Abstract
Recombination is a well-known phenomenon for enteroviruses. However, the actual extent of recombination in circulating nonpoliovirus enteroviruses is not known. We have analyzed the phylogenetic relationships in four genome regions, VP1, 2A, 3D, and the 5′ nontranslated region (NTR), of 40 enterovirus B strains (coxsackie B viruses and echoviruses) representing 11 serotypes and isolated in 1981 to 2002 in the former Soviet Union states. In the VP1 region, strains of the same serotype expectedly grouped with their prototype strain. However, as early as the 2A region, phylogenetic grouping differed significantly from that in the VP1 region and indicated recombination within the 2A region. Moreover, in the 5′ NTR and 3D region, only 1 strain of 40 grouped with its prototype strain. Instead, we observed a major group in both the 5′ NTR and the 3D region that united most (in the 5′ NTR) or all (in the 3D region) of the strains studied, regardless of the serotype. Subdivision within that major group in the 3D region correlated with the time of virus isolation but not with the serotype. Therefore, we conclude that a majority, if not all, circulating enterovirus B strains are recombinants relative to the prototype strains, isolated mostly in the 1950s. Moreover, the ubiquitous recombination has allowed different regions of the enterovirus genome to evolve independently. Thus, a novel model of enterovirus genetics is proposed: the enterovirus genome is a stable symbiosis of genes, and enterovirus species consist of a finite set of capsid genes responsible for different serotypes and a continuum of nonstructural protein genes that seem to evolve in a relatively independent manner.
Human enteroviruses, members of the Picornaviridae family, comprise about 70 serotypes, which were only very recently divided into species, on the basis of their molecular, serological, and pathogenetic characteristics. Enterovirus B species, which were the object of this study, include 28 serotypes of echoviruses, 6 serotypes of coxsackie B viruses, and several other viruses (http://www.ncbi.nlm.nih.gov/Taxonomy/). These viruses are important human pathogens, responsible for a wide range of clinical manifestations, from asymptomatic or very mild infections to serious disorders, such as meningitis, myocarditis, encephalitis, and hemorrhagic disease of newborns (34).
An important property of enteroviruses is their capability for extensive recombination. Recombination in poliovirus was discovered as early as 1962 (18, 25) and has been extensively studied since then. Recombination presumably occurs by a copy choice mechanism, in which the viral polymerase switches from one template RNA molecule to another in the course of minus-strand RNA synthesis (22). Recombination is known to occur readily between the three strains of live poliovirus vaccine upon oral administration (4, 29, 31) and between vaccine and/or wild poliovirus strains (10, 13-15, 17, 27, 29). Up to 79% of poliovirus strains secreted after vaccination were found to be recombinants (8). Recombination was most often reported to occur in the nonstructural protein (NSP) genome region and, less frequently, in the 5′ nontranslated region (NTR)-VP4 junction region. Only a few reports indicated recombination in the structural protein region (2, 11). Much less attention has been devoted to recombination in the nonpoliovirus enteroviruses (NPEVs). Several completely sequenced prototype NPEVs (CAV21, EV70, SVDV, and EV18) have been found to be recombinants, in comparison to other prototype enterovirus strains (1, 19, 35, 39). In addition, evidence of recombination in the phylogenetic history of the entire enterovirus genus has led to the formation of the four enterovirus species (37). The molecular epidemiology of coxsackievirus A9 also suggests recombination in the NSP genome region in circulating strains (36). In contrast, a recent EV7 isolate was found to be a direct descendant of the prototype EV7 strain (7). Recombination was recently demonstrated between circulating heterologous echoviruses (EV4, EV7, and EV30) during an extensive meningitis outbreak (33). However, little is known about the actual prevalence of recombination among circulating enteroviruses, and no attempts have been made to consider simultaneously multiple enterovirus serotypes in similar studies.
In this work, we have analyzed four distinct genome regions (Fig. 1) of 40 enterovirus B isolates representing 10 serotypes in order to elucidate the occurrence of recombination in all of the enterovirus B species.
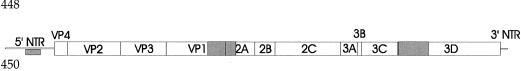
FIG. 1.
Schematic representation of the enterovirus genome. Genome regions studied are indicated by grey boxes.
