Abstract
Tacaribe virus (TV) is the prototype of the New World group of arenaviruses. The TV genome encodes four proteins, the nucleoprotein (N), the glycoprotein precursor, the polymerase (L), and a small RING finger protein (Z). Using a reverse genetic system, we recently demonstrated that TV N and L are sufficient to drive transcription and full-cycle RNA replication mediated by TV-like RNAs and that Z is a powerful inhibitor of these processes (N. López, R. Jácamo, and M. T. Franze-Fernández, J. Virol. 65:12241-12251, 2001). In the present study we investigated whether Z might interact with either of the proteins, N and L, required for RNA synthesis. To that end, we used coimmunoprecipitation with monospecific antibodies against the viral proteins and coimmunoprecipitation with serum against glutathione S-transferase (GST) and binding to glutathione-Sepharose beads when Z was expressed as a fusion protein with GST. We demonstrated that Z interacted with L but not with N and that Z inhibitory activity was dependent on its ability to bind to L. We also evaluated the contribution of different Z regions to its binding ability and functional activity. We found that integrity of the RING structure is essential for Z binding to L and for Z inhibitory activity. Mutants with deletions at the N and C termini of Z showed that amino acids within the C-terminal region and immediately adjacent to the RING domain N terminus contribute to efficient Z-L interaction and are required for inhibitory activity. The data presented here provide the first evidence of an interaction between Z and L, suggesting that Z interferes with viral RNA synthesis by direct interaction with L. In addition, coimmunoprecipitation studies revealed a previously unreported interaction between N and L.
Tacaribe virus (TV) is the prototype of the New World group of arenaviruses. Within this group the viruses form three phylogenetically distinct clades, one of which includes TV together with the four known South American pathogens that produce severe hemorrhagic disease (Junin, Machupo, Guanarito, and Sabia viruses) (5). TV, however, does not seem to be a human pathogen.
TV, like all arenaviruses, is an enveloped virus containing two single-stranded RNA segments called S and L. The S RNA contains two genes, one encoding the nucleoprotein (N; 64 kDa) and one encoding the glycoprotein precursor (55 kDa) (10), while the L RNA encodes the RNA-dependent RNA polymerase (L; 240 kDa) (15) and a small protein with a RING finger motif (Z; 11 kDa) (16). In both S and L RNAs, the genes are arranged in opposite orientation and are separated by noncoding sequences that have the potential to form stable secondary structures (11). The 5′ regions of arenavirus genomes and antigenomes, though positively stranded, are not translated directly into proteins. Rather, genomes and antigenomes are found only as nucleocapsids tightly bound to N, and the coding sequences are expressed from mRNAs transcribed from the 3′ regions of the genomes or antigenomes (1, 11, 17, 19, 26). These mRNAs contain short stretches of nontemplated nucleotides at their 5′ ends and appear to be capped, as they reacted with anticap antibodies (19). By analogy with other negative-strand viruses, L together with N constitutes the viral RNA polymerase when N is associated with RNA in nucleocapsid structures. Using a reconstituted transcription and replication system based on plasmid-supplied TV RNAs and proteins, we recently demonstrated that L and N proteins are sufficient to support full-cycle RNA replication and transcription mediated by minireplicons and by an S-genome analog in which the N open reading frame (ORF) is replaced by the chloramphenicol acetyltransferase (CAT) ORF. Using this system, we also demonstrated that Z is a powerful inhibitor of RNA replication and transcription (21). Lymphocytic choriomeningitis virus (LCMV) Z protein exhibits a similar inhibitory effect when transcription and replication are analyzed in the homologous (LCMV) minigenome system (6).
Arenavirus Z proteins contain a conserved 37-amino-acid (aa) RING domain that comprises about 40% of their amino acid residues (9, 16, 27, 29). The RING domain binds two zinc ions in a cross-brace arrangement and is involved in protein-protein interactions (2). It has been shown that LCMV Z interacts with different cellular proteins, such as PML, and translation factors, such as the ribosomal P proteins and translation initiation factor e1F4E (3, 4, 18).
To determine the mechanism of Z protein inhibition of RNA replication and transcription, we investigated possible interactions between TV Z and the viral N and L proteins. We present evidence that TV Z interacts with L in a dose-dependent manner that correlates with Z-mediated inhibition of minigenome expression. We also investigated the contribution of different regions of Z protein to its binding ability and functional activity. In addition, these studies revealed a previously unreported interaction between N and L proteins.
MATERIALS AND METHODS
Cells and viruses.
CV1 cells were grown in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum (GIBCO-BRL, Gaithersburg, Md.). Recombinant vaccinia virus vTF7-3, which expresses the T7 RNA polymerase, was kindly provided by B. Moss (National Institutes of Health, Bethesda, Md.) (12). Cells were routinely infected with 3 to 5 PFU of vTF7-3 per cell so that more than 90% of the cells were infected.
Plasmids.
Plasmids expressing the minigenome (pGenCAT), the TV N protein (pN), and the TV L protein (pL) were described previously (21). A plasmid expressing TV Z protein (pZ) was obtained by introducing into the BamHI-EcoRI sites of pGEM4 (Promega, Madison, Wis.) a DNA fragment covering the entire Z ORF and 43 nucleotides (nt) of the noncoding region 5′ of the Z AUG that was excised from pTACV-Z (kindly provided by D. Kolakofsky [13]) with BamHI-EcoRI. Plasmid pgstZ contained the glutathione S-transferase (GST) gene in frame with the 5′ end at nt 1 of the Z ORF. To generate pgstZ, we first constructed the plasmid pgst by inserting the GST gene, excised from pGEX 2T (Amersham Pharmacia Biotech, Uppsala, Sweden) by digestion with HincII and BamHI enzymes, into the same restriction sites of pGEM4. The entire Z gene devoid of the 5′ noncoding sequence was obtained from pZ by PCR amplification with a forward primer containing a BamHI site followed by nt 70 to 85 of the L genome and a reverse primer containing a sequence complementary to the EcoRI-to-KpnI sites of the pGEM4 polylinker (RevpGEM4). The PCR product was digested with BamHI and EcoRI, and the DNA fragment was inserted into pgst previously digested with the same enzymes.
Plasmids pZ(1-35) and pZ(1-77) express Z mutants with deletions in the last 60 or 18 aa, respectively (see Fig. 6). The Z genes for mutants with C-terminal deletions were obtained by PCR from pZ using a forward primer containing a BamHI site 5′ of nt 27 to 42 of the L genome (FwpZ) and different reverse primers. The reverse primer for the pZ(1-35) construct contained a sequence complementary to nt 194 to 169 of the L genome, with a single change (C to G) at nt 183 that generated a stop codon immediately after aa 35. For pZ(1-77) construction, the reverse primer harbored a sequence complementary to nt 300 to 282 of the L genome preceded by the sequence 5′ TTACTA 3′, which generated two stop codons immediately after aa 77. The PCR products were digested with BamHI and inserted into the BamHI-SmaI sites of pGEM4. The GST-tagged Z mutants with deletions at the C terminus [pgstZ(1-35) and pgstZ(1-77)] were obtained as follows. DNA fragments encoding aa 17 to 35 or 17 to 77 of the Z protein were excised with XhoI and EcoRI from pZ(1-35) and pZ(1-77), respectively. These fragments were inserted into pgstZ digested with the same enzymes to generate pgstZ(1-35) and pgstZ(1-77), respectively. For pZ(36-95) and pgstZ(36-95) construction, a DNA fragment encoding aa 36 to 95 was generated by PCR using pZ as template, a forward primer containing a BamHI site and an AUG followed by nt 175 to 201 of the L genome, and RevpGEM4 as reverse primer. The PCR product was digested with BamHI and EcoRI and the excised DNA fragment was inserted into either pZ or pgstZ digested with the same enzymes, generating pZ(36-95) and pgstZ(36-95), respectively. Construction of pgstZ(36-77) was performed as follows. A DNA fragment encoding aa 38 to 77 of the Z protein was obtained from pgstZ(1-77) by cleavage with MfeI and SacI. This fragment was inserted into pgstZ(36-95) digested with the same enzymes.
