loop blocked replication. The results directly implicate this loop in Flavivirus replication.
Comparisons of conservation in the 5′- and 3′-terminal structures and nucleotide sequences among the Flaviviridae.
As in the Flavivirus species, conservation of terminal nucleotides is found in all genera. The genomic terminal sequences 5′GUAU/CCCC3′ and the internal ribosomal entry site are conserved in RNA of Pestivirus species BVDV (3, 13, 53; GenBank accession no. AF220247 and NC_002514) and hog cholera virus RNA (13; GenBank accession no. J04358 and AF091661). Results of experiments with synthetic templates and the recombinant NS5B RdRp of BVDV (19), with mutations in the conserved 5′GUAU of the BVDV replicon (3), or with 5′GUAU added to a substituted HCV 5′ UTR in a chimeric BVDV genome (14) are all compatible with a role in replication for a 3′-penultimate or -terminal cytidine in either RNA strand as a template. HCV genomic RNA (infectious in chimpanzees) and HCV replicon RNA (self-amplifying in Huh-7 cells) have conserved terminal sequences 5′GCCAG//CAA/U GU3′ (28, 32; GenBank accession no. AF009606 and AJ242654); hence only the RNA negative strand terminates with a 3′ cytidine residue and it is a more efficient template in vitro than the RNA positive strand (40). The failure by HCV to normally employ a penultimate or terminal 3′ cytidine in genomic RNA so as to improve efficiency of RNA negative-strand synthesis may represent a strategy to restrict the level of virus replication to below the required level for immunosurveillance during some persistent infections.
In summary, these comparisons indicate a strong preference either for a 3′-penultimate cytidine followed by a pyrimidine or for a 3′-terminal cytidine for efficient replication of both RNA positive and negative strands of all of the species of Flaviviridae: e.g., RNA positive strand, -CU3′, -CC3′; and RNA negative strand, -CU3′, -CC3′, -AC3′, or -GC3′.
Is the model for initiation of RNA synthesis by the RNA polymerase of dsRNA bacteriophage φ6 RdRp applicable to Flavivirus RNA synthesis?
The replication strategy of flaviviruses has some striking parallels with that of the segmented double-stranded RNA (dsRNA) bacteriophage φ6 (35). In both cases the polymerase can function without a primer on a single-stranded RNA template and use dsRNA as a recycling template for semiconservative and asymmetric replication of RNA positive strands that are destined to function as mRNAs and also as the single-stranded template for RNA negative-strand synthesis, thus reforming the genomic dsRNA (φ6) or RF (flaviviruses). Thus, the models proposed independently for KUN (11, 37, 51) and φ6 phage (35) are virtually identical in their major features of replication strategy. The determination of the crystal structure of the φ6 polymerase protein L2 (9) and of the flavivirus HCV RdRp NS5B (5, 6, 29) and the associated comparative analyses have indicated the commonality of the molecular architecture of these enzymes, as emphasized by Butcher et al. (9), who suggested an evolutionary link. Their common features include the roughly spherical topology and superimposition of 418 Cα atoms, such that “they resemble each other much more than either resembles any other known polymerase structure” (9). Comparisons with the φ6 polymerase show that there are 12 fully conserved amino acid residues for KUN NS5 in the sequence comprising the C, D, and E polymerase motifs (E. G. Westaway, unpublished observations) compared with only 3 in the corresponding conserved palm region of HCV NS5B (9). The RNA positive and negative strands of the φ6 L, M, and S segments all terminate similarly to KUN RNA with a 3′ pyrimidine and a penultimate cytidine (GenBank accession no. NC_003715, NC_003716, and NC_003714) other than one relatively inefficient exception. Butcher et al. (9) proposed that the 3′ terminus of the RNA negative strand is separated from dsRNA and fed directly into the template channel past the catalytic site and thence temporarily into a specific binding pocket (the “S” pocket) that is too small to accommodate a 3′-terminal purine. Initiation of φ6 RNA positive-strand synthesis is apparently facilitated by fraying of the ends of the dsRNA template associated with a low melting point of the first 9 bp in the helix (7 of 9 bp are AU) (9). Similarly, at the 3′ end of the RNA negative strand in the KUN RF on which initiation of RNA positive-strand synthesis must occur, five of the seven terminal nucleotides can form AU pairs with the 5′ terminus of the cRNA positive strand 5′AGUAGUU3′ (21), and there are only 2 well-separated GC bp in the first 8 bp in the predicted 3′-terminal stem-loop of KUN replicon RNA (free energy, −9.7 kcal) (Fig. 1).