MATERIALS AND METHODS
The enterovirus strains used in this work (Table 1) were isolated from clinical (stool samples) and environmental (sewage) specimens collected in accordance with the WHO Polio Eradication Initiative in the European Region and in some countries of the former Soviet Union (Table 1). Some of the strains were obtained for identification from the National Laboratories for Polio Investigation in the countries of the former Soviet Union. Two cell culture lines were used for isolation—RD (human rhabdomyosarcoma) and HEp-2 (human epidermoid carcinoma). Most strains underwent two to four passages before being used for this study. Serotypes of isolates were determined by the neutralization test with RIVM (Bilthoven, The Netherlands) antiserum pools. The procedures for isolation and identification were implemented according to standard protocols (WHO Manual for the Virological Investigation of Polio; World Health Organization, Geneva, Switzerland). Throughout the text, the names of strains are given in the following format: serotype_strain name_year of isolation.
TABLE 1.
Strains used in this work
| Strain | Serotypea | Country of sampling | Date of sampling (day.mo.yr) | Sourceb |
|---|---|---|---|---|
| 08477 | EV30 | Russia | 08.98c | Human, meningitis |
| 09025 | EV11 | Russia | 14.07.98 | Sewage |
| 09039 | EV11 | Russia | 18.10.98 | Human, facial nerve paresis |
| 10887 | EV6 | Russia | 24.09.99 | Human, facial nerve paresis |
| 11059 | CBV (3) | Uzbekistan | 29.09.99 | Human, AFP |
| 11202 | EV30 | Ukraine | 02.10.99 | Human, meningoencephalitis |
| 11267 | EV12 | Georgia | 19.11.99 | Human, healthy |
| 11292 | EV11 | Armenia | 22.09.99 | Human, healthy |
| 11665 | EV12 | Russia | 2000 | Human, healthy |
| 12167 | EV30 | Ukraine | 18.10.99 | Human, healthy |
| 12647 | EV13 | Tajikistan | 28.04.00 | Human, AFP |
| 13162 | EV30 | Georgia | 13.07.00 | Human |
| 13991 | EV2 | Kyrgyzstan | 24.11.00 | Human, AFP |
| 13998 | EV11 | Kyrgyzstan | 25.11.00 | Human, AFP |
| 14103 | EV6 | Russia | 26.12.00 | Human, healthy |
| 14125 | EV30 | Ukraine | 30.11.00 | Human, healthy |
| 15529 | CBV (5) | Ukraine | 13.07.01 | Human, AFP |
| 15819 | EV9 | Russia | 10.09.01 | Human, AFP |
| 15936 | EV7 | Azerbaijan | 16.09.01 | Sewage |
| 16228 | EV7 | Russia | 2001 | Sewage |
| 16272 | EV7 | Georgia | 13.09.01 | Sewage |
| 16274 | EV12 | Georgia | 13.09.01 | Sewage |
| 17763 | EV20 | Uzbekistan | 12.05.02 | Human, AFP |
| 17891 | EV30 | Russia | 02.06.02 | Human, healthy |
| 18214 | CBV (3) | Moldavia | 07.11.02 | Sewage |
| 18219 | CBV (3) | Moldavia | 18.06.02 | Human, meningitis |
| 18311 | EV6 | Kyrgyzstan | 09.09.02 | Human, AFP |
| 18515 | CBV (5) | Russia | 01.08.02 | Human, healthy |
| 18594 | CBV (3) | Kyrgyzstan | 10.07.02 | Human, healthy |
| 18706 | EV6 | Georgia | 01.08.02 | Sewage |
| 18711 | EV11 | Russia | 02.07.02 | Human, healthy |
| 18733 | EV30 | Moldavia | 16.07.02 | Human, meningitis |
| 18744 | EV11 | Moldavia | 11.07.02 | Sewage |
| K452_81d | EV19 | Russia | 12.04.81 | Human, uveitis |
| Kar_87d | EV11 | Russia | 14.01.88 | Human, uveitis |
| Kh1_94d | EV19 | Russia | 16.08.94 | Human, gastroenteritis |
| Kh3_97d | EV11 | Russia | 20.06.97 | Human, gastroenteritis |
| Kh6_80d | EV11 | Russia | 14.08.80 | Human, meningitis |
| Kust_86d | EV11 | Russia | 14.12.86 | Human, uveitis |
| MorM_82d | EV11 | Russia | 21.04.82 | Human, uveitis |
Numbers in parentheses indicate the exact serotype identified by using the VP1 sequence for CBV strains serotyped only by using group-specific antiserum.
AFP, acute flaccid paralysis.
Month. year.
Strains sequenced and studied previously (28; Lukashev et al., submitted).
