Abstract
Here we present the complete genomic sequence of bovine herpesvirus 5 (BHV-5), an alphaherpesvirus responsible for fatal meningoencephalitis in cattle. The 138,390-bp genome encodes 70 putative proteins and resembles the α2 subgroup of herpesviruses in genomic organization and gene content. BHV-5 is very similar to BHV-1, the etiological agent of infectious bovine rhinotracheitis, as reflected by the high level of amino acid identity in their protein repertoires (average, 82%). The highest similarity to BHV-1 products (≥95% amino acid identity) is found in proteins involved in viral DNA replication and processing (UL5, UL15, UL29, and UL39) and in virion proteins (UL14, UL19, UL48, and US6). Among the least conserved (≤75%) are the homologues of immediate-early (IE) proteins BICP0, BICP4, and BICP22, the three proteins being longer in BHV-5 than in BHV-1. The structure of the BHV-5 latency-related (LR) region departs markedly from that of BHV-1 in both coding and transcriptional regulatory regions. Given the potential significance of IE genes and the LR region in virus-neuron interactions, it is likely these differences contribute to BHV-5 neuropathogenicity.
Bovine herpesvirus 5 (BHV-5) is a pathogen of cattle responsible for sporadic epizootics of fatal meningoencephalitis (6, 37). Due to similarities in virion morphology, cytopathic effects in cell culture, and antigenic properties (37, 38), BHV-5 was formerly regarded as a neuropathogenic variant of bovine herpesvirus 1 (BHV-1), the etiological agent of infectious bovine rhinotracheitis and vulvovaginitis. Subsequent comparative studies based on restriction site mapping of viral DNA (28, 32, 93), cross-neutralization tests, and monoclonal antibody reactivity (24, 63) indicated that the viruses differ in genomic and antigenic properties. In 1992 BHV-5 was recognized as a distinct virus by the International Committee on Taxonomy of Viruses (78).
Both BHV-5 and BHV-1 are neurotropic viruses, but only BHV-5 is capable of significant replication in the central nervous system (CNS) and induction of neurological disease (5, 10). Outbreaks of meningoencephalitis caused by BHV-5 have been reported in Australia (37), North and South America (8, 15, 42, 77), and Europe (9, 66). The course of the disease after experimental infection with BHV-5 depends on the virus isolate, the route of inoculation, and the immunological status and age of the animal. Calves up to 4 months of age are most susceptible. During the first week following intranasal inoculation of virus, the animals either present signs of mild rhinitis and conjunctivitis or they remain asymptomatic (5, 16, 66, 73). At this point, animals can recover from infection or, alternatively, progress to neurological disease and die. Regardless of the appearance of clinical neurological disease in infected calves, BHV-5 invades the CNS and causes various degrees of pathology (16, 65). The neural route followed by BHV-5 to invade the CNS in cattle has not been defined, although both the trigeminal and the olfactory pathways have been implicated (6, 65). In a rabbit model for BHV-5, the olfactory pathway is the main route for neural dissemination (19, 57).
At times when no infectious virus can be isolated from peripheral sites, surviving animals exhibit BHV-5 viral sequences in their trigeminal ganglia (TG), indicating that, as in BHV-1, BHV-5 remains latent in the TG (5, 16, 65, 91). Latent virus can be reactivated after treatment of latently infected animals with dexamethasone (10, 16). Although recrudescence of clinical disease has been observed after experimental virus reactivation, its occurrence in nature remains unknown (73). Previous vaccination of cattle against BHV-1 resulted in protection against BHV-5-induced neuropathology and clinical signs of disease (16). This is partially explained by the cross-reactivity of induced BHV-1 neutralizing antibodies (24). Vaccination with BHV-1, however, does not prevent establishment of latency by BHV-5 (16).
The marked neuroinvasiveness (and often neurovirulence) displayed by BHV-5 contrasts with the inability of BHV-1 to invade the CNS and cause neurological disease to any significant degree. During infection with other neurotropic herpesviruses (e.g., pseudorabies virus [PRV] and herpes simplex virus [HSV]), neuroinvasiveness and local virus spread in the CNS are properties which largely rely on certain viral envelope glycoproteins (46, 67). However, other viral functions could contribute to a successful infection of the CNS.
Comparative genomics has proven useful in identifying genes involved in virulence. The complete BHV-1 genomic sequence, a composite including sequences of five different virus strains, is available (GenBank accession number AJ004801). However, less than 15% of the BHV-5 genome (mostly representing envelope glycoproteins) has been sequenced (1, 18, 20, 22, 31, 40, 64, 80, 86). Here we present the complete sequence of a neurovirulent strain of BHV-5, with analysis and comparison to BHV-1.
MATERIALS AND METHODS
DNA isolation, cloning, and sequencing.
BHV-5 strain SV507/99 was originally isolated from bovine brain tissue from a case of fatal encephalitis in southern Brazil (91).
To obtain viral DNA, BHV-5-infected MDBK cells were pelleted, washed in NTE buffer (10 mM Tris [pH 8.0], 150 mM NaCl, 5 mM EDTA), resuspended in KTE buffer (10 mM Tris [pH 8.0], 10 mM KCl, 5 mM EDTA), and incubated in KTE buffer containing 0.25% β-mercaptoethanol and 10% Triton X-100 for 10 min on ice. After centrifugation at 3,000 rpm (Sorvall), 5 min, 4°C, virus in the supernatant was pelleted onto a 30% sucrose cushion (13,000 rpm, 1 h, 4°C), resuspended in Tris-EDTA (TE; pH 8.0), incubated in the presence of sodium dodecyl sulfate (0.5% [wt/vol]) and proteinase K (10 mg/ml) for 2 h at 37°C, and phenol extracted. Viral DNA was precipitated with ethanol and resuspended in TE buffer (pH 8.0). Random 1- to 8-kbp DNA fragments were obtained by incomplete enzymatic digestion with AciI endonuclease (New England Biolabs, Beverly, Mass.). DNA fragments of 1.5 to 3 kbp were isolated after separation in size exclusion columns (Clontech), cloned into the dephosphorylated AciI site of pUC19 plasmids, and grown in Escherichia coli DH10B cells (Gibco BRL, Gaithersburg, Md.). Plasmids were purified by alkaline lysis as instructed by the manufacturer (Eppendorf 5 Prime; Boulder, Colo.). DNA templates were sequenced from both ends with M13 forward and reverse primers and from selected plasmids with transposon insertion (EZ::TN<KAN-2> transposon insertion kit; Epicentre Tech., Madison, Wis.), using dideoxy-chain terminator sequencing chemistries (82) and an Applied Biosystems PRISM 3700 automated DNA sequencer (PE Biosystems, Foster City, Calif.). Bases were called from chromatogram traces with Phred (35), which also produced a quality file containing a predicted probability error at each base position.
DNA sequence analysis.