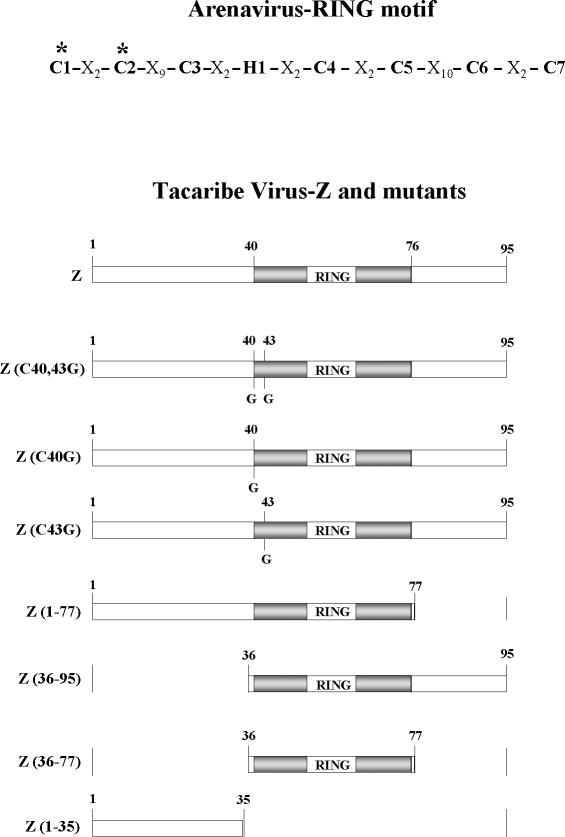
FIG. 6.
Schematic representation of arenavirus RING motif, domains of TV Z protein, and mutants used in this study. The asterisks indicate the first two ligands of the first zinc-binding site of the RING domain, which in TV Z correspond to Cys residues 40 and 43. The RING domain is indicated by the shaded box, and the N- and C-terminal domains are indicated by open boxes. The amino acid numbers limiting each domain are indicated. Cys residues that were changed to Gly are marked “G.” The names of the Z deletion mutants indicate the N- and C-terminal amino acid numbers in the remaining sequence.
In order to introduce single nucleotide changes into the RING motif of the Z protein, we carried out PCR site-directed mutagenesis (23). For construction of pZ(C43G), the Z gene was amplified from pZ by use of a forward primer containing an MfeI site followed by nt 179 to 206 of the L genome with a single change (T to G) at position 196 (codon 43) and RevpGEM4 as reverse primer. The PCR product was digested with MfeI and EcoRI and inserted into pZ digested with the same enzymes. Two rounds of PCR amplification were performed to change Cys residue 40 to Gly. First, the Z ORF was amplified from pZ by use of FwpZ as forward primer and a reverse primer containing the complementary sequence to nt 194 to 175 of the L genome with a single change (A to C) at position 187 (codon 40). The PCR product was used as forward primer for a second round of amplification, in which pZ was used as template and RevpGEM4 was used as reverse primer. The amplification product was digested with BamHI and EcoRI and inserted into the BamHI and EcoRI sites of pGEM4 to yield pZ(C40G). When pZ(C43G) was used as template for the second round of amplification, the PCR product contained changes at positions 187 and 196 (codons 40 and 43). The amplification product digested with BamHI and EcoRI and inserted into the same restriction sites of pGEM4 generated pZ(C40,43G). Digestion of pZ(C43G), pZ(C40G), and pZ(C40,43G) with XhoI and EcoRI and insertion of each fragment into pgstZ excised with the same enzymes yielded plasmids pgstZ(C43G), pgstZ(C40G), and pgstZ(C40,43G), respectively.
In plasmid pgstN, the N ORF was fused in frame at the end encoding the amino terminus to the GST ORF. For pgstN construction, the N gene was excised from plasmid p2b2 (10) with SacI and SmaI and the DNA fragment was inserted into pBluescript II KS(+/−) vector (Stratagene, La Jolla, Calif.) digested with the same enzymes. The resulting plasmid cleaved with EcoRI and SacI gave a DNA fragment containing the entire N ORF that was inserted in frame into the EcoRI and SacI sites of pgst.
Plasmid constructions were verified by DNA sequencing determination of the junctions. DNA fragments obtained by PCR were totally sequenced. All plasmids were purified by Tip-100 (Qiagen Inc., Valencia, Calif.).
DNA transfections.
Subconfluent monolayers of CV1 cells were infected with vTF7-3 for 1 h at 37°C. The inoculum was removed, and the cells were washed and transfected using Lipofectamine 2000 reagent according to the manufacturer's (GIBCO-BRL) specifications. Unless otherwise indicated, the amount of pGenCAT, pN, and pL added to approximately 4 × 105 cells grown in a 12-well dish was, respectively, 2 μg, 2 μg, and 50 ng per well, as these amounts are optimal for RNA replication and transcription (21). The amounts of other plasmids added are indicated in each figure. In all cases, the total amount of transfected DNA was kept constant by the addition of an appropriate amount of pGEM-3 (Promega) DNA.
Protein analysis.
Cells grown in 12-well dishes were radiolabeled for 1 h at 5 h posttransfection (hpt) by the addition of [35S]methionine-cysteine mix (150 μCi/ml; NEG 772; New England Nuclear) in cysteine- and methionine-free medium (GIBCO). After the labeling period, cell monolayers were lysed by the addition of 200 μl of TNEN (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.2% Nonidet P-40) containing protease inhibitors (2 μg of aprotinin per ml, 20 μg of phenylmethylsulfonyl fluoride per ml, and 50 μg of N-α-p-tosyl-l-lysine chloro-methyl ketone [Sigma-Aldrich, St. Louis, Mo.]). Cell lysates were clarified at 13,000 × g for 5 min at 4°C, and aliquots were used for immunoprecipitation or bead binding. For immunoprecipitation, samples of the labeled cell lysates, corresponding to about 1 × 105 to 2 × 105 cells, were incubated with the indicated serum for 1 h at 4°C. The antigen-antibody complex was incubated for 1 h at 4°C with protein A-Sepharose CL-4B beads (Sigma) and precipitated by centrifugation at 7,000 × g. Beads were washed three times. All immunoprecipitation steps were performed in TNEN buffer. For bead binding, glutathione-Sepharose 4B beads (15 μl per reaction) were prepared as indicated by the manufacturer (Amersham Pharmacia Biotech) and resuspended in TNEN buffer. Aliquots of the labeled cell lysates (about 1 × 105 to 2 × 105 cells) were incubated with beads for 1 h at 4°C. Beads were pelleted by centrifugation at 7,000 × g and washed three times with TNEN buffer. Labeled proteins bound to protein A-Sepharose CL-4B beads or glutathione-Sepharose 4B beads were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) together with 14C-labeled markers and autoradiography as indicated previously (22). When indicated, quantitation of the bands was performed on a PhosphorImager (Molecular Dynamics). Monospecific sera against the recombinant TV proteins L, N, and Z were raised in rabbits as indicated before (25). Serum against GST was obtained from Amersham Pharmacia Biotech.