Assuming the analogy with the φ6 replicase is reasonable, the results described above for the effects of mutations at the 5′ and 3′ termini on replication of the KUN replicon are readily interpreted. The 3′-penultimate cytidine and 3′-terminal uridine in the RNA positive and negative strands are essential recognition sites in a promoter sequence for the replicase complex to efficiently initiate RNA synthesis at the catalytic site in the template tunnel of NS5. Their mutations had drastic effects on synthesis of either strand (Fig. 2 and Table 1). Other sites, such as the conserved pentanucleotide at the top of the 3′-terminal stem-loop, may also be essential as attachment sites for components of the RC as mentioned above. Mutations at the 5′ terminus of the KUN replicon RNA probably blocked synthesis of RNA positive strands (i) by causing loss of the 3′-penultimate cytidine in the recognition site of the RNA negative template strand of the RF, thus preventing the correct positioning of the template in the replicase; (ii) by causing a change from 3′-terminal uridine to the bulky purine adenosine in the RNA negative template that was thus too large to initially enter the putative binding pocket; or (iii) by increasing the GC base pairing at the terminus of the helix, thus inhibiting fraying of the ends that normally facilitates initiation.
In summary, replication by flaviviruses exhibits a very strong preference for a 3′-penultimate or 3′-terminal cytidine in both RNA positive and negative template strands, and a penultimate cytidine is invariably adjacent to a 3′-terminal pyrimidine.
ADDENDUM IN PROOF
Using recombinant NS5 of dengue-2 virus in an in vitro RNA-dependent RNA assay, Nomaguchi et al. (M. Nomaguchi, M. Ackermann, C. Yon, S. You, and R. Padmanabhan, J. Virol. 77: 8831-8842, 2003) measured elongation (hairpin) and de novo RNA synthesis by using a subgenomic template with mutations in the 3′-terminal dinucleotide. The inhibition of mutant de novo RNA synthesis relative to that of wild-type RNA was similar to our data for in vivo synthesis of transfected Kunjin virus genomic RNA at 45 h, as shown in Table 1, for 3′-terminal mutations of -CU to -CC, -CA, or -CG (the mutation to -CC was the most inhibited in both systems). However, the effect of mutations in the penultimate C was strikingly different for the two systems: a C-to-U mutation did not affect the template activity for de novo dengue-2 virus RNA synthesis in vitro, whereas all three mutations of the Kunjin virus RNA penultimate C (to A, U, or G) caused a complete block of RNA synthesis in vivo (Table 1). It should be noted that the environment for replication of transfected Kunjin virus genomic RNA in cell culture differed from that of the in vitro RNA replication system involving only a single protein (purified recombinant NS5), a small 3′-terminal dengue-2 virus RNA template (770 nucleotides), excess of nucleoside triphosphates, and no cellular or other viral components.
Acknowledgments
This work was supported by grants 102481 and 142983 from the National Health and Medical Research Council of Australia.
Footnotes
This is publication no. 181 from the Sir Albert Sakzewski Virus Research Centre.