DNA sequences were assembled with Phrap (34), using the quality files and default settings to produce a consensus sequence which was manually edited with Consed (41). An identical sequence was assembled using the Cap3 assembler with quality files and clone length constraints (45). The final DNA consensus sequence represented an average eightfold redundancy at each base position. Gap closure was achieved by primer walking of gap-spanning clones and sequencing of PCR products. A total of 7,835 usable traces were assembled into a 127,162-bp contig by bidirectional sequencing of random clones and 61 PCR products, including those that crossed the terminal junction sequences from concatemeric replicative intermediates. The assembled contig had an estimated error rate of <0.03/10 kbp and showed no evidence of polymorphism using Polyphred analysis (34), including within the single, complete unique short (US) repeat sequence which assembled with double average redundancy at each base position, consistent with a bimolar representation of repeat sequences derived from two identical copies, as found in other alphaherpesviruses. Thus, the assembled contig contained 570 bp of the terminal repeat (TR) at the left terminus, all of the unique long (UL), internal repeat (IR), and US sequences, and 738 bp of the TR at the right terminus. IR and TR sequences were also assembled separately with clones containing the unique-repeat junctions and overlapping clones, using length constraints and position as provided by the computer assembly programs. These assemblies were manually joined at the unique-repeat boundaries, thus providing the complete genome. For descriptive purposes, we have presented BHV-5 in a linearized fashion as described by Dolan et al. (29). Genome DNA composition, structure, repeats, and restriction enzyme patterns were analyzed as previously described (2). Open reading frames (ORFs) encoding proteins of ≥60 amino acids with a methionine start codon (88) were evaluated for coding potential using the Hexamer (ftp.sanger.ac.uk/pub/rd) and Glimmer (81) computer programs. Other criteria included similarity to other herpesvirus and compact gene arrangements with little gene overlap. Homology searches were conducted using BLAST (3), PSIBLAST (4), FASTA (72), BLIMPS (92), and HMMER (87) programs with the Prosite, Pfam, Prodom, Sbase, Blocks, Domo, and GenBank databases (14). GCG (26), MEMSAT (54), and SAPS (12) programs were used for gene analysis. The coding potential and splicing patterns in the latency-related (LR) region of BHV-5 and BHV-1 were analyzed with Glimmer, Hexamer, Splice (a neural network program for eukaryotic splice site prediction; ftp://genome.lbl.gov/pub/reese/SPLICE), NNPP (eukaryotic promoter prediction; ftp://genome.lbl.gov/pub/reese/NNPP), TIGR GeneSplicer (74), HMM gene trained on human and Drosophila gene sets (55), and Eponine for transcription start site detection (30). We also compared BHV-1 mapping data to the BHV-5 LR region.
Nucleotide sequence accession number.
The BHV-5 genome sequence has been deposited in GenBank under accession no. AY261359.
RESULTS AND DISCUSSION
Genome organization.
The BHV-5 genome is 138,390 bp long, 2,518 bp longer than the BHV-1 genome, and contains a 75% G+C base composition. The genome consists of two unique sequences, long or UL (104,054 bp) and short or US (9,548 bp), with the latter being flanked by inverted IR and TR elements of 12,109 bp each. This arrangement corresponds to the D-type herpesviral genome (62).
The BHV-5 origins of DNA replication (ORI) are located in the repeat regions from nucleotide positions 113206 to 113418 and 129595 to 129807. ORI sequences consist of two imperfect AT-rich direct repeats (ORIa and ORIb) which contain herpesvirus consensus sites for the origin-binding protein (84). As in BHV-1, an additional truncated repeat (ORIc) is located 130 bp downstream from ORIb.
Gene characterization.
BHV-5 contains 72 genes (Table 1), of which 68 are present as single copies within the unique regions and 2 initiate and are completely located within the repeat regions (BICP4 and BICP22). BHV-5 proteins are most similar to homologues from BHV-1, averaging 82% amino acid identity. All BHV-5 ORFs are present in BHV-1; however, BHV-5 lacks a homologue of UL0.5. Among nonbovine herpesviruses, ORFs of equine herpesviruses (EHV) 1 and 4 are the most similar to those of BHV-5 (28 to 69% amino acid identity). The similarity in gene arrangement and the high percentage of amino acid identity between BHV-5 and previously sequenced alphaherpesviruses support the inclusion of BHV-5 in the α2 subgroup of herpesviruses (62), as was previously suggested by envelope glycoprotein B sequence analysis (80).
TABLE 1.
Characterization of BHV-5 genes
| ORF no. | ORF name | Position (nt)a | Length (aa)a | BHV-5 accession no.b | BHV-1
|
Closest non-BHV-1 species
|
Predicted product and/or functiond | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (aa) | % Identity | Accession no.b | Namec | Length (aa) | % Identity | Accession no.b | ||||||
| BHV5-01 | circ | 1167-1901 | 245 | 247 | 84 | M96453 | EHV-1 | 257 | 43 | P28988 | Myristylated virion protein | |
| BHV5-02 | UL54 | 3482-2274 | 403 | 400 | 82 | M96453 | EHV-1 | 470 | 47 | Q05906 | Regulates and transports RNA | |
| BHV5-03 | UL53 | 4699-3707 | 331 | 332 | 85 | U34593 | EHV-1 | 343 | 39 | AF030027 | Glycoprotein K | |
| BHV5-04 | UL52 | 7931-4677 | 1,085 | 1,074 | 83 | Z54206 | EHV-1 | 1,081 | 44 | P28962 | Component of DNA helicase-primase complex | |
| BHV5-05 | UL51 | 7930-8715 | 262 | 243 | 81 | Z54206 | PRV | 236 | 49 | X87246 | Palmitoylated protein (cytoplasm) | |
| BHV5-06 | UL50 | 9821-8859 | 321 | 325 | 83 | S62816 | EHV-1 | 326 | 40 | P28892 | Deoxyuridine triphosphatase (dUTPase) | |
| BHV5-07 | UL49.5 | 9772-10056 | 95 | 96 | 81 | S62816 | PRV | 98 | 36 | U38547 | Glycoprotein N | |
| BHV5-08 | UL49 | 10193-10993 | 267 | 258 | 72 | U211137 | EHV-1 | 304 | 41 | X17684 | Tegument protein | |
| BHV5-09 | UL48 | 11189-12634 | 482 | AY034598 | 505 | 98 | Z11610 | EHV-4 | 448 | 52 | AF030027 | trans-Inducing factor (tegument) |
| BHV5-10 | UL47 | 12808-15030 | 741 | 739 | 93 | P36338 | EHV-4 | 871 | 35 | P28929 | Tegument phosphoprotein | |
| BHV5-11 | UL46 | 15166-17367 | 734 | 748 | 82 | Z54206 | EHV-1 | 747 | 40 | P28937 | Tegument protein | |
| BHV5-12 | UL44 | 19051-17594 | 486 | Z49224 | 508 | 75 | Z54206 | CaHV-1 | 521 | 59 | Z49225 | Glycoprotein C |
| BHV5-13 | UL43 | 20382-19243 | 380 | 378 | 88 | Z54206 | EHV-4 | 403 | 28 | AF030027 | Virion protein (membrane) | |
| BHV5-14 | UL42 | 21671-20436 | 412 | 408 | 79 | Z54206 | PRV | 384 | 38 | P36702 | Processivity factor for DNA polymerase | |
| BHV5-15 | UL41 | 21714-23288 | 525 | 459 | 91 | Z54206 | EHV-1 | 497 | 48 | P28957 | Virion host shutoff factor (tegument) | |