CAT assay.
CAT activity was assayed in extracts from vTF7-3-infected-transfected CV1 cells as described previously (21). CAT activity was calculated by determining the percentage of radioactivity associated with monoacetylated chloramphenicol species relative to total radioactivity. Quantitation was performed on a PhosphorImager.
RESULTS
TV Z protein interacts with L protein.
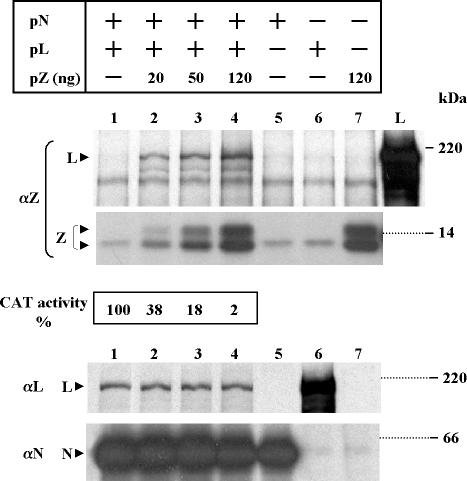
Using a reconstituted system based on plasmid-supplied TV RNAs and TV proteins, we previously demonstrated that Z inhibited full-cycle RNA replication and transcription, as analyzed by CAT expression (21). To assess whether Z interacted with L and/or N TV protein, which are required for viral RNA synthesis (21), we used coimmunoprecipitation with monospecific antibodies to viral proteins in cytoplasmic extracts from cells transfected with plasmids expressing the minigenome, N and L, with or without the addition of increasing amounts of the plasmid expressing Z (Fig. 1). The amounts of Z plasmid used were those previously found to produce potent inhibition of replication and transcription (21). At 5 hpt, the transfected cells were labeled for 1 h and lysed, the cell lysate was divided into three aliquots, and each aliquot was immunoprecipitated with monospecific serum against either Z, L, or N. To relate eventual complex formation with Z functional activity, cell extracts prepared from dishes transfected in parallel were used to assay CAT activity. As seen in Fig. 1, SDS-PAGE analysis of the extracts immunoprecipitated with serum against Z (panel αZ) revealed that L coimmunoprecipitated with Z and that the level of L increased in parallel with that of expressed Z. L was undetectable when the Z plasmid was not included in the transfection (compare lane 1 with lanes 2 to 4). Moreover, the dose-response curve of Z-mediated inhibition of CAT activity correlated with the increasing level of L coimmunoprecipitating with Z. It should be noted that in this experiment Z was resolved into two bands, which were occasionally observed with both plasmid-expressed and TV-expressed Z protein (R. Jácamo, unpublished observations). Immunoprecipitation with serum against L or N confirmed previous findings (21) indicating that cotransfection with pZ at amounts producing potent inhibition of transcription and replication do not affect L and N protein synthesis (lanes 1 to 4). The amount of L expressed alone was higher than that when coexpressed with N (panel αL, compare lanes 1 to 4 with lane 6). Nevertheless, the amounts of transfected L and N plasmids were those previously found to be optimal for TV-like RNA transcription and replication (21).
FIG. 1.
TV L protein coimmunoprecipitates with Z in the reconstituted transcription and replication system. Subconfluent CV1 cell monolayers were transfected with pGenCAT with (+) or without (−) the addition of pN and pL as indicated. For experiments in which pZ was included, the amount used is indicated. At 5 hpt, cells were labeled and lysed, the cell lysates were divided into three aliquots, and each aliquot was immunoprecipitated with monospecific serum against either Z (αZ), L (αL), or N (αN). Immunoprecipitated proteins were resolved by SDS-PAGE on gels containing 12% (αZ and αN) or 7% (αL) polyacrylamide. Lane L (panel αZ) corresponds to plasmid-expressed L immunoprecipitated with serum against L. Cells transfected in parallel as in lanes 1 to 4 were used to assay CAT activity as indicated in Materials and Methods. Molecular masses of 14C-labeled markers are indicated on the right. The positions of the L, Z, and N proteins are indicated on the left by arrowheads. In this experiment, Z was resolved into two bands, and a nonspecifically immunoprecipitated protein migrates slightly above the lower Z band (lanes 1, 5, and 6). Films were exposed for 40 or 96 h (αZ, upper and lower panels, respectively) or for 8 h (αL and αN).
We were unable to analyze Z coimmunoprecipitation by using L antibodies or to detect Z-N complexes by using serum against either Z or N, as the results were jeopardized by nonspecifically immunoprecipitated proteins (data not shown). To overcome these problems and the possibility that antibodies against the viral proteins might interfere with their sites of interaction, we used cell extracts expressing a GST-tagged Z protein (gstZ) and analyzed complex formation by cobinding of candidate proteins to glutathione-Sepharose beads or coimmunoprecipitation with GST antiserum.
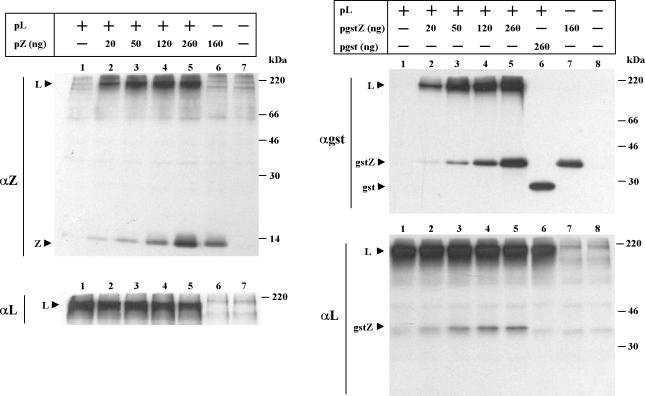
We then compared the ability of Z and gstZ to interact with L in the absence of N and minigenome expression (Fig. 2). Cells were transfected with a fixed amount of the plasmid expressing L, with or without the addition of increasing amounts of either pZ or pgstZ. Cells were labeled, cell extracts were divided into two aliquots, each aliquot was immunoprecipitated with serum against L and either Z (left panel) or GST (right panel), and the immunoprecipitated proteins were analyzed as described above. Immunoprecipitation with serum against Z showed that the level of coimmunoprecipitating L increased in parallel with that of the expressed Z, indicating that the interaction between Z and L does not require the expression of other viral components (left panel). Similarly, immunoprecipitation with serum against GST showed that when gstZ and L were coexpressed, increasing amounts of L coimmunoprecipitated along with the increasing level of gstZ (right panel). In contrast, when L was coexpressed with a plasmid expressing GST, L did not coimmunoprecipitate, indicating that the interaction with L is mediated through the Z portion of the fusion protein (lane 6). Conversely, gstZ, but not GST, coimmunoprecipitated with L antiserum when either of these proteins was coexpressed with L (right panel αL, compare lanes 2 to 5 with lane 6). It can also be seen that although L expression was not affected in the range of pgstZ used (right panel), a slight decrease of L synthesis was observed with the inclusion of 260 ng of pZ (left panel). This last amount of pZ, however, is well beyond the range used to produce potent inhibitory activity (Fig. 1) (21). Using serum against L, we were unable to detect Z coimmunoprecipitation, as nonspecifically immunoprecipitated proteins migrated together with Z in the SDS-PAGE gels (not shown).
FIG. 2.