REFERENCES
- 1.Ali, N., K. D. Tardif, and A. Siddiqui. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J. Virol. 76:12001-12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartholomeusz, A. I., and P. J. Wright. 1993. Synthesis of dengue virus RNA in vitro: initiation and the involvement of proteins NS3 and NS5. Arch. Virol. 128:111-121. [DOI] [PubMed] [Google Scholar]
- 3.Becher, P., M. Orlich, and H.-J. Thiel. 2000. Mutations in the 5′ nontranslated region of bovine viral diarrhea virus result in altered growth characteristics. J. Virol. 74:7884-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blight, K. J., and C. M. Rice. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinton, M. A., and J. H. Dispoto. 1988. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology 162:290-299. [DOI] [PubMed] [Google Scholar]
- 8.Brinton, M. A., A. V. Fernandez, and J. H. Dispoto. 1986. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 153:113-121. [DOI] [PubMed] [Google Scholar]
- 9.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C.-J., M.-D. Kuo, L.-J. Chien, S.-L. Hsu, Y.-M. Wang, and J.-H. Lin. 1997. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 71:3466-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu, P. W., and E. G. Westaway. 1985. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology 140:68-79. [DOI] [PubMed] [Google Scholar]
- 12.Chu, P. W., and E. G. Westaway. 1987. Characterization of Kunjin virus RNA-dependent RNA polymerase: reinitiation of synthesis in vitro. Virology 157:330-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng, R., and K. V. Brock. 1993. 5′ and 3′ untranslated regions of pestivirus genome: primary and secondary structure analyses. Nucleic Acids Res. 21:1949-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frolov, I., M. S. McBride, and C. M. Rice. 1998. cis-acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5′ nontranslated region chimeras. RNA 4:1418-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong, Y., R. Trowbridge, T. B. Macnaughton, E. G. Westaway, A. D. Shannon, and E. J. Gowans. 1996. Characterization of RNA synthesis during a one-step growth curve and of the replication mechanism of bovine viral diarrhoea virus. J. Gen. Virol. 77:2729-2736. [DOI] [PubMed] [Google Scholar]
- 16.Guyatt, K. J., E. G. Westaway, and A. A. Khromykh. 2001. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J. Virol. Methods 92:37-44. [DOI] [PubMed] [Google Scholar]
- 17.Heinz, F. X., M. S. Collet, R. H. Purcell, E. A. Gould, C. R. Howard, M. Houghton, R. J. M. Moormann, C. M. Rice, and H.-J. Thiel. 2000. Family Flaviviridae, p. 859-879. In M. H. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop et al. (ed.), Virus taxonomy, classification and nomenclature of viruses. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 18.Ito, T., and M. M. C. Lai. 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 71:8698-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao, C. C., A. M. Del Vecchio, and W. Zhong. 1999. De novo initiation of RNA synthesis by a recombinant Flaviviridae RNA-dependent RNA polymerase. Virology 253:1-7. [DOI] [PubMed] [Google Scholar]
- 20.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khromykh, A. A., and E. G. Westaway. 1994. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J. Virol. 68:4580-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khromykh, A. A., M. T. Kenney, and E. G. Westaway. 1998. trans-Complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J. Virol. 72:7270-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 1999. trans-Complementation analysis of the flavivirus Kunjin ns5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J. Virol. 73:9247-9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 2000. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 74:3253-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, M., H. Kim, S.-P. Cho, and M.-K. Min. 2002. Template requirements for de novo RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase on the viral X RNA. J. Virol. 76:6944-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 29.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 30.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In B. N. Fields, D. M. Knipe, P. M. Howley et al. (ed.), Fields virology, 4th ed. Lippincott-Raven, New York, N.Y.