| BHV5-16 | UL40 | 24363-23419 | 315 | 314 | 91 | Q01319 | PRV | 303 | 76 | X72087 | Ribonucleotide reductase small subunit | |
| BHV5-17 | UL39 | 26785-24386 | 800 | 787 | 97 | Z54206 | PRV | 835 | 67 | X72087 | Ribonucleotide reductase large subunit | |
| BHV5-18 | UL38 | 28787-27153 | 545 | 474 | 87 | Z54206 | EHV-4 | 462 | 47 | AF030027 | Capsid protein | |
| BHV5-19 | UL37 | 28869-32030 | 1,078 | 1,024 | 88 | Z54206 | EHV-4 | 1,021 | 41 | AF030027 | Tegument protein | |
| BHV5-20 | UL36 | 32133-41744 | 3,204 | 3,247 | 80 | Z78205 | EHV-1 | 3,421 | 40 | P28955 | Very large tegument protein | |
| BHV5-21 | UL35 | 42351-41977 | 125 | 124 | 87 | Z78205 | PRV | 103 | 54 | AJ276165 | Capsid protein | |
| BHV5-22 | UL34 | 43226-42399 | 276 | 260 | 83 | Z78205 | PRV | 260 | 58 | AF301599 | Virion protein (membrane) | |
| BHV5-23 | UL33 | 43639-43310 | 110 | 108 | 89 | Z78205 | EHV-1 | 162 | 59 | P28953 | Capsid packaging protein | |
| BHV5-24 | UL32 | 43620-45413 | 598 | 601 | 89 | Z78205 | EHV-1 | 620 | 53 | P28952 | Cleavage and packaging protein | |
| BHV5-25 | UL31 | 45409-46545 | 379 | 361 | 84 | Z78205 | EHV-4 | 326 | 64 | AF030027 | UL34-associated nuclear protein | |
| BHV5-26 | UL30 | 50224-46475 | 1,250 | 1,246 | 90 | X94677 | EHV-1 | 1,220 | 61 | P28858 | DNA polymerase, catalytic subunit | |
| BHV5-27 | UL29 | 50495-54118 | 1,208 | 1,203 | 95 | X94677 | EHV-1 | 1,209 | 62 | P28932 | Single-stranded DNA binding protein | |
| BHV5-28 | UL28 | 54358-56805 | 816 | 826 | 90 | X94677 | EHV-1 | 775 | 55 | M86664 | Cleavage and packaging protein | |
| BHV5-29 | UL27 | 56661-59501 | 947 | AF359759 | 932 | 93 | P12640 | FHV-1 | 948 | 58 | S49775 | Glycoprotein B |
| BHV5-30 | UL26.5 | 60960-60016 | 315 | 308 | 80 | 431809 | HSV-2 | 329 | 38 | L37443 | Capsid scaffolding protein | |
| BHV5-31 | UL26 | 61872-60016 | 619 | 621 | 83 | U31809 | EHV-1 | 646 | 46 | P28936 | Capsid maturation serine protease | |
| BHV5-32 | UL25 | 63788-61980 | 603 | 598 | 94 | AJ004801 | EHV-1 | 587 | 58 | P28928 | DNA packaging virion protein | |
| BHV5-33 | UL24 | 64609-63773 | 279 | 293 | 74 | L39072 | EHV-1 | 272 | 45 | P28927 | Putative membrane-associated protein | |
| BHV5-34 | UL23 | 64608-65675 | 356 | S56149 | 359 | 83 | P36226 | EHV-4 | 352 | 46 | AF030027 | Thymidine kinase |
| BHV5-35 | UL22 | 65795-68338 | 848 | AF113752 | 842 | 86 | P27599 | EHV-4 | 855 | 33 | A21045 | Glycoprotein H |
| BHV5-36 | UL21 | 70346-68538 | 603 | 574 | 77 | Z48053 | EHV-1 | 530 | 38 | P28972 | Tegument protein | |
| BHV5-37 | UL20 | 70386-71144 | 253 | 231 | 93 | Z48053 | EHV-1 | 239 | 35 | P28971 | Virion protein (membrane) | |
| BHV5-38 | UL19 | 71227-75399 | 1,391 | 1,385 | 97 | Z48053 | EHV-1 | 1,376 | 69 | P28920 | Major capsid protein | |
| BHV5-39 | UL18 | 75509-76456 | 316 | 316 | 92 | Z48053 | EHV-1 | 314 | 60 | P28921 | Capsid protein | |
| BHV5-40 | UL17 | 77927-80047 | 707 | 701 | 90 | Z48053 | EHV-1 | 706 | 44 | P28950 | Tegument protein | |
| BHV5-41 | UL16 | 80077-81105 | 343 | 339 | 93 | Z48053 | EHV-1 | 370 | 47 | P28970 | Virion protein | |
| BHV5-42 | UL15 | 82277-76670 | 737 | 735 | 95 | Z48053 | EHV-1 | 734 | 63 | P28969 | DNA cleavage-packaging protein (terminase) | |
| BHV5-43 | UL14 | 82340-83011 | 224 | 222 | 97 | Z48053 | EHV-4 | 321 | 48 | AF030027 | Minor tegument protein | |
| BHV5-44 | UL13 | 82923-84407 | 495 | 492 | 90 | Z48053 | EHV-1 | 594 | 38 | P28966 | Virion serine/threonine protein kinase | |
| BHV5-45 | UL12 | 84407-85867 | 487 | 487 | 88 | Z48053 | PRV | 483 | 51 | X97257 | Alkaline exonuclease | |
| BHV5-46 | UL11 | 85822-86121 | 100 | 89 | 73 | Z48053 | EHV-4 | 75 | 48 | AF030027 | Myristylated protein (tegument) | |
| BHV5-47 | UL10 | 87526-86270 | 419 | 438 | 85 | AJ004801 | EHV-1 | 450 | 38 | P28948 | Glycoprotein M | |
| BHV5-48 | UL9 | 87647-90115 | 823 | 859 | 89 | Z48053 | EHV-4 | 887 | 57 | AF030027 | Ori-binding protein | |
| BHV5-49 | UL8 | 90216-92486 | 757 | 748 | 89 | AJ004801 | EHV-4 | 751 | 40 | AF030027 | Component of DNA helicase-primase complex | |
| BHV5-50 | UL7 | 93451-92549 | 301 | 299 | 88 | X91751 | EHV-1 | 303 | 40 | P28945 | Virion-associated protein | |
| BHV5-51 | UL6 | 95795-93336 | 820 | 688 | 84 | Z48053 | PRV | 643 | 62 | X97257 | Virion protein | |
| BHV5-52 | UL5 | 95551-98064 | 838 | 838 | 99 | Z48053 | EHV-1 | 861 | 64 | P28934 | Component of DNA helicase-primase complex | |
| BHV5-53 | UL4 | 98088-98651 | 188 | 185 | 86 | Z48053 | EHV-4 | 227 | 39 | AF030027 | Nuclear protein | |
| BHV5-54 | UL3.5 | 99127-98684 | 148 | 126 | 69 | U32173 | EHV-4 | 133 | 29 | AF030027 | Virion protein | |
| BHV5-55 | UL3 | 99786-99136 | 217 | 204 | 75 | U32173 | EHV-1 | 212 | 53 | P28942 | Phosphoprotein | |
| BHV5-56 | UL2 | 100729-99836 | 298 | 301 | 76 | AJ004801 | PRV | 339 | 58 | L13855 | Uracil-DNA glycosylase | |
| BHV5-57 | UL1 | 101201-100716 | 162 | 158 | 81 | AJ004801 | FHV-1 | 147 | 39 | AF022391 | Glycoprotein L | |
| BHV5-58 | UL0.7 | 101005-101607 | 201 | 97 | 41 | AJ004801 | HHV-6b | 1,520 | 31 | AF157706 | Unknown | |
| UL0.5 | Not present | 87 | AJ004801 | Unknown | ||||||||
| BHV5-59a | LRORF2 | 102159-102311 | 51e | 181 | 82 | M61143 | LR region | |||||
| BHV5-59b | LRORF1 | 102553-103632 | 360 | 336 | 66 | M61143 | LR region | |||||
| BHV5-60 | BICP0 | 104408-102249 | 720 | 676 | 70 | P29836 | PRV | 410 | 32 | A40505 | IE transactivator protein with Zn finger | |
| BHV5-61 | BICP4 | 109909-105686 | 1,406 | 1,343 | 75 | L14320 | EHV-1 | 1,487 | 42 | P17473 | Positive and negative gene regulator | |
| BHV5-62 | BICP22 | 114776-115717 | 314 | 300 | 68 | X76943 | EHV-1 | 278 | 40 | Z67986 | Transcription factor | |
| BHV5-63 | US1.67 | 117249-116509 | 247 | 243 | 79 | AJ004801 | EHV-1 | 272 | 35 | P28984 | Virion protein (EHV-1) | |
| BHV5-64 | US2 | 117987-117310 | 226 | 220 | 69 | AJ004801 | EHV-1 | 303 | 37 | P32517 | Tegument protein | |
| BHV5-65 | US3 | 118110-119441 | 444 | 468 | 79 | AJ004801 | EHV-4 | 384 | 42 | AF030027 | Virion serine/threonine protein kinase | |
| BHV5-66 | US4 | 119552-120871 | 440 | X99755 | 444 | 72 | AJ004801 | FHV-1 | 435 | 32 | S72415 | Glycoprotein G |
| BHV5-67 | US6 | 121129-122379 | 417 | U14656 | 417 | 98 | A25177 | CeHV-1 | 179 | 84 | AF078735 | Glycoprotein D |
| BHV5-68 | US7 | 122532-123692 | 387 | 382 | 78 | AJ004801 | EHV-1 | 424 | 30 | P18553 | Glycoprotein I | |
| BHV5-69 | US8 | 123984-125780 | 597 | AF208294 | 575 | 74 | AJ004801 | EHV-1 | 552 | 41 | P24380 | Glycoprotein E |
| BHV5-70 | US9 | 125875-126276 | 134 | AY064172 | 144 | 79 | Z23068 | PRV | 106 | 48 | U27487 | Virion protein (tegument) |
| BHV5-71 | BICP22 | 128237-127296 | 314 | 300 | 67 | X76943 | EHV-1 | 272 | 40 | Z67986 | Transcription factor | |
| BHV5-72 | BICP4 | 133104-137327 | 1,406 | 1,343 | 75 | L14320 | EHV-1 | 1,487 | 42 | P17473 | Positive and negative gene regulator | |
aa, amino acids; nt, nucleotides.