Comparison of Z and gstZ ability for interacting with L. CV1 cells were transfected with (+) or without (−) the addition of pL as indicated. For experiments in which pZ (left) or pgstZ or pgst (right) was included, the amount used is indicated. At 5 hpt, cells were labeled, cell extracts were prepared, and the proteins were immunoprecipitated with serum against either Z (αZ) or L (αL) (left) or either GST (αgst) or L (αL) (right). Immunoprecipitated proteins were subjected to SDS-12% PAGE. Molecular masses of 14C-labeled markers are indicated on the right. The positions of the L, Z, GST, and gstZ proteins are indicated on the left. Films were exposed for 24 h (left, αZ), 12 h (left, αL), or 7 h (right, αgst and αL).
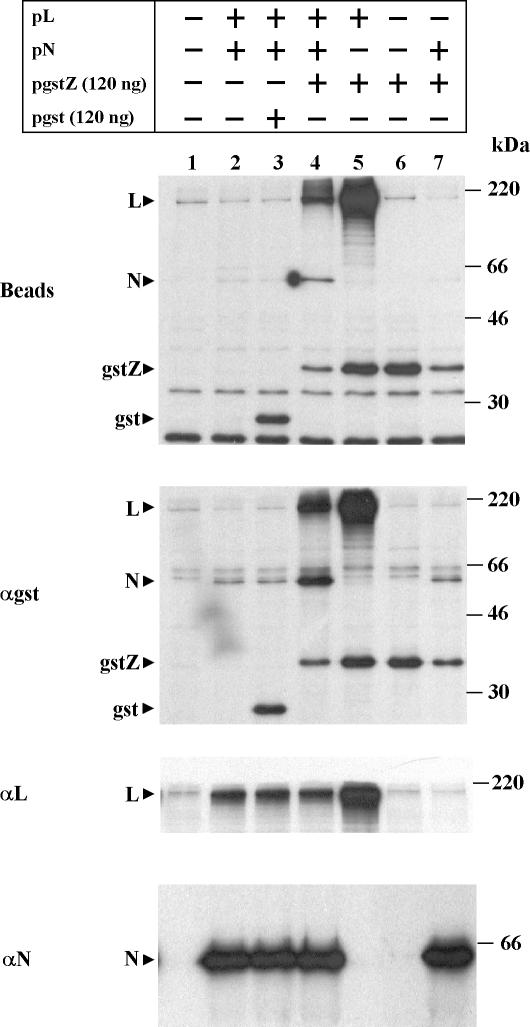
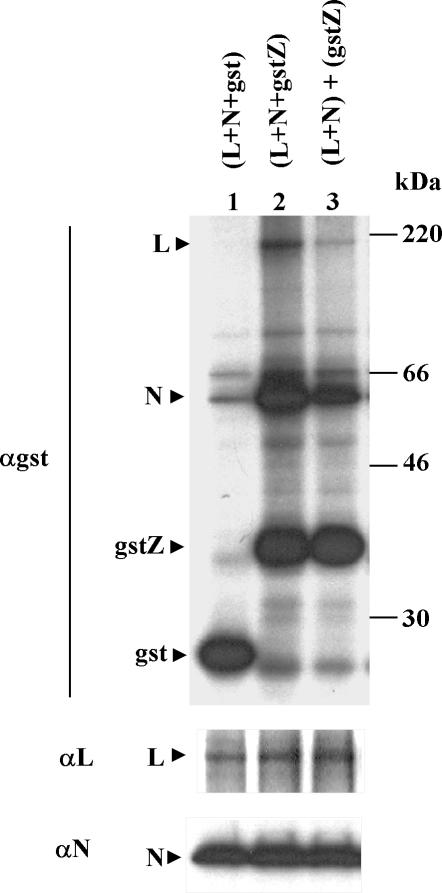
Since the gstZ protein could be used to demonstrate the binding of L to Z, we used the fusion protein to assess whether Z interacted with N. To this end, we transfected cells with the minigenome-expressing plasmid together with the combination of plasmids indicated in Fig. 3. Cells were labeled and processed as described above, and aliquots of the cell lysates were incubated with glutathione-Sepharose beads and immunoprecipitated with serum against either GST, L, or N. Gel analysis showed that proteins from all the transfected plasmids were synthesized (lower three panels). When the extracts were incubated with beads (upper panel), gstZ and GST bound in every case. Coexpression of gstZ, L, and N resulted in cobinding of both L and N to gstZ (lane 4). The specificity of the interaction was demonstrated by the fact that neither L nor N cobound to GST (lane 3). L coexpressed with gstZ cobound to gstZ (lane 5), whereas there was no cobinding of N coexpressed with gstZ (lane 7). Similar results were obtained by coimmunoprecipitation with serum against GST (panel αgst), confirming that N did not coimmunoprecipitate with gstZ unless L was coexpressed (compare lanes 4 and 7; the faint band of N in lane 7 appears to be nonspecifically immunoprecipitated N, as a similar band was detected in lanes 2 and 3 where gstZ was not coexpressed). Note that coexpression with gstZ or GST did not affect the level of expression of L or N (panels αL and αN, lanes 2 to 4). However, cotransfection with pN affected both gstZ and L expression (panel αgst, compare lane 4 with lane 5 and lane 6 with lane 7; panel αL, compare lanes 2 to 4 with lane 5). Thus, the effect of cotransfection with a high amount of pN appears to be nonspecific (see below).
FIG. 3.
Complex formation between L, N, and gstZ in the minigenome expression system. Subconfluent CV1 cell monolayers were transfected with pGenCAT, with (+) or without (−) the addition of the plasmids indicated. Cells were labeled and lysed as indicated in Materials and Methods, cell extracts were divided into four aliquots, and each aliquot was bound to glutathione-Sepharose beads or immunoprecipitated with serum against either GST, L, or N. The precipitated proteins were analyzed by SDS-12% PAGE. Molecular masses of 14C-labeled markers are indicated on the right. The positions of the L, N, GST, and gstZ proteins are indicated on the left. Films were exposed for 30 h (beads and αgst) or for 4 h (αL and αN).
To determine whether gstZ-L interaction required coexpression of the proteins, a mixing experiment was performed. Coexpression of gstZ with L and N gave significant amounts of both L and N coimmunoprecipitating with gstZ but not with GST (Fig. 4, lanes 1 and 2). In contrast, when separate cell extracts containing L plus N and gstZ were incubated together and then immunoprecipitated with GST antibodies, a reduced amount of L and N coimmunoprecipitated with gstZ (lane 3). Quantitation of the coimmunoprecipitated L and N bands indicated that the amounts of L and N in the mixed lysates were 19 and 22% of those in the cotransfected cell lysate, respectively.
FIG. 4.
Binding between gstZ and L requires coexpression of the proteins. CV1 cell monolayers grown in a 12-well dish were transfected with pGenCAT, together with the plasmids expressing the following proteins: L, N, and GST (lane 1); L, N, and gstZ (lane 2); and L and N (lane 3). An independent dish was transfected with gstZ plasmid. The amount of pgst and pgstZ added was 140 ng, and the amounts of the other plasmids are indicated in Materials and Methods. At 5 hpt, cells were labeled, lysed with 200 μl of TNEN, and incubated for 30 min at 30°C (lanes 1 and 2). Cells expressing L plus N and gstZ alone were lysed with 100 μl of TNEN, and the cell extracts were mixed and incubated for 30 min at 30°C (lane 3). Cell extracts were divided into three parts, and each part was immunoprecipitated with serum against either GST, L, or N. Proteins were resolved by SDS-PAGE on 12% polyacrylamide gels. Molecular masses of 14C-labeled markers are indicated on the right. The positions of the L, N, GST, and gstZ proteins are indicated on the left. Films were exposed for 3 days (αgst) or for 5 h (αL and αN).
In summary, by using coimmunoprecipitation with Z antiserum as well as cobinding to glutathione-Sepharose beads and coimmunoprecipitation with serum against GST and GST-tagged Z, we demonstrated that Z formed complexes with L. The results also indicated that the nucleocapsid protein N did not form complexes with gstZ. Conversely, when the GST-tagged N protein (gstN; see below) and Z were coexpressed, we could not detect Z binding to beads or Z coimmunoprecipitating with gstN (not shown). However, when gstZ, L, and N were coexpressed, both L and N cobound to beads and coimmunoprecipitated with GST antibodies.
Nucleocapsid protein N and polymerase protein L interact with each other.
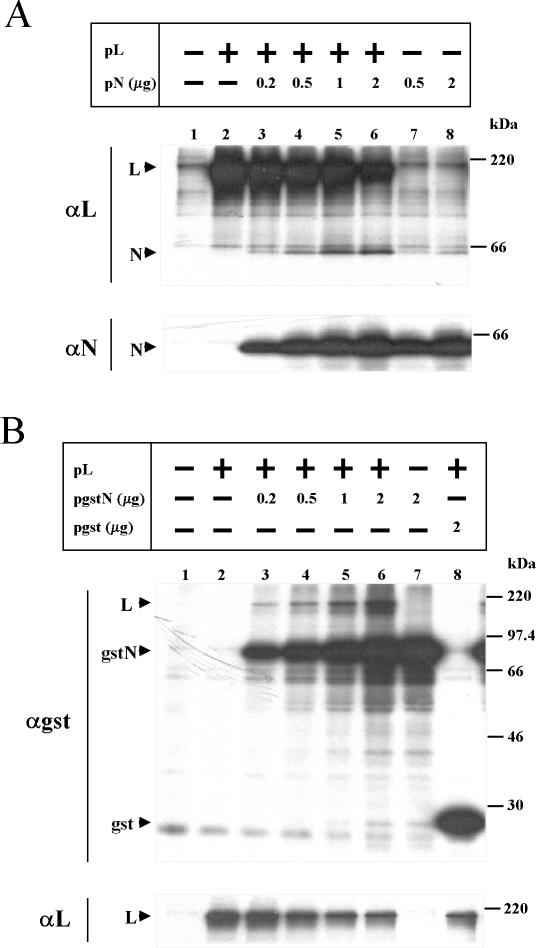
The observation that L formed complexes with gstZ, whereas N did not unless L was coexpressed, suggested to us that N and L interact with each other. Since, to our knowledge, there are no reports describing interactions between L and N in viruses of the Arenaviridae family, we investigated whether this interaction readily occurs. To that end, cells were transfected with plasmids expressing L and N as indicated in Fig. 5A, and cell extracts were processed as described earlier and analyzed using L or N antiserum. Immunoprecipitation with anti-L antibodies revealed that N coimmunoprecipitated with L, with the level of N increasing in parallel with that of the expressed N (compare lanes 3 to 6 with lanes 1, 2, 7, and 8). Maximal N-L interaction was observed with amounts of cotransfected N (1 to 2 μg of pN) and L plasmids previously found to be optimal for minigenome transcription and replication (21) (lanes 5 and 6). With anti-N antibody, we were unable to detect coimmunoprecipitated L when N and L were expressed together. Since monospecific serum against N was raised against the whole N protein (25), it is possible that antibodies to N might interfere with the sites of N-L interactions. To overcome this problem, we constructed a plasmid expressing a GST-tagged N protein (pgstN; see Materials and Methods) and analyzed N-L complex formation by immunoprecipitation with anti-GST antibody. For the experiment shown in Fig. 5B, cells were transfected with the plasmids expressing L, gstN, or GST as indicated and cell extracts were immunoprecipitated with serum against either GST or L. Immunoprecipitation with GST antibody showed that when gstN and L were coexpressed, increasing amounts of L coimmunoprecipitated along with the increasing levels of gstN (compare lanes 3 to 6 with lanes 1, 2, and 7). In contrast, when L was coexpressed with a plasmid expressing GST, L did not coimmunoprecipitate, indicating that the interaction with L is mediated through the N portion of the fusion protein (compare lanes 3 to 6 with lane 8). The eventual small amount of gstN that might coimmunoprecipitate with anti-L antibody was undetectable over the background of nonspecific proteins migrating together with gstN (not shown). Note that L expression decreased when cotransfected with larger amounts of either pN, pgstN, or pgst (A and B, panel αL), suggesting a nonspecific plasmid effect on protein expression.
FIG. 5.
Interaction between L and N proteins. Cells were transfected with (+) or without (−) the addition of the plasmid expressing L. For experiments in which pN (A) or pgstN or pgst (B) was added, the amount used is indicated. All dishes were cotransfected with pGenCAT. Labeling of the cells and preparation of cell extracts were done as described in the text. (A) Cell extracts immunoprecipitated with serum against either L or N. (B) Cell extracts immunoprecipitated with serum against GST or L. Immunoprecipitated proteins were resolved by SDS-PAGE in gels containing 12% polyacrylamide. Molecular masses of 14C-labeled markers are indicated on the right. The positions of the L, N, GST, and gstN proteins are indicated on the left. Films for panel A were exposed for 16 h and for panel B were exposed for either 40 h (αgst) or 4 h (αL).
Thus, we have identified an interaction between L and N by use of coimmunoprecipitation with serum against L as well as serum against GST when the GST-tagged N and L proteins were coexpressed.
TV Z requires an intact first Zn binding site in its RING domain for both its interaction with L and its inhibitory activity.
It has been shown that an intact first zinc-binding site in the Z RING motif is required for efficient interaction between LCMV Z protein and the eukaryotic translation initiation factor e1F4E, as mutation of its first two ligands (Cys1 and Cys2 of the RING motif; Fig. 6) reduced the association of Z with its partner (18). Therefore, we sought to determine whether mutation of Cys1 and Cys2 of the TV Z RING motif alters its ability to interact with L. To that end, Cys residues at positions 40 and/or 43 of TV Z were replaced with Gly. The mutants were named Z(C40,43G), Z(C40G), and Z(C43G) (Fig. 6). Since both gstZ and Z interact equally with L, the mutant forms of Z were tagged with GST at the N terminus. In addition, the use of serum against the tag for analysis of the GST-tagged proteins was chosen to overcome the possible differential reactivity of the mutant proteins to Z antibodies. The tagged Z mutants were then analyzed for the ability to interact with L and to inhibit transcription under the experimental conditions of the minigenome expression system. vTF7-3-infected cells were transfected with the minigenome-expressing plasmid, together with combinations of the indicated plasmids (Fig. 7), and the 35S-labeled cytoplasmic cell lysates were immunoprecipitated with serum against either GST, L, or N to analyze protein-protein interaction and to determine the level of protein expression (Fig. 7A). To relate complex formation with functional activity, nonlabeled cell extracts from cells transfected in parallel were used to assay CAT activity (Fig. 7B).
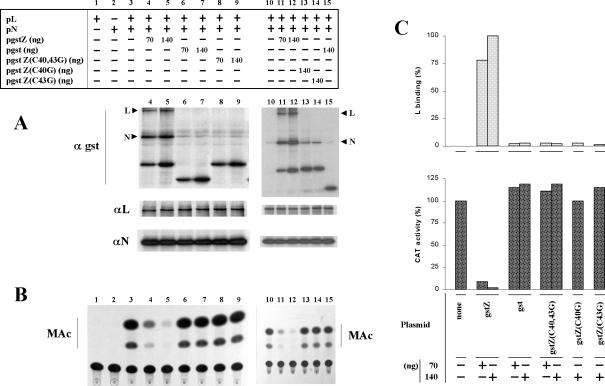
FIG. 7.
Effect of mutations in the first zinc-binding site of TV Z RING domain on Z-L interaction and Z-mediated inhibitory activity. Duplicate sets of dishes containing subconfluent CV1 cell monolayers were transfected with pGenCAT and, when indicated (+), with pL and pN. For experiments in which another plasmid was cotransfected, the amount used is indicated. (A) Cells in one set of dishes were radiolabeled and lysed, and aliquots of the cell lysates were immunoprecipitated with serum against either GST, L, or N as indicated in Materials and Methods. The immunoprecipitated proteins were resolved by SDS-12% PAGE. The positions of the L and N proteins are indicated on the left. (B) Cells in the other set of dishes were used to assay CAT activity as indicated in Materials and Methods. MAc, monoacetylated chloramphenicol species. (C) The L bands coimmunoprecipitating with serum against GST were quantitated, and the values were corrected for minor differences in the expression of the corresponding GST-tagged Z protein and plotted as percentages of the L band coimmunoprecipitating in cells transfected with 140 ng of pgstZ (upper panel). CAT activity was quantitated as indicated in Materials and Methods, and the results, shown as the percentage of the corresponding value without the addition of plasmids expressing GST-tagged proteins or GST, were plotted (lower panel). For panels A and B, results shown in lanes 1 to 9 and lanes 10 to 15 correspond to two independent experiments. For panel C, the combined results of the two experiments are shown.
Immunoprecipitation with serum against GST (panel A) showed that the GST-tagged proteins were all expressed to similar levels and confirmed the results shown above indicating that both L and N coimmunoprecipitated with gstZ but not with GST (compare lanes 6 and 7 with lanes 4 and 5 and lane 15 with lanes 11 and 12). Mutations at either or both Cys residues at positions 40 and 43 abolished the ability of Z to interact with L (compare lanes 8 and 9 with lanes 4 and 5 and lanes 13 and 14 with lanes 11 and 12; the results shown in lanes 4 to 9 and 10 to 15 correspond to two independent experiments; the coimmunoprecipitated L band from the two experiments was quantitated and the combined results were plotted in panel C, upper panel). The expression levels of L and N were not affected by cotransfection with 70 and 140 ng of plasmids expressing the GST-tagged proteins (Fig. 7A, panels αL and αN).
As shown in panel B, CAT activity was undetectable in cells expressing N and L individually, whereas in cells expressing L and N together, CAT activity was readily detected (lanes 1 to 3), confirming previous findings (21). Inclusion of 70 ng of pgstZ in cells expressing L and N reduced CAT activity to less than 10% of that exhibited without the gstZ plasmid, and cotransfection with 140 ng of pgstZ almost abolished CAT expression (compare lanes 4 and 5 with lane 3 and lanes 11 and 12 with lane 10; the average CAT expression from the two experiments is represented in Fig. 7C, lower panel). When cells were cotransfected with the plasmid expressing GST, no inhibition of CAT activity was detected even at the highest amount of plasmid used (compare lanes 6 and 7 with lanes 4 and 5 and lane 15 with lanes 11 and 12; see Fig. 7C, lower panel). These results indicated that the GST portion of the fusion protein has no effect on transcription and does not interfere with Z functional activity, as the dose-response curve of gstZ-mediated inhibition was similar to that with coexpression with Z (compare with data in Fig. 1 and with previously published results [21]).
We then used the tagged form of Z protein to analyze the effect of mutations in the RING motif on Z inhibitory activity. The results showed that mutations at either Cys 40, Cys 43, or both abolished Z-mediated inhibition of CAT expression (Fig. 7B, compare lanes 8 and 9 with lanes 4 and 5 and lanes 13 and 14 with lanes 11 and 12; Fig. 7C, lower panel). Thus, TV Z requires an intact first zinc-binding site in its RING domain for Z-L protein interaction and Z functional activity.
Regions of Z protein required for binding L protein and inhibitory activity.
Analysis of the TV Z amino acid sequence (16) reveals three defined regions (schematically represented in Fig. 6): a hydrophilic N-terminal region comprising aa 1 to 39, followed by the 37-aa RING domain and a 19-aa C-terminal region. Comparison of the deduced protein sequence of all arenavirus Z genes sequenced thus far reveals a similar organization, with a conserved ring domain (51% homology) and less conserved N- and C-terminal regions (22 and 31% homology, respectively) (9, 16, 27, 29). To assess whether the Z protein N- and C-terminal regions contribute to L binding and inhibition, we prepared Z mutants containing deletions at the C terminus, the N terminus, both the N and C termini, and the RING-plus-C-terminal regions. The mutants were designated, respectively, Z(1-77), Z(36-95), Z(36-77), and Z(1-35) (Fig. 6). The Z deletion mutants were then fused to GST at the N terminus, and the resulting proteins were tested for the ability both to bind to L and to inhibit CAT activity (Fig. 8). vTF7-3-infected cells were transfected with plasmids expressing the minigenome, L and N, with or without the addition of the indicated amounts of plasmids expressing the GST-tagged form of either Z or the Z-deletion mutants. After incubation, cells were labeled and lysed, and the cell lysates were used to analyze protein-protein interaction and the level of protein expression (panel A). Nonlabeled cell extracts from dishes that were infected and transfected in parallel were used to assay CAT activity (panel B).
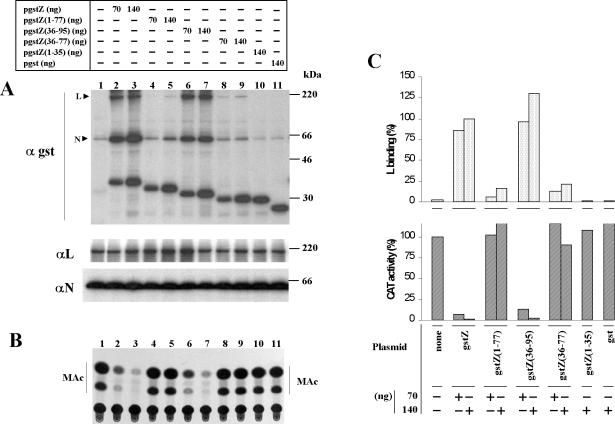
FIG. 8.
Regions of Z protein required for binding to L protein and for inhibitory activity. Duplicate sets of dishes containing CV1 cell monolayers were transfected with pGenCAT, pL, and pN as indicated in Materials and Methods and, when indicated, were cotransfected with 70 or 140 ng of the plasmid expressing gstZ, GST, or one of the GST-tagged Z deletion mutants. Cells from one of the sets of dishes were used to analyze binding to L (A) and cell extracts from the other set were used to assay CAT activity (B). Procedures were carried out as described for Fig. 7. Molecular masses of 14C-labeled markers are indicated on the right. The positions of the L and N proteins are indicated on the left. (C) Results of three experiments quantitated as described in the legend for Fig. 7, with the average results represented.
Immunoprecipitation with either GST, L, or N antibody (panel A) showed that the use of the same amount of each plasmid expressing the GST-tagged proteins resulted in similar levels of expression of each of the proteins and that L and N were expressed to similar levels in all cases. It was also found that both L and N coimmunoprecipitated with gstZ but not with GST or with the fusion protein containing the first 35 N-terminal Z aa [gstZ(1-35)] (panel αgst). The binding results are consistent with the inhibitory effect on CAT activity exhibited by gstZ but not by the mutant gstZ(1-35) or GST (panels A and B, compare lanes 10 and 11 with lanes 1, 2, and 3; panel C). Deletion of the C-terminal region [gstZ(1-77)] or both the C-terminal region and the first 35 N-terminal aa [gstZ(36-77)] resulted in a reduced ability to interact with L compared with wild-type Z (Fig. 8A, panel αgst, compare lanes 4, 5, 8, and 9 with lanes 2 and 3). Considering the amount of L coimmunoprecipitating with gstZ in cells cotransfected with 140 ng of the plasmid to be 100%, inclusion of the same amount of either mutant gave values which, in different experiments, varied from 12 to 21% for gstZ(1-77) and from 21 to 22% for gstZ(36-77) (the average of independent experiments is represented in Fig. 8C, upper panel). Neither gstZ(1-77) nor gstZ(36-77) exhibited a detectable inhibition of CAT activity, although the expression levels of the mutants were similar to those at which gstZ gave potent inhibition of CAT expression (panel B, compare lanes 4, 5, 8, and 9 with lanes 1, 2, and 3; panel C, bottom). The Z fusion mutant with a deletion in the first 35 N-terminal aa [gstZ(36-95)] retained full binding to L, and its ability to inhibit transcription was comparable to that of wild-type Z (panels A and B, compare lanes 6 and 7 with lanes 1, 2, and 3). Quantitation of the results indicated that the addition of 70 and 140 ng of pgstZ reduced CAT activity to 7 and 2% of that without gstZ plasmid, respectively, whereas inclusion of the same amounts of pgstZ(36-95) gave 13 and 3% activity, respectively (Fig. 8C, lower panel).
In summary, deletion of the first 35 N-terminal aa [Z(36-95)] has no effect on the ability of Z to interact with L or on its inhibitory activity. Deletion of the C-terminal region [Z(1-77)] or both the C-terminal region and the first 35 aa at the N-terminal region [Z(36-77)] gave mutants with a reduced ability to bind to L that were devoid of inhibitory activity.
DISCUSSION
We have previously reported the establishment of a transcription and replication system based on plasmid-supplied TV-like RNAs and TV proteins. Using this system, we demonstrated that coexpression of L and N proteins is sufficient to support full-cycle RNA replication and transcription and that Z is an inhibitor of these processes (21). For the present report we investigated whether Z might interact with one or both of the proteins (L and N) that are required for RNA synthesis. For these studies we used the reconstituted system, as its context should reflect true protein-protein interactions occurring during replication and transcription. In addition, the use of this system allowed us to relate Z binding ability with Z functional activity which, for this study, was evaluated as the inhibitory effect on CAT expression. Since in our reconstituted system RNA replication involves extensive amplification of the plasmid-supplied template by the TV polymerase, Z-mediated inhibition of CAT activity most probably reflects reduced template availability for transcription due to reduced RNA replication (21). By means of coimmunoprecipitation with Z antibodies we showed that L was bound to Z and that the binding was related to CAT activity inhibition (Fig. 1). With a GST-tagged Z protein, which retains inhibitory function, and cobinding to glutathione-Sepharose beads or coimmunoprecipitation with GST antibodies, Z was shown to specifically bind L and not N (Fig. 2 and 3). Further, the gstZ inhibitory effect was related to its binding ability (Fig. 7 and 8). These data provide the first evidence of an interaction between Z and L, suggesting that Z interferes with replication and transcription by direct action on the L polymerase protein.
The interaction between Z or gstZ and L does not require coexpression of other viral components (Fig. 2); however, maximal interaction requires coexpression of the proteins (Fig. 4). If Z inhibitory activity on transcription and replication is related to its interaction with the L protein, then this last result might explain the discrepancy between the observed inhibitory effect of Z in the reconstituted system, in which Z and L were coexpressed (21), and the reported lack of inhibition of viral RNA synthesis when Z is added to TV-infected cell extracts (13).
To identify TV Z domains involved in Z interaction with the L protein (Fig. 6), we mutated zinc-coordinating Cys residues in the first zinc-binding site of the Z RING domain and prepared Z mutants with N- and C-terminal deletions. Since LCMV Z protein was described as a potential translational inhibitor (18), we exerted care to control that coexpression with gstZ and its mutants did not affect the level of expression of the proteins (L and N) required for RNA synthesis. In neither case did the fusion proteins exhibit an inhibitory effect on L and N synthesis. The results demonstrated (Fig. 7 and 8) that integrity of the RING structure is essential for Z binding to L and for Z inhibitory activity, as mutations of Cys1 and/or Cys2 in the RING motif abolished both Z-mediated functions [mutants Z(C40,43G), Z(C40G), and Z(C43G)]. Mutant Z(36-95) behaved like wild-type Z in its binding ability and inhibitory activity, indicating that the first 35 N-terminal aa are not required for these functions. Mutants with an intact RING domain that were devoid of the C-terminal region [Z(1-77)] or both the C-terminal region and the first 35 aa of the N-terminal region [Z(36-77)] retained, albeit at a reduced level, the ability to bind to L but exhibited no inhibitory activity. Thus, amino acids within the C-terminal region and possibly within 4 aa adjacent to the RING domain N terminus contribute to efficient Z-L interaction and are required for Z inhibitory function. Further mutational analysis will be required to define the regions and individual amino acids associated with the Z-L interaction and with Z functional activity. A recent report on LCMV Z regions required for inhibitory activity demonstrated that the RING domain is necessary but not sufficient for Z-mediated inhibition of CAT activity and that amino acid residues surrounding the RING domain at its N and C termini may contribute to the inhibitory effect (7).
Although the data presented here suggest that Z inhibits RNA synthesis through its interaction with L, the poor understanding of basic aspects of arenavirus gene expression renders it difficult to speculate on molecular mechanisms underlying this effect. It appears that Z is not acting through an increased degradation of the L protein, since in a pulse-chase experiment in which L was either expressed alone or coexpressed with gstZ, 61 and 64% of the L remained, respectively, after a 16-h chase period. The gstZ-L complex showed a similar decay (R. Jácamo, unpublished observations). Considering that the interaction between Z and L is more effective if both proteins are expressed in the same cell and that the effect of Z was evaluated on multiple rounds of RNA replication (21), we suggest that Z may affect the proper folding of the L protein and therefore alter its ability to initiate RNA replication.
The results just described indicate that gstZ, at an expression level producing a potent inhibition of CAT activity, interacts with L but not with N. However, when gstZ, N, and L were coexpressed, both L and N cobound to beads and coimmunoprecipitated with GST antibodies (Fig. 3). Further, when Z mutants were analyzed for the ability to bind to L, there was a correlation with the levels of coimmunoprecipitating L and N (Fig. 7 and 8). These results suggested that the cobinding of N occurred through its interaction with L and led to evidence, for the first time, that L and N interact with each other (Fig. 5). We would like to point out that the observation that the binding of N to L does not interfere with the Z-L complex suggests that the Z and N binding sites on L are independent of each other.
Arenavirus nucleocapsid protein N is critical for RNA replication (10, 24, 28). The replication process should involve cap-independent initiation and elongation of the RNA chain that becomes encapsidated by N. We suggest that the observed binding between N and L facilitates cap-independent initiation and keeps N in a suitable form for specific encapsidation of the nascent RNA chain. It is worth noting that whereas the P protein plays a key role during RNA encapsidation by complexing with the N protein in rhabdoviruses and paramyxoviruses (8, 14), there is no counterpart of P in arenaviruses, where the viral proteins N and L are sufficient to accomplish RNA replication and transcription (20, 21).
The results in this study demonstrate, for the first time, interactions between the viral proteins implicated in arenavirus transcription and replication. Understanding these interactions and identifying the protein domains involved will contribute to elucidating the mechanism of arenavirus genome expression and may provide potential targets for antiviral strategies for these highly pathogenic hemorrhagic fever viruses.
Acknowledgments
We thank Lucia Rothman-Denes (Department of Molecular Genetics and Cell Biology, University of Chicago) for critical review of the manuscript.
N.L. and M.T.F.-F. are research investigators of CONICET. R.J. is a recipient of a fellowship from CONICET. M.W. is a recipient of a fellowship from ANPCyT (grant no. 6285). M.T.F.-F. thanks Laboratorios Bago (Argentina) for its support.
REFERENCES
- 1.Bishop, D. H. L., and D. D. Auperin. 1987. Arenavirus gene structure and organization. Curr. Top. Microbiol. Immunol. 133:5-17. [DOI] [PubMed] [Google Scholar]
- 2.Borden, K. L. 2000. RING domains: master builders of molecular scaffolds? J. Mol. Biol. 295:1103-1112. [DOI] [PubMed] [Google Scholar]
- 3.Borden, K. L., E. J. Campbell Dwyer, G. W. Carlile, M. Djavani, and M. S. Salvato. 1998. Two RING finger proteins, the oncoprotein PML and the arenavirus Z protein, colocalize with the nuclear fraction of the ribosomal P proteins. J. Virol. 72:3819-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borden, K. L., E. J. Campbell Dwyer, and M. S. Salvato. 1998. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J. Virol. 72:758-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowen, M. D., C. J. Peters, and S. T. Nichol. 1996. The phylogeny of New World (Tacaribe complex) arenaviruses. Virology 219:285-290. [DOI] [PubMed] [Google Scholar]
- 6.Cornu, T. I., and J. C. de la Torre. 2001. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J. Virol. 75:9415-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornu, T. I., and J. C. de la Torre. 2002. Characterization of the arenavirus RING finger Z protein regions required for Z-mediated inhibition of viral RNA synthesis. J. Virol. 76:6678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das, T., and A. K. Banerjee. 1992. Role of the phosphoprotein (P) in the encapsidation of presynthesized and de novo synthesized vesicular stomatitis virus RNA by the nucleocapsid protein (N) in vitro. Cell. Mol. Biol. 38:875-884. [PubMed] [Google Scholar]
- 9.Djavani, M., I. S. Lukashevich, A. Sanchez, S. T. Nichol, and M. S. Salvato. 1997. Completion of the Lassa fever virus sequence and identification of a RING finger open reading frame at the L RNA 5′ end. Virology 235:414-418. [DOI] [PubMed] [Google Scholar]
- 10.Franze-Fernández, M. T., C. Zetina, S. Iapalucci, M. A. Lucero, C. Bouissou, R. López, O. Rey, M. Daheli, G. N. Cohen, and M. M. Zakin. 1987. Molecular structure and early events in the replication of Tacaribe arenavirus S RNA. Virus Res. 7:309-324. [DOI] [PubMed] [Google Scholar]
- 11.Franze-Fernández, M. T., S. Iapalucci, N. López, and C. Rossi. 1993. Subgenomic RNAs of Tacaribe virus, p. 113-132. In M. S. Salvato (ed.), The Arenaviridae. Plenum Press, New York, N.Y.
- 12.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcin, D., S. Rochat, and D. Kolakofsky. 1993. The Tacaribe arenavirus small zinc finger protein is required for both mRNA synthesis and genome replication. J. Virol. 67:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horikami, S. M., J. Curran, D. Kolakofsky, and S. A. Moyer. 1992. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iapalucci, S., R. López, O. Rey, N. López, M. T. Franze-Fernández, G. N. Cohen, M. Lucero, A. Ochoa, and M. M. Zakin. 1989. Tacaribe virus L gene encodes a protein of 2210 aa residues. Virology 170:40-47. [DOI] [PubMed] [Google Scholar]
- 16.Iapalucci, S., N. López, O. Rey, M. M. Zakin, G. N. Cohen, and M. T. Franze-Fernández. 1989. The 5′ region of Tacaribe virus L RNA encodes a protein with a potential metal binding domain. Virology 173:357-361. [DOI] [PubMed] [Google Scholar]
- 17.Iapalucci, S., N. López, and M. T. Franze-Fernández. 1991. The 3′ end termini of the Tacaribe arenavirus subgenomic RNAs. Virology 182:269-278. [DOI] [PubMed] [Google Scholar]
- 18.Kentsis, A., E. Campbell Dwyer, J. M. Perez, M. Sharma, A. Chen, Z. Q. Pan, and K. L. B. Borden. 2001. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol. 312:609-623. [DOI] [PubMed] [Google Scholar]
- 19.Kolakofsky, D., and D. Garcin. 1993. The unusual mechanism of arenavirus RNA synthesis, p. 103-112. In M. S. Salvato (ed.), The Arenaviridae. Plenum Press, New York, N.Y.
- 20.Lee, K. J., I. S. Novella, M. N. Teng, M. B. A. Oldstone, and J. C. de la Torre. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 74:3470-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López, N., R. Jácamo, and M. T. Franze-Fernández. 2001. Transcription and RNA replication of Tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J. Virol. 65:12241-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López, N., L. Scolaro, C. Rossi, R. Jácamo, N. Candurra, C. Pujol, E. B. Damonte, and M. T. Franze-Fernández. 2000. Homologous and heterologous glycoproteins induce protection against Junin virus challenge in guinea pigs. J. Gen. Virol. 81:1273-1281. [DOI] [PubMed] [Google Scholar]
- 23.Perrin, S., and G. Gilliland. 1990. Site-specific mutagenesis using asymmetric polymerase chain reaction and a single mutant primer. Nucleic Acids Res. 18:7433-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinschewer, D. D., M. Perez, and J. C. de la Torre. 2003. Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication. J. Virol. 77:3882-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi, C., O. Rey, P. Jenik, and M. T. Franze-Fernández. 1996. Immunological identification of Tacaribe virus proteins. Res. Virol. 147:203-211. [DOI] [PubMed] [Google Scholar]
- 26.Salvato, M. S. 1993. Molecular biology of the prototype arenavirus, lymphocytic choriomeningitis virus, p 133-156. In M. S. Salvato (ed.), The Arenaviridae. Plenum Press, New York, N.Y.
- 27.Salvato, M., and E. M. Shimomaye. 1989. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology 173:1-10. [DOI] [PubMed] [Google Scholar]
- 28.Tortorici, M. A., C. G. Albariño, D. M. Posik, P. D. Ghiringhelli, M. E. Lozano, R. Rivera Pomar, and V. Romanowski. 2001. Arenavirus nucleocapsid protein displays a transcriptional antitermination activity in vivo. Virus Res. 73:41-45. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, L., K. A. Marriott, D. G. Harnish, and J. F. Aronson. 2001. Reassortant analysis of guinea pig virulence of Pichinde virus variants. Virology 290:30-38. [DOI] [PubMed] [Google Scholar]