RNA was isolated from infected cell cultures by guanidine thiocyanate lysis and absorption on silica sorbent (3). Reverse transcription was carried out with Moloney murine leukemia virus reverse transcriptase and random hexamer primers. The genome fragments amplified by PCR were as follows: a segment of the 5′ NTR (nucleotides [nt] 313 to 641), the VP1-2A junction region (nt 2957 to 3587), and part of the 3D polymerase gene (nt 5837 to 6506). All nucleotide positions throughout the text are given relative to those of the EV11 strain Gregory genome (9). Primers used to amplify and sequence these regions are listed in Table 2. After amplification, the DNA was run in an agarose gel, and the PCR product was extracted with a QIAquick kit (Qiagen) and sequenced directly with PCR primers. Each fragment was sequenced in two directions with a MegaBACE DNA sequencer and a DYEnamic ET terminator kit (Amersham).
TABLE 2.
Oligonucleotides used in this work
| Name | Sequence | Nucleotide positiona |
|---|---|---|
| 0340F | TAGATCAGGCYGATGAGTCACCGC | 313-336 |
| 0640R | GATGGCCAATCCAATAGCTATATG | 641-618 |
| 3000F | AACCCYAGYATTTTYTGGACIGARGGGAACGC | 2957-2988 |
| 3600R | GACTGGTAYCTYTTIGGGTARTAYTCRCTCTCCTG | 3621-3587 |
| 5850F | CAGTGYGGIGGIGTICTCATGTC | 5837-5859 |
| 6500R | AGRTTGCCAAAYGTYTGYCTCATTGC | 6531-6506 |
All nucleotide positions are given relative to those of the EV11 strain Gregory genome (9).
In several ambiguous cases that might suggest contamination, sequencing experiments were performed twice, starting from a new PCR. In all such cases, the results were reproduced, although in a few cases, there were minor sequence deviations not exceeding 0.25% of the nucleotide sequence. Such sequence variations can be readily explained by enzyme errors and the heterogeneity of the virus population. Most sequences, however, were not additionally verified in a separate PCR sequencing experiment, as the sequences of unrelated strains in these cases were different enough to exclude cross-contamination. The use of high-titer virus stocks (106 to 108 tissue culture infective doses/ml) additionally reduced the probability of contamination at different stages of sequencing.
The resulting DNA sequences (GenBank accession numbers AY271379 to AY271512) were aligned with the corresponding regions of the available full enterovirus genome sequences: EV1 strain Farouk (AF029859), EV5 strain Noyce (AF083069) (26), EV6 strain Charles (U16283) (16), EV7 strain Wallace (AY036579), EV9 strain Barty (X92886) (40), EV11 strain Gregory (X80059) (9), EV18 strain Metcalf (AF317694) (1), EV30 strain Bastianni (AF311938), CBV1 (M16560) (20), CBV2 strain Ohio (AF081485), CBV3 (M33854) (23), CBV4 strain Benschoten (X05690) (21), CBV5 (X67706) (39), CBV6 strain Schmitt (AF105342) (30), and CAV9 strain Griggs (D00627) (6). Additionally, we used the corresponding sequence fragments of seven echovirus strains that had been sequenced previously (28; A. N. Lukashev, V. A. Lashkevich, G. A. Koroleva, J. Ilonen, and A. E. Hinkkanen, submitted for publication) (GenBank accession numbers AF447479, AF446125, AF447484, AF446114, AF447487, AF446117, AY167107, AY167106, AY167105, and AY167104).
The sequences were aligned by the neighbor-joining (NJ) algorithm as implemented in ClustalX software (38), with manual corrections to match the open reading frame. For technical considerations, the sequences were slightly truncated, and the genome fragments analyzed were as follows: 5′ NTR (nt 337 to 600), VP1 (nt 3017 to 3343), 2A (nt 3344 to 3552), and 3D (nt 5885 to 6481). Then, two different algorithms were used to build the phylogenetic trees. First, the neighbor-joining (NJ) algorithm was used as implemented in ClustalX. Gap-containing positions were excluded, and correction for multiple substitutions was used. The trees were bootstrapped 1,000 times. Second, phylogenetic trees were created with a maximum-likelihood (ML) algorithm implemented in the DNAML module of the PHYLIP software package (12), with 1,000 bootstrap pseudoreplicates. As the ML and NJ trees were very similar and no significant differences between trees produced by the two methods were observed, only one (NJ) tree for each genome region is given, with ML bootstrap values manually added to the trees. The trees were drawn with the Ngraph module of ClustalX, and the in-tree comments were added with Corel Draw 10.
We did not attempt to analyze the sequences obtained in any aspect other than phylogenetically, since the fragments studied were relatively short and there are almost no reliable data on the genetic determinants of pathogenicity in human enteroviruses.
Nucleotide sequence accession numbers.
The DNA sequences determined here were submitted to GenBank under accession numbers AY271379 to AY271512.
RESULTS
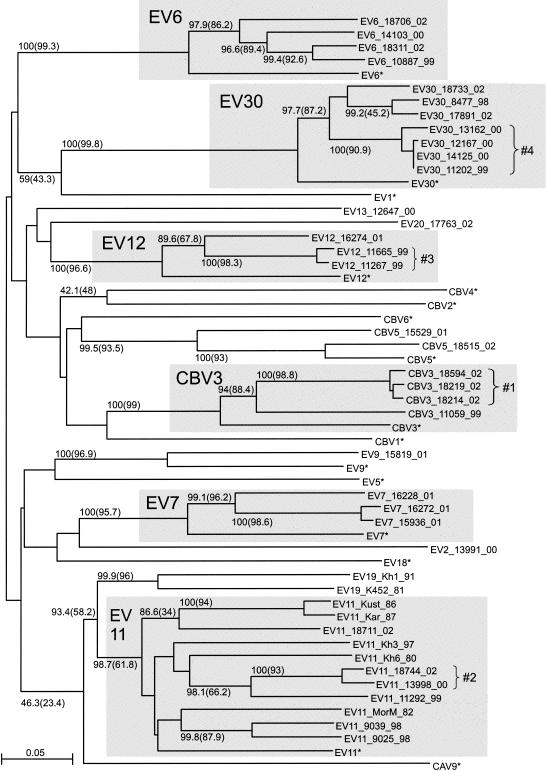
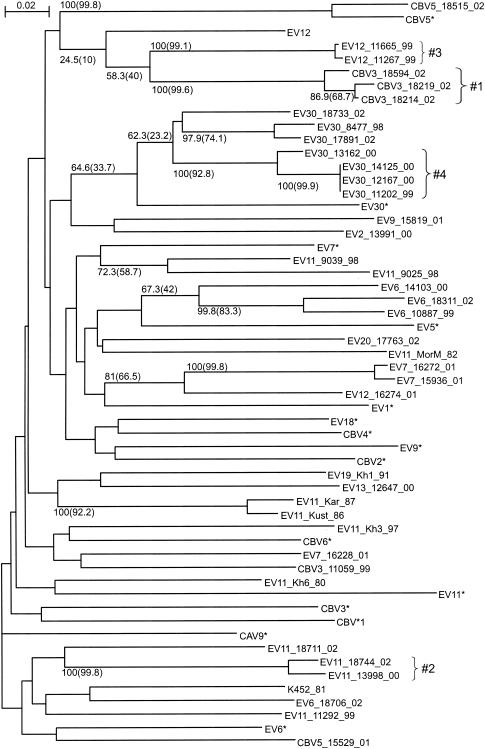
We have sequenced and analyzed four genome regions (Fig. 1) of 40 recently isolated (1980 to 2002) enterovirus B strains representing 11 different enterovirus B serotypes (Table 1). For the VP1 genome region (nt 3017 to 3343), the phylogenetic grouping of the viruses studied correlated perfectly with the serotype. All strains clustered with the corresponding prototype strain of that serotype (Fig. 2). Additionally, using a partial VP1 sequence, we identified the serotype for six CBV strains that had been serotyped by use of antiserum pools as coxsackie B viruses: strains 11059, 18214, 18219, and 18594 proved to be CBV3, and strains 15529 and 18515 were CBV5. In general, for the VP1 region, the grouping of strains studied was in accordance with that found in other phylogenetic studies (5, 32). The consistency of the results for the VP1 region with conventional serotyping provided a form of internal control and excluded occasional jumbling of samples in the course of our work.
FIG. 2.
Phylogenetic tree for the VP1 region (nt 3017 to 3343), created with ClustalX software. Numbers at tree nodes represent the numbers of bootstrap pseudoreplicates that support the group below, determined with NJ (ClustalX) and (in parentheses) ML (PHYLIP DNAML) algorithms. Prototype strains are indicated by an asterisk. The scale bar indicates nucleotide sequence distances. Minor groups discussed in the main text are indicated to the right of braces. Shaded areas show groupings of strains of the same serotype.
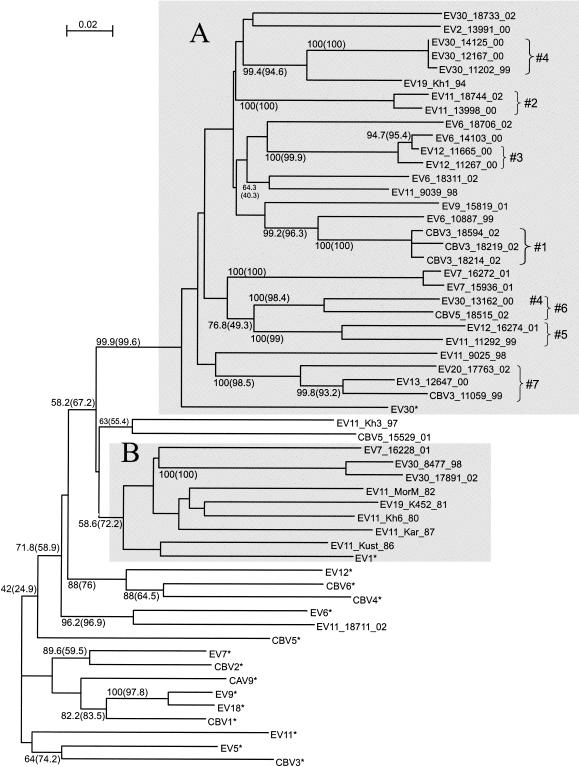
For the 3D region (nt 5885 to 6481), the phylogenetic grouping of the strains studied did not correlate with the serotype (Fig. 3). Strikingly, not a single strain studied grouped with its prototype strain. Instead, grouping clearly was correlated with the time of virus isolation. A majority (14 of 16) of the prototype enterovirus strains were clearly discrete from most of the strains studied, except for EV11_18711_02. Many prototype strains of different serotypes grouped together reliably (e.g., CBV1, EV9, and EV18 or EV12, CBV4, and CBV6). Most strains (isolated in 1981 to 2002) formed a major group (bootstrap values of 58.2% [NJ] and 67.2% [ML]) that was distinct from most prototype echovirus strains, except for EV1 and EV30. Within that major group, a highly reliable group (near 100% bootstrap values) included 29 of 34 strains isolated in 1994 to 2002, regardless of the serotype, and the prototype strain EV30 (Fig. 3A). In addition, all five enterovirus strains isolated in 1980 to 1987 grouped together (bootstrap values of 58.6% [NJ] and 72.2% [ML]); however, this group also included the prototype strain EV1 and three newer strains isolated in 1998 to 2002 (Fig. 3A).
FIG. 3.
Phylogenetic tree for the 3D region (nt 5885 to 6481). See the legend to Fig. 2 for details. Shaded areas show groupings of strains isolated mostly in the 1990s (A) and 1980s (B).
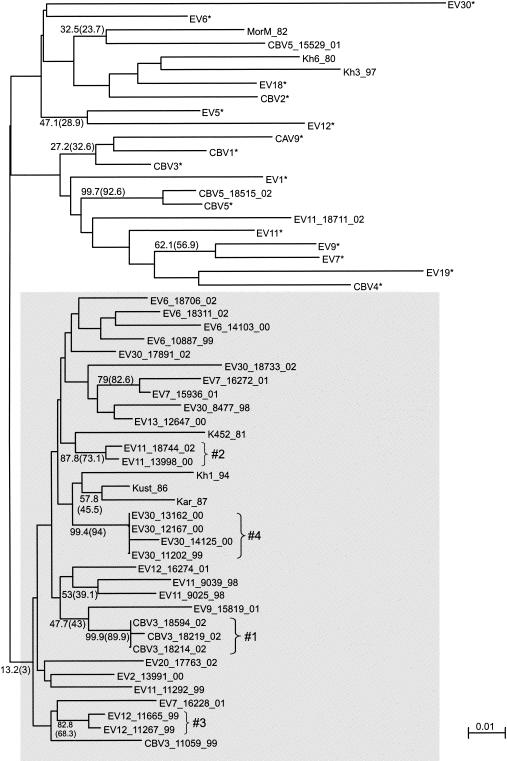
The phylogenetic tree for the 5′ NTR (nt 337 to 600) demonstrated very low bootstrap values, likely due to the relatively small size of the alignment and the high degree of sequence similarity in this highly conserved genome region. Sequencing of the hypervariable region of the 5′ NTR could yield better results, but we decided to avoid this procedure, as the possibility that recombination takes place within that region could not be excluded. For the 5′ NTR, only one strain, CBV5-18515, grouped with its prototype strain. As for the 3D region, 35 of 40 enterovirus strains studied formed a group that did not include a single prototype enterovirus strain (Fig. 4, shaded area). The bootstrap values for this group were only 13.2% (NJ) and 3% (ML). Despite the understandably low bootstrap values, we still regard the phylogenetic grouping observed for the 5′ NTR with confidence, as it is in principle supported by much more reliable results for the 3D region. An alternative, less significant grouping observed as a result of bootscan analysis (not shown on the tree) was also in line with the described grouping of modern strains. Thus, the tendency of most modern enterovirus B strains of different serotypes to show clustering for the 5′ NTR was obvious.
FIG. 4.
Phylogenetic tree for the 5′ NTR (nt 337 to 600). See the legend to Fig. 2 for details. The shaded area shows a grouping including a majority of modern strains.
We chose to sequence the VP1-2A junction region in our study in order to obtain information on fragments of both structural protein (VP1)- and NSP (2A)-coding regions. The phylogenetic tree for the 5′ end of the 2A region (Fig. 5) (nt 3344 to 3552) turned out to be strikingly different from the tree for the adjacent VP1 region (Fig. 2). The correlation observed for the VP1 region between phylogenetic grouping and serotype was seriously disturbed. While many minor groups of closely related strains of the same serotype could be seen, these did not group any more with the corresponding prototype strains, with the exception of the EV30 group. Furthermore, the regular ladder-like topology seen with the VP1 tree was not observed. Instead, the 2A tree was very star-like, with long branches of similar lengths leading almost from the root to single strains or distinct groups of closely related strains. Explanations for this observation could include a low signal-to-noise ratio of the phylogenetic signal resulting from low variability and/or multiple reversions or, much more likely, recombination within the region studied, as the 2A region is a well-known recombination hot spot in enteroviruses.
FIG. 5.
Phylogenetic tree for the 2A region (nt 3344 to 3552). See the legend to Fig. 2 for details.
One example of recombination could be unambiguously detected within the VP1-2A junction region, despite high sequence variability and, probably, numerous recombinations in the past. Strains EV12_11665_99, EV12_11267_99, CBV3_18214_02, CBV3_18219_02, and CBV3_18594_02 grouped together in the 2A region with moderate reliability (58.3% [NJ] and 40% [ML]). Using similarity plot, bootscan, and informative sight analyses as described elsewhere (Lukashev et al., submitted) for four sequences, those from EV12_11267_99, CBV3_18214_02, and the prototype strains EV12 and CBV3, a recombination point for this group was identified around nt 3400 (±10 nt) and supported by bootstrap values of over 95% on the bootscan graphs (data not shown).
In addition to evaluating the entire tree topology and major groups, we tracked the consistency of minor groups, which were reliably observed for at least one genome region, in trees for other genome regions. Several minor groups were observed for all four genome regions studied and thus did not indicate recombination. These were as follows: 1—CBV3_18214_02, CBV3_18219_02, and CBV3_18594_02; 2—EV11_13998_00 and EV11_18744_02; and 3—EV12_11267_00 and EV12_11665_00 (Fig. 2 to 5). However, the majority of reliable minor groups observed were inconsistent throughout the genome, including, of course, all serotype-related groups in the VP1 region, but also many others (Fig. 2 to 5). For example, group 1 clustered more or less reliably with CBV3 strains for the VP1 region, with EV12_11267_99 and EV12_11665_99 for the 2A region, with EV6_10887_99 for the 3D region, and with EV9_15819_01 for the 5′ NTR. Four strains, EV30_11202_99, EV30_12167_00, EV30_13162_00, and EV30_14125_00, clustered reliably for the 5′ NTR, VP1 region, and 2A region (group 4); however, for the 3D region, strain EV30_13162_00 clustered with CBV5_18515_02. Especially for the 3D region, several reliably observed clusters included highly similar sequences of strains of different serotypes, for example, EV12_16274_01 and EV11_11292_99 (group 5), CBV5_18515_02 and EV30_13162_00 (group 6), and EV13_12647_00, CBV3_11059_99, and EV20_17763_02 (group 7).
DISCUSSION
Recombination is a well-known phenomenon for enteroviruses. However, the prevalence of recombination in circulating NPEVs has