Accession numbers are from GenBank or SwissProtein.
Names for viruses: EHV-1, equine herpesvirus 1; PRV, pseudorabies virus; EHV-4, equine herpesvirus 4; CaHV-1, caprine herpesvirus 1; HSV-2, herpes simplex virus type 2; FHV-1, feline herpesvirus 1; HHV-6b, human herpesvirus 6B; CeHV-1, cervid herpesvirus 1.
Function was deduced from the degree of amino acid similarity to products of known genes.
The remaining homologous region is present but contains an in-frame stop and two in-frame frameshifts.
UL region.
The UL region, extending from nucleotide positions 570 to 104623, contains 60 putative genes. Starting from the left end of the genome, the first 58 genes are colinear with their BHV-1 counterparts and represent 73% of the BHV-5 genome. Similarly, the first 53 BHV-5 genes (with the exception of circ) are largely colinear with genes UL54 to UL4 of HSV type 1 (HSV-1). Predicted UL proteins average 84% amino acid identity to BHV-1 homologues, with the most similar (≥95% amino acid identity) involving viral DNA replication and processing (UL5, UL15, UL29, and UL39), tegument (UL14 and UL48), and capsid (UL19). Compared with nonbovine herpesviruses, all BHV-5 capsid proteins and six of eight proteins involved in viral DNA replication or processing are the most conserved (≥60%).
BHV-5 UL49, UL44, UL24, UL11, UL3.5, UL3, UL0.7, LR, and BICP0 are the least conserved UL genes (≤75%) relative to BHV-1 and are discussed below. Homologues of UL49 and UL3 in HSV-1 and of UL49 in BHV-1 are not essential for virus growth in cultured cells, suggesting a role for these genes in viral pathogenesis and host range (7, 59, 75).
BHV-5 UL44 encodes glycoprotein C (gC), which is not essential for neurovirulence in strain TX89; however, it affects neurotropism and is important for high levels of virus replication and full expression of virulence in the rabbit CNS (21). As alphaherpesvirus gC mediates primary attachment of virus to target cells via binding to surface glycans, variability in the heparin binding sites of BHV-5 gC (gC5) and BHV-1 gC (gC1) likely account for differences in their heparin-binding phenotypes (60). Although gC5 and gC1 are 75% identical, the amino-terminal third of the proteins (amino acids 1 to 102 and 1 to 123, respectively) differ significantly. Notably, there is a 35-amino-acid deletion in gC5 which removes two potential N-linked glycosylation sites present in gC1 (amino acids 93 and 111) (36) and a gC1-specific epitope (amino acids 103 to 122) (18). Comparison of gC sequences between strain SV507/99 (this report) and strain TX89 (18) reveals substantial differences at the amino-terminal third. However, a more detailed analysis shows that the proteins are 97% identical and that the discrepancy likely results from a missing base at position 480 in the published TX89 sequence.
The homologue of UL24 in HSV-1 is required for efficient replication in TG of mice (50). The homologue of UL11 in HSV-1 encodes a tegument protein with roles in virion envelopment and egress (7). BHV-5 and BHV-1 UL3.5 and UL0.7 are not found in other herpesviruses, and their functions remain unknown. BHV-5 lacks a homologue of BHV-1 UL0.5, which is predicted to encode an 87-amino-acid protein of unknown function.
The latency-related (LR) gene is postulated to contribute via alternative splicing to the LR protein(s) (27, 44), whereas BICP0 encodes a homologue of BHV-1 BICP0, an immediate-early (IE) and early (E) transactivator (95). Given the potential roles of LR and BICP0 genes in virulence and host range, these genes are treated in more detail below (see “IE genes” and “The LR region,” below).
US region.
The US region, extending from positions 116733 to 126280, contains eight genes (US1.67, 2 to 4, and 6 to 9), four of which have been previously sequenced (1, 20, 22, 31). BHV-5 US genes exhibit 69 to 98% amino acid identity (average, 79%) and overall less conservation to BHV-1 homologues than those within the UL region (84% average amino acid identity) (58). BHV-5 US2 to US9 are syntenic with homologues in HSV-1, with the exception of the US5 homologue, which is lacking in BHV-5.
The two BHV-5 genes located at the ends of the US are likely significant for virus-host interactions. The gene at the US-IR boundary, US1.67, contains the 75 carboxy-terminal amino acids within the IR and is homologous to ORFs found in other members of the α2 herpesvirus subgroup. Notably, the homologue of US1.67 in EHV-1 is a virulence determinant and is involved in egress of viral nucleocapsids (70, 71, 89). The gene at the US-TR boundary, US9, is essential for neurovirulence in the TX89 strain. Following intranasal inoculation of rabbits, a TX89 strain US9 deletion mutant failed to invade the CNS mainly due to an inability of the virus to spread to the olfactory bulb via anterograde transport (22).
BHV-5 US2, US4, and US8 are the least-conserved US ORFs (≤75%) in comparison with BHV-1. The US2 homologue in HSV-1 and HSV-2 seems dispensable in tissue culture and is not involved in HSV-2 neuropathogenesis in mice (52, 61). BHV-5 US4 encodes a glycosaminoglycan-associated protein (31) and is 72% identical to BHV-1 gG, which is involved in cell-cell virus transmission in vitro (69) and in prevention of apoptosis in certain cell lines (68). BHV-5 US8 encodes gE, which is important for neurovirulence in rabbits. Deletion of BHV-5 US8 or its substitution by BHV-1 gE resulted in viruses that replicated and spread much less efficiently in the rabbit brain than revertant or wild-type viruses (20).
Repeats.
The IR and TR regions, located at nucleotide positions 104624 to 116732 and 126281 to 138389, respectively, are 12,109 bp in length. Each repeat contains two genes, BICP4 and BICP22, which are 75 and 68% identical to BHV-1 BICP4 and BICP22, respectively (see “IE genes,” below). Among nonbovine herpesviruses, genes of EHV-1 are the most similar to BICP4 (42% amino acid identity) and BICP22 (40% amino acid identity). Interestingly, a homologue of the γ1 34.5 gene, a neurovirulence determinant of HSV-1, is not present in BHV-5 (79).
A 4,867-bp noncoding region separates BICP4 from BICP22 start codons. Overall, this region is 56% identical to the homologous BHV-1 intergenic region and contains transcriptional regulatory elements for the flanking IE genes and the origins of DNA replication.
IE genes.
Herpesvirus IE genes are critical regulators of viral gene expression. BHV-5 BICP0, BICP4, and BICP22, the homologues of BHV-1 IE genes BICP0, BICP4, and BICP22, are relatively less conserved than other classes of viral genes (Table 1).
In BHV-1, the BICP0 gene is transcribed at IE and E times postinfection from two separate promoters, leading to accumulation of 2.9- and 2.6-kb RNAs, respectively (94, 95). The IE promoter also controls BICP4 expression at IE times postinfection. A BICP0-specific transcript of similar size has been detected in BHV-5-infected cells at IE times (96). Several sequence elements involved in IE transcription and RNA processing of BHV-1 BICP0, including the TATA box (BHV-5 position 110799), major transcription start site (position 110766), donor and acceptor splice sites (positions 110417 and 104413, respectively), and polyadenylation signal (position 102150), are conserved in BHV-5. Further, potential TATA and SP1 sites are located at −39 and −71 relative to the first BICP0 codon, respectively, suggesting a similar promoter may direct E expression of BHV-5 BICP0 as in BHV-1 (95). Taken together, these features suggest that common mechanisms control BICP0 transcription in the two viruses.
BHV-5 BICP0 shares 70% amino acid identity with BHV-1 BICP0. Most amino acid differences are found at the carboxy half of the protein, a region which plays a role in subcellular localization of ICP0-like proteins (33, 47). BHV-5 BICP0 is 44 amino acids longer that its BHV-1 counterpart, including several small amino acid insertions and a 30-amino-acid extension at the carboxy terminus. This difference is likely responsible for the slightly larger size of IE transcripts originated from BHV-5 BICP0 relative to those from BHV-1 (96). Both BHV-5 and BHV-1 BICP0s are most similar in the amino half, which contains a conserved acidic cluster (amino acids 273 to 323 in BHV-5) and a C3HC4 zinc ring finger (amino acids 22 to 59). However, the region between amino acids 119 and 145 (110 to 155 in BHV-1) is poorly conserved and includes a 20-amino-acid deletion in BHV-5. In BHV-1, the BICP0 ring finger has been implicated in promoting transactivation, stimulation of productive infection, and cytotoxicity (47). It is not known if BHV-1 BICP0 contributes to virus pathogenesis; however, a point mutation in HSV-1 ICP0 significantly reduced neuroinvasiveness of an HSV-1 neurovirulent strain after peripheral inoculation of mice (90).
BHV-5 BICP4 encodes the homologue of BHV-1 BICP4, an IE transactivator and transrepressor (84). Based on sequence homologies, alphaherpesvirus ICP4 orthologs are divided into regions I to V from the amino to the carboxy terminus (13, 17, 43, 84). BHV-5 and BHV-1 ICP4 are most similar (90 to 100% amino acid identity) within regions II and IV. When compared with HSV-1, BHV-5 BICP4 regions II and IV are also the most similar (>50% amino acid identity). HSV-1 ICP4 region II is involved in homodimerization and DNA binding, whereas region IV is required for efficient transactivation (see reference 13 and references therein). BHV-5 BICP4 is 63 amino acids longer than BHV-1 BICP4, and this may be responsible for the slightly larger size of IE transcripts originated from BHV-5 BICP4 relative to their BHV-1 counterparts (96). The difference in length is partially due to a 47-amino-acid insertion in BHV-5 BICP4 region III, which includes a unique stretch of the dipeptide DG between two glutamic acid-rich clusters unique to bovine alphaherpesvirus ICP4s. BHV-5 BICP22 encodes the homologue of the BHV-1 IE and late transrepressor BICP22 (85). Most differences are observed in the carboxy halves of the proteins (48% amino acid identity versus 82% in the amino termini). This region contains 30 serines in BHV-5 and 16 in BHV-1 in a tract of 35 residues.
The LR region.
The BHV-1 LR region is the only transcriptionally active region in latently infected neurons and is hypothesized to play a role in latent infections (44, 76). In vitro, the BHV-1 LR gene inhibits cell cycle progression (83), BICP0-dependent activation of productive infection (39), and apoptosis (23). Within the LR gene there exist two major ORFs designated ORF1 and ORF2, where ORF1 is completely overlapped by BICP0 (56). Using an antibody against the amino terminus of ORF2, an LR protein was detected in lytically infected cells and in latently infected TG neurons (44, 51). Infection of cattle with mutant BHV-1 containing three stop codons that should prevent LR protein expression leads to a reduced ability of virus to replicate in the eye during acute infection (48), lower levels of viral DNA in latently infected TG, and a severe defect in virus reactivation after dexamethasone treatment of latently infected animals (49).
A complex pattern of alternative splicing in the LR transcript (LRT) has been described (27). LRT splicing varied according to the source of LR RNA (cell lines or TG), the time postinfection, and the type of RNA [poly(A)+ or poly(A)−] used for the experiments. The splicing pattern allows for different combinations between methionine-initiated ORFs and between methionine-initiated and non-methionine-initiated ORFs which may result in different protein isoforms.
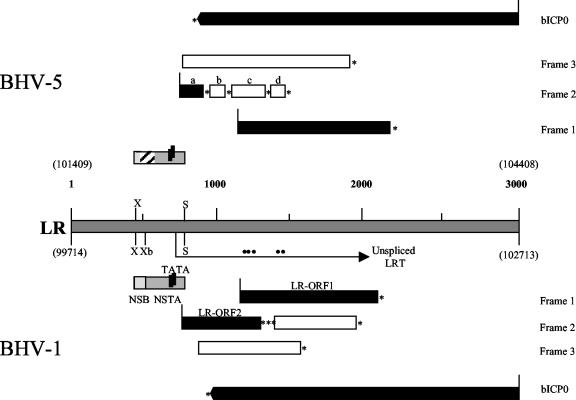
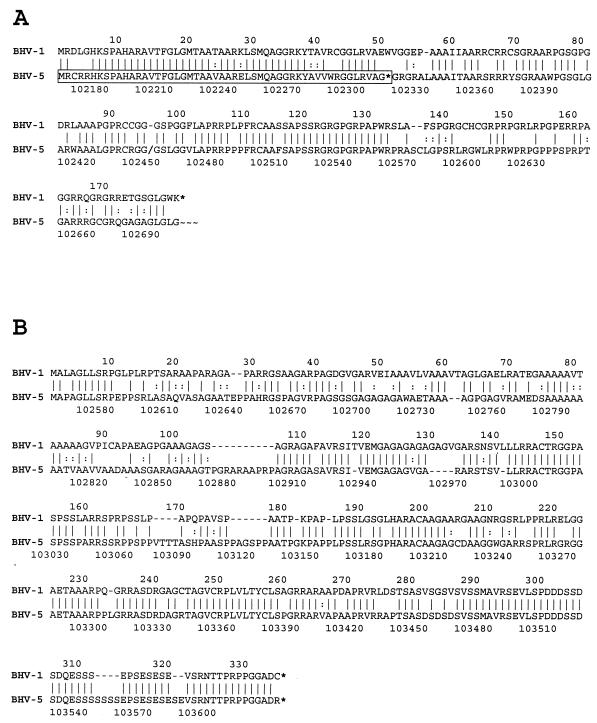
Figure 1 summarizes major differences between BHV-5 and BHV-1 in the LR region. Whereas both viruses contain ORF1 in frame 1 (66% amino acid identity), the structure of BHV-5 frame 2 departs from that of BHV-1. In BHV-1, frame 2 contains an ORF (LRORF2) which could encode a 181-amino-acid protein and a downstream reading frame without an initiating methionine. In BHV-5, frame 2 is interrupted four times, resulting in reading frames a to d (Fig. 1), with reading frame a being the only one to be methionine initiated. Reading frame a, which is truncated at 51 amino acids, and reading frame b are 82 and 75% identical to the corresponding regions in BHV-1 ORF2, respectively, while reading frames c and d are frameshifted. Interestingly, all splicing donor sites mapped in the BHV-1 LR transcript fall downstream of the carboxy terminus of frame b, suggesting that if BHV-5 proteins were made from this region they would differ substantially from BHV-1 LR products. A detailed comparison of LRORF2 and LRORF1 amino acid sequences between the two viruses is shown in Fig. 2A and B.
FIG. 1.
Comparison of the LR regions of BHV-5 and BHV-1. The LR nucleotide positions are given above (BHV-5) and below (BHV-1) the central stippled box, and the relative nucleotide positions are in bold (1 to 3,000 bp). Restriction sites are XhoI (X), XbaI (Xb), and SphI (S). The dots above the BHV-1 LRT represent the positions of splice donor signals (27). Methionine- and non-methionine-initiated ORFs are represented by black and open boxes, respectively. Asterisks indicate the positions of in-frame stop codons. BHV-1 neuron-specific transcriptional regulatory regions NSB and NSTA are represented by boxes between XhoI and SphI sites. The hatched box in the BHV-5 regulatory region represents deleted sequences. The structure of BHV-5 frames 1 and 2 is discussed in the text and shown in detail below in Fig. 2A and B.
FIG. 2.
(A) Alignment of BHV-1 LRORF2 with translated BHV-5 genomic sequence using tfastx, indicating the homologous BHV-5 ORF in frame 2 (boxed), stop codons (*), frameshift to BHV-5 frame 3 (/), conserved substitutions (:), identities (|), and remaining open sequence in BHV-5 frame 3 (∼∼∼). Total aligned sequence (182 amino acids) is 69% identical with BHV-1 LRORF2. (B) Alignment of BHV-1 LRORF1 with translated BHV-5 sequence using tfastx. Identities and substitutions are labeled as for panel A. Total aligned sequence (367 amino acids) is 66% identical with BHV-1 LRORF1. (C) Alignment of BHV-1 and BHV-5 LR regulatory sequences represented by the hatched box in Fig. 1, using FASTA. In BHV-1, the underlined sequence 1 was specifically protected from exonuclease digestion by ganglionic nuclear factors (25); underlined sequence 2 was protected from exonuclease digestion by neuroblastoma nuclear factors (11). Potential TATA and CAT boxes are shown in bold.
The promoter which drives BHV-1 LR transcription is contained near the 5′ terminus of the LR gene (53) and includes two neuron-specific regulatory regions designated the neuron-specific binding protein region and neuron-specific transcription activator (NSB and NSTA, respectively) (Fig. 1) (11, 25). While regions in the vicinity of the TATA box are conserved between BHV-5 and BHV-1, nucleotide sequences 5′ of NSTA are poorly conserved or lacking in BHV-5 (Fig. 2C). These sequences were shown to be important for efficient enhancer activity of the BHV-1 LR promoter in neuroblastoma cells and for binding of nuclear factors (11). The NSB region is 72 bp in length, specifically binds bovine ganglionic nuclear factors, and is required for maximal BHV-1 LR promoter activity in neuroblastoma cells (25). Almost 70% of the NSB region is deleted in BHV-5, including a 20-bp region responsible for binding to neuron-specific factors (25) (Fig. 2C) and a latency-specific transcription start site (44).
In summary, the BHV-5 LR region differs substantially from the BHV-1 LR region in both coding and transcriptionally regulatory regions. Given the potential significance of this region in viral latency and/or host range and the differential pathogenesis of these closely related viruses, it is likely that these differences are of biological significance for aspects of virus-neuron-host interactions.
Conclusions.
Although BHV-5 and BHV-1 are closely related viruses, only BHV-5 causes CNS disease. The molecular basis for the differential pathogenesis remains unknown but probably arises from multiple genetic contributions. The complete BHV-5 genome provides a framework from which comparisons between BHV-5 and BHV-1 pathogenesis may be made. Strategies based on the construction of chimeric viruses will certainly contribute to our overall understanding of pathogen-host interactions and the evolution of herpesvirus virulence.
REFERENCES
- 1.Abdelmagid, O. Y., H. C. Minocha, J. K. Collins, and S. I. Chowdhury. 1995. Fine mapping of bovine herpesvirus-1 (BHV-1) glycoprotein D (gD) neutralizing epitopes by type-specific monoclonal antibodies and sequence comparison with BHV-5 gD. Virology 206:242-253. [DOI] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2000. The genome of fowlpox virus. J. Virol. 74:3815-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashbaugh, S. E., K. E. Thompson, E. B. Belknap, P. C. Schultheiss, S. Chowdhury, and J. K. Collins. 1997. Specific detection of shedding and latency of bovine herpesvirus 1 and 5 using a nested polymerase chain reaction. J. Vet. Diagn. Investig. 9:387-394. [DOI] [PubMed] [Google Scholar]
- 6.Bagust, T. J., and L. Clark. 1972. Pathogenesis of meningo-encephalitis produced in calves by infectious bovine rhinotracheitis herpesvirus. J. Comp. Pathol. 82:375-383. [DOI] [PubMed] [Google Scholar]
- 7.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barenfus, M., C. A. Delli Quadri, R. W. McIntyre, and R. J. Schroeder. 1963. Isolation of infectious bovine rhinotracheitis virus from calves with meningoencephalitis. J. Am. Vet. Med. Assoc. 143:725-728. [PubMed] [Google Scholar]
- 9.Bartha, A., G. Haidu, P. Aldassy, and G. Paczolay. 1969. Occurrence of encephalomyelitis caused by infectious bovine rhinotracheitis virus in calves in Hungary. Acta Vet. Acad. Sci. Hungaricae 19:145-151. [PubMed] [Google Scholar]
- 10.Belknap, E. B., J. K. Collins, V. K. Ayers, and P. C. Schultheiss. 1994. Experimental infection of neonatal calves with neurovirulent bovine herpesvirus type 1.3. Vet. Pathol. 31:358-365. [DOI] [PubMed] [Google Scholar]
- 11.Bratanich, A. C., and C. J. Jones. 1992. Localization of cis-acting sequences in the latency-related promoter of bovine herpesvirus 1 which are regulated by neuronal cell type factors and immediate-early genes. J. Virol. 66:6099-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brendel, V., P. Bucher, I. R. Nourbakhsh, B. E. Blaisdell, and S. Karlin. 1992. Methods and algorithms for statistical analysis of protein sequences. Proc. Natl. Acad. Sci. USA 89:2002-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruce, J. W., and K. W. Wilcox. 2002. Identification of a motif in the C terminus of herpes simplex virus regulatory protein ICP4 that contributes to activation of transcription. J. Virol. 76:195-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burks, C. 1999. Molecular Biology Database list. Nucleic Acids Res. 27:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrillo, B. J., A. Ambrosi, A. A. Schudel, M. Vazquez, E. Dahme, and A. Pospischil. 1983. Meningoencephalitis caused by IBR virus in calves in Argentina. Zentbl. Vet. Med. B 30:327-332. [DOI] [PubMed] [Google Scholar]
- 16.Cascio, K. E., E. B. Belknap, P. C. Schultheiss, A. D. Ames, and J. K. Collins. 1999. Encephalitis induced by bovine herpesvirus 5 and protection by prior vaccination or infection with bovine herpesvirus 1. J. Vet. Diagn. Investig. 11:134-139. [DOI] [PubMed] [Google Scholar]
- 17.Cheung, A. K. 1989. DNA nucleotide sequence analysis of the immediate-early gene of pseudorabies virus. Nucleic Acids Res. 17:4637-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chowdhury, S. I. 1995. Molecular basis of antigenic variation between the glycoproteins C of respiratory bovine herpesvirus 1 (BHV-1) and neurovirulent BHV-5. Virology 213:558-568. [DOI] [PubMed] [Google Scholar]
- 19.Chowdhury, S. I., B. J. Lee, D. Mosier, J.-H. Sur, F. A. Osorio, G. Kennedy, and M. L. Weiss. 1997. Neuropathology of bovine herpesvirus type 5 (BHV-5) meningo-encephalitis in a rabbit seizure model. J. Comp. Pathol. 117:295-310. [DOI] [PubMed] [Google Scholar]
- 20.Chowdhury, S. I., B. J. Lee, A. Ozkul, and M. L. Weiss. 2000. Bovine herpesvirus 5 glycoprotein E is important for neuroinvasiveness and neurovirulence in the olfactory pathway of the rabbit. J. Virol. 74:2094-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chowdhury, S. I., B. J. Lee, M. Onderci, M. L. Weiss, and D. Mosier. 2000. Neurovirulence of glycoprotein C (gC)-deleted bovine herpesvirus type-5 (BHV-5) and BHV-5 expressing BHV-1 gC in a rabbit seizure model. J. Neurovirol. 6:284-295. [DOI] [PubMed] [Google Scholar]
- 22.Chowdhury, S. I., M. Onderci, P. S. Bhattacharjee, A. Al-Mubarak, M. L. Weiss, and Y. Zhou. 2002. Bovine herpesvirus 5 (BHV-5) Us9 is essential for BHV-5 neuropathogenesis. J. Virol. 76:3839-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciacci-Zanella, J., M. Stone, G. Henderson, and C. Jones. 1999. The latency-related gene of bovine herpesvirus 1 inhibits programmed cell death. J. Virol. 73:9734-9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins, J. K., V. K. Ayers, C. A. Whetstone, and S. van Drunen Little-van den Hurk. 1993. Antigenic differences between the major glycoproteins of bovine herpesvirus type 1.1 and bovine encephalitis herpesvirus type 1.3. J. Gen. Virol. 74:509-1517. [DOI] [PubMed] [Google Scholar]
- 25.Delhon, G., and C. Jones. 1997. Identification of DNA sequences in the latency related promoter of bovine herpes virus type 1 which are bound by neuronal specific factors. Virus Res. 51:93-103. [DOI] [PubMed] [Google Scholar]
- 26.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devireddy, L. R., and C. Jones. 1998. Alternative splicing of the latency-related transcript of bovine herpesvirus 1 yields RNAs containing unique open reading frames. J. Virol. 72:7294-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.d'Offay, J. M., R. E. Mock, and R. W. Fulton. 1993. Isolation and characterization of encephalitic bovine herpesvirus type 1 isolates from cattle in North America. Am. J. Vet. Res. 54:534-539. [PubMed] [Google Scholar]
- 29.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Down, T. A., and T. J. P. Hubbard. 2002. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 12:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engelhardt, T., and G. M. Keil. 1996. Identification and characterization of the bovine herpesvirus 5 US4 gene and gene products. Virology 225:126-135. [DOI] [PubMed] [Google Scholar]
- 32.Engels, M., C. Giuliani, P. Wild, T. M. Beck, E. Loepfe, and R. Wyler. 1986. The genome of bovine herpesvirus 1 (BHV-1) strains exhibiting a neuropathogenic potential compared to known BHV-1 strains by restriction site mapping and cross-hybridization. Virus Res. 6:57-73. [DOI] [PubMed] [Google Scholar]
- 33.Everett, R. D. 1988. Analysis of the functional domains of herpes simplex virus type 1 immediate early polypeptide Vmw110. J. Mol. Biol. 202:87-96. [DOI] [PubMed] [Google Scholar]
- 34.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 35.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 36.Fitzpatrick, D. R., L. A. Babiuk, and T. J. Zamb. 1989. Nucleotide sequence of bovine herpesvirus type 1 glycoprotein gIII, a structural model for gIII as a new member of the immunoglobulin superfamily, and implications for the homologous glycoproteins of other herpesviruses. Virology 173:46-57. [DOI] [PubMed] [Google Scholar]
- 37.French, E. L. 1962. A specific virus encephalitis in calves: isolation and characterization of the causal agent. Aust. Vet. J. 38:216-221. [Google Scholar]
- 38.French, E. L. 1962. Relationship between infectious rhinotracheitis (IBR) virus and a virus isolated from calves with encephalitis. Aust. Vet. J. 38:555-556. [Google Scholar]
- 39.Geiser, V., M. Inman, Y. Zhang, and C. Jones. 2002. The latency-related gene of bovine herpesvirus-1 can inhibit the ability of bICP0 to activate productive infection. J. Gen. Virol. 83:2965-2971. [DOI] [PubMed] [Google Scholar]
- 40.Gillette, K., V. Misra, and A. Bratanich. 2002. Sequence analysis of the alpha trans-inducing factor of bovine herpesvirus type 5 (BHV-5). Virus Genes 24:149-152. [DOI] [PubMed] [Google Scholar]
- 41.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:192-202. [DOI] [PubMed] [Google Scholar]
- 42.Gough, A., and D. James. 1975. Isolation of IBR virus from heifer with meningo-encephalitis in calves. Can. Vet. J. 16:313-314. [PMC free article] [PubMed] [Google Scholar]
- 43.Grundy, F. J., R. P. Baumann, and D. J. O'Callaghan. 1989. DNA sequence and comparative analysis of the equine herpesvirus type 1 immediate early gene. Virology 172:223-236. [DOI] [PubMed] [Google Scholar]
- 44.Hossain, A., L. M. Schang, and C. Jones. 1995. Identification of gene products encoded by the latency-related gene of bovine herpesvirus 1. J. Virol. 69:5345-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang, X., and A. Madam. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Husak, P. J., T. Kuo, and L. W. Enquist. 2000. Pseudorabies virus membrane proteins gI and gE facilitate anterograde spread of infection in projection-specific neurons in the rat. J. Virol. 74:10975-10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inman, M., Y. Zhang, V. Geiser, and C. Jones. 2001. The zinc finger in the bICP0 protein encoded by bovine herpesvirus-1 mediates toxicity and activates productive infection. J. Gen. Virol. 82:483-492. [DOI] [PubMed] [Google Scholar]
- 48.Inman, M., L. Lovato, A. Doster, and C. Jones. 2001. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J. Virol. 75:8507-8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inman, M., L. Lovato, A. Doster, and C. Jones. 2002. A mutation in the latency-related gene of bovine herpesvirus 1 disrupts the latency-reactivation cycle in calves. J. Virol. 76:6771-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacobson, J. G., S. H. Chen, W. J. Cook, M. F. Kramer, and D. M. Coen. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161-169. [DOI] [PubMed] [Google Scholar]
- 51.Jiang, Y., A. Hossain, M. T. Winkler, T. Holt, A. Doster, and C. Jones. 1998. A protein encoded by the latency-related gene of bovine herpesvirus 1 is expressed in trigeminal ganglionic neurons of latently infected cattle and interacts with cyclin-dependent kinase 2 during productive infection. J. Virol. 72:8133-8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang, Y. M., H. Yamada, F. Goshima, T. Daikoku, S. Oshima, K. Wada, and Y. Nishiyama. 1998. Characterization of the herpes simplex virus type 2 (HSV-2) US2 gene product and a US2-deficient HSV-2 mutant. J. Gen. Virol. 79:2777-2784. [DOI] [PubMed] [Google Scholar]
- 53.Jones, C., G. Delhon, A. Bratanich, G. Kutish, and D. Rock. 1990. Analysis of the transcriptional promoter which regulates the latency-related transcript of bovine herpesvirus 1. J. Virol. 64:1164-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038-3049. [DOI] [PubMed] [Google Scholar]
- 55.Krogh, A. 1997. Two methods for improving performance of an HMM and their application for gene finding, p. 179-186. In T. Gaasterland et al. (ed.), Proceedings of the Fifth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 56.Kutish, G., T. Mainprize, and D. L. Rock. 1990. Characterization of the latency-related transcriptionally active region of the bovine herpesvirus 1 genome. J. Virol. 64:5730-5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee, B. J., M. L. Weiss, D. Mosier, and S. I. Chowdhury. 1999. Spread of bovine herpesvirus type 5 (BHV-5) in the rabbit brain after intranasal inoculation. J. Neurovirol. 5:474-484. [DOI] [PubMed] [Google Scholar]
- 58.Leung-Tack, P., J.-C. Audonnet, and M. Riviere. 1994. The complete DNA sequence and the genetic organization of the short unique region (Us) of the bovine herpesvirus type 1 (ST strain). Virology 199:409-421. [DOI] [PubMed] [Google Scholar]
- 59.Liang, X., B. Chow, Y. Li, C. Raggo, D. Yoo, S. Attah-Poku, and L. A. Babiuk. 1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J. Virol. 69:3863-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liman, A., M. Engels, G. Meyer, and M. Ackermann. 2000. Glycoprotein C of bovine herpesvirus 5 (BHV-5) confers a distinct heparin-binding phenotype to BHV-1. Arch. Virol. 145:2047-2059. [DOI] [PubMed] [Google Scholar]
- 61.Longnecker, R., and B. Roizman. 1987. Clustering of genes dispensable for growth in culture in the S component of the HSV-1 genome. Science 236:573-576. [DOI] [PubMed] [Google Scholar]
- 62.McGeoch, D. J., and A. J. Davison. 1999. The molecular evolutionary history of the herpesviruses, p. 441. In E. Domingo, R. Webster, and J. Holland (ed.), Origin and evolution of viruses. Academic Press, London, England.
- 63.Metzler, A. E., A. A. Schudel, and M. Engels. 1986. Bovine herpesvirus 1: molecular and antigenic characteristics of variant viruses isolated from calves with neurological disease. Arch. Virol. 87:205-217. [DOI] [PubMed] [Google Scholar]
- 64.Meyer, G., O. Bare, and E. Thiry. 1999. Identification and characterization of bovine herpesvirus type 5 glycoprotein H and gene products. J. Gen. Virol. 80:2849-2859. [DOI] [PubMed] [Google Scholar]
- 65.Meyer, G., M. Lemaire, C. Ros, K. Belak, A. Gabriel, D. Cassart, F. Coignoul, S. Belak, and E. Thiry. 2001. Comparative pathogenesis of acute and latent infections of calves with bovine herpesvirus types 1 and 5. Arch. Virol. 146:633-652. [DOI] [PubMed] [Google Scholar]
- 66.Moretti, B., Z. Orfei, G. Mondino, and A. Perschino. 1964. Infectious bovine rhinotracheitis clinical observations and isolation of virus. Vet. Italiana 15:676. [PubMed] [Google Scholar]
- 67.Mulder, W. A. M., L. Jacobs, J. Priem, G. L. Kok, F. Wagenaar, T. G. Kimman, and J. M. A. Pol. 1994. Glycoprotein gE-negative pseudorabies virus has a reduced capability to infect second- and third-order neurons of the olfactory and the trigeminal routes in the porcine central nervous system. J. Gen. Virol. 75:3095-3106. [DOI] [PubMed] [Google Scholar]
- 68.Nakamichi, K., D. Kuroki, Y. Matsumoto, and H. Otsuka. 2001. Bovine herpesvirus 1 glycoprotein G is required for prevention of apoptosis and efficient viral growth in rabbit kidney cells. Virology 279:488-498. [DOI] [PubMed] [Google Scholar]
- 69.Nakamichi, K., Y. Matsumoto, and H. Otsuka. 2002. Bovine herpesvirus 1 glycoprotein G is necessary to maintain cell-to-cell junctional adherence among infected cells. Virology 294:22-30. [DOI] [PubMed] [Google Scholar]
- 70.Osterrieder, N., A. Neubauer, C. Brandmüller, O.-R. Kaaden, and D. J. O'Callaghan. 1996. The equine herpesvirus 1 IR6 protein influences virus growth at elevated temperature and is a major determinant of virulence. Virology 226:243-251. [DOI] [PubMed] [Google Scholar]
- 71.Osterrieder, N., A. Neubauer, C. Brandmüller, O.-R. Kaaden, and D. J. O'Callaghan. 1998. The equine herpesvirus 1 IR6 protein that colocalizes with nuclear lamins is involved in nucleocapsid egress and migrates from cell to cell independently of virus infection. J. Virol. 72:9806-9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 73.Perez, S. E., G. Bretschneider, M. R. Leunda, E. A. Osorio, E. F. Flores, and A. C. Odeon. 2002. Primary infection, latency, and reactivation of bovine herpesvirus type 5 in the bovine nervous system. Vet. Pathol. 39:437-444. [DOI] [PubMed] [Google Scholar]
- 74.Pertea, M., X. Lin, and S. L. Salzberg. 2001. A new computational method for splice site prediction. Nucleic Acids Res. 29:1185-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pomeranz, L. E., and J. A. Blaho. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J. Virol. 73:6769-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rock, D. L., S. L. Beam, and J. E. Mayfield. 1987. Mapping bovine herpesvirus type 1 latency-related RNA in trigeminal ganglia of latently infected rabbits by in situ hybridization. J. Virol. 61:3827-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roehe, P. M., T. C. Silva, N. B. Nardi, D. G. Oliveira, and J. C. A. Rosa. 1997. Diferenciaco entre o vírus da rinotraqueite infecciosa bovina (BHV-1) e o herpesvirus da encefalite bovina (BHV-5). Pesq. Vet. Bras. 17:41-44. [Google Scholar]
- 78.Roizman, B., R. C. Desrosiers, B. Fleckenstein, C. Lopez, A. C. Minson, and M. J. Studdert. 1992. The family Herpesviridae: an update. Arch. Virol. 123:425-449. [DOI] [PubMed] [Google Scholar]
- 79.Roizman, B., and N. Markovitz. 1997. Herpes simplex virus virulence: the functions of the γ1 34.5 gene. J. Neurovirol. 3(Suppl. 1):S1-S2. [PubMed] [Google Scholar]
- 80.Ros, C., and S. Belák. 2002. Characterization of the glycoprotein B gene from ruminant alphaherpesviruses. Virus Genes 24:99-105. [DOI] [PubMed] [Google Scholar]
- 81.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schang, L. M., A. Hossain, and C. Jones. 1996. The latency-related gene of bovine herpesvirus 1 encodes a product which inhibits cell cycle progression. J. Virol. 70:3807-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schwyzer, M., C. Vlcek, O. Menekse, C. Fraefel, and V. Paces. 1993. Promoter, spliced leader, and coding sequence for BICP4, the largest of the immediate-early proteins of bovine herpesvirus 1. Virology 197:349-357. [DOI] [PubMed] [Google Scholar]
- 85.Schwyzer, M., U. V. Wirth, B. Vogt, and C. Fraefel. 1994. BICP22 of bovine herpesvirus 1 is encoded by a spliced 1.7 kb RNA which exhibits immediate early and late transcription kinetics. J. Gen. Virol. 75:1703-1711. [DOI] [PubMed] [Google Scholar]
- 86.Smith, G. A., P. L. Young, and J. S. Mattick. 1991. Nucleotide and amino acid sequence analysis of the thymidine kinase gene of a bovine encephalitis herpesvirus. Arch. Virol. 119:199-210. [DOI] [PubMed] [Google Scholar]
- 87.Sonnhammer, E. L. L., S. R. Eddy, E. Birney, A. Bateman, and R. Durbin. 1998. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 26:320-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Staden, A., and A. D. McLachlan. 1982. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 10:141-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Telford, E. A. R., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 90.Van Sant, C., Y. Kawaguchi, and B. Roizman. 1999. A single amino acid substitution in the cyclin D binding domain of the infected cell protein 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc. Natl. Acad. Sci. USA 96:8184-8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vogel, F. S. F., I. Oldoni, E. F. Flores, R. Weiblen, S. V. Mayer, and N. Winkelmann. 2002. Investigation of neural and non-neural sites of latency by bovine herpesvirus type 5 in experimentally infected calves. Virus Rev. Res. 7:63. [Google Scholar]
- 92.Wallace, J. C., and S. Henikoff. 1991. PATMAT: a searching and extraction program for sequence, pattern, and block queries and databases. Comput. Appl. Biol. Sci. 8:249-254. [DOI] [PubMed] [Google Scholar]
- 93.Whetstone, C. A., B. S. Seal, and J. M. Miller. 1993. Variability occurs in the inverted repeat region of genomic DNA from bovine herpesvirus 1 respiratory, genital and bovine herpesvirus 5 encephalitic isolates. Vet. Microbiol. 38:181-189. [DOI] [PubMed] [Google Scholar]
- 94.Wirth, U. V., B. Vogt, and M. Schwyzer. 1991. The three major immediate-early transcripts of bovine herpesvirus 1 arise from two divergent and spliced transcription units. J. Virol. 65:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wirth, U. V., C. Fraefel, B. Vogt, C. Vlcek, V. Paces, and M. Schwyzer. 1992. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivation protein. J. Virol. 66:2763-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wirth, U. V. 1993. Comparison of immediate-early transcripts among bovine herpesvirus type 1 and type 5 strains differing in neurovirulent potential. Virus Res. 27:1-12. [DOI] [PubMed] [Google Scholar]