- 31.Liu, W. J., P. L. Sedlak, N. Kondratieva, and A. A. Khromykh. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis in virus assembly. J. Virol. 76:10766-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 33.Mackenzie, J. M., A. A. Khromykh, and E. G. Westaway. 2001. Stable expression of noncytopathic Kunjin replicons simulates both ultrastructural and biochemical characteristics observed during replication of Kunjin virus. Virology 279:161-172. [DOI] [PubMed] [Google Scholar]
- 34.Mackenzie, J. M., A. A. Khromykh, M. K. Jones, and E. G. Westaway. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203-215. [DOI] [PubMed] [Google Scholar]
- 35.Makeyev, E. V., and D. H. Bamford. 2000. The polymerase subunit of a dsRNA virus plays a central role in the regulation of viral RNA metabolism. EMBO J. 19:6275-6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandl, C. W., H. Holzmann, C. Kunz, and F. X. Heinz. 1993. Complete genomic sequence of Powassan virus: evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology 194:173-184. [DOI] [PubMed] [Google Scholar]
- 37.Oh, J. W., G. T. Sheu, and M. M. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 38.Palmenberg, A. C., and J.-Y. Sgro. 1997. Topological organization of picornaviral genomes: statistical prediction of RNA structural signals. Semin. Virol. 8:231-241. [Google Scholar]
- 39.Proutski, V., E. A. Gould, and E. C. Holmes. 1997. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 25:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reigadas, S., M. Ventura, L. Sarih-Cottin, M. Castroviejo, S. Litvak, and T. Astier-Gin. 2001. HCV RNA-dependent RNA polymerase replicates in vitro the 3′ terminal region of the minus-strand viral RNA more efficiently than the 3′ terminal region of the plus RNA. Eur. J. Biochem. 268:5857-5867. [DOI] [PubMed] [Google Scholar]
- 41.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229:726-733. [DOI] [PubMed] [Google Scholar]
- 42.Shi, P. Y., M. Tilgner, and M. K. Lo. 2002. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology 296:219-233. [DOI] [PubMed] [Google Scholar]
- 43.Shi, P. Y., M. A. Brinton, J. M. Veal, Y. Y. Zhong, and W. D. Wilson. 1996. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry 35:4222-4230. [DOI] [PubMed] [Google Scholar]
- 44.Steffens, S., H. J. Thiel, and S. E. Behrens. 1999. The RNA-dependent RNA polymerases of different members of the family Flaviviridae exhibit similar properties in vitro. J. Gen. Virol. 80:2583-2590. [DOI] [PubMed] [Google Scholar]
- 45.Tan, B. H., J. Fu, R. J. Sugrue, E. H. Yap, Y. C. Chan, and Y. H. Tan. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216:317-325. [DOI] [PubMed] [Google Scholar]
- 46.Varnavski, A. N., P. R. Young, and A. A. Khromykh. 2000. Stable high-level expression of heterologous genes in vitro and in vivo by noncytopathic DNA-based Kunjin virus replicon vectors. J. Virol. 74:4394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warrilow, D., W. B. Lott, S. Greive, and E. J. Gowans. 2000. Properties of the bovine viral diarrhoea virus replicase in extracts of infected MDBK cells. Arch. Virol. 145:2163-2171. [DOI] [PubMed] [Google Scholar]
- 48.Wengler, G., and E. Castle. 1986. Analysis of structural properties which possibly are characteristic for the 3′-terminal sequence of the genome RNA of flaviviruses. J. Gen. Virol. 67:1183-1188. [DOI] [PubMed] [Google Scholar]
- 49.Westaway, E. G., A. A. Khromykh, and J. M. Mackenzie. 1999. Nascent flavivirus RNA colocalized in situ with double-stranded RNA in stable replication complexes. Virology 258:108-117. [DOI] [PubMed] [Google Scholar]
- 50.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westaway, E. G., J. M. Mackenzie, and A. A. Khromykh. 2002. Replication and gene function in Kunjin virus. Curr. Top. Microbiol. Immunol. 267:323-351. [DOI] [PubMed] [Google Scholar]
- 52.Yamschikov, V. F., G. Wengler, A. A. Perelygin, M. A. Brinton, and R. W. Compans. 2001. An infectious clone of the West Nile flavivirus. Virology 281:294-304. [DOI] [PubMed] [Google Scholar]
- 53.Yu, H., C. W. Grassmann, and S.-E. Behrens. 1999. Sequence and structural elements at the 3′ terminus of bovine viral diarrhea virus genomic RNA: functional role during RNA replication. J. Virol. 73:3638-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong, W., A. S. Uss, E. Ferrari, J. Y. N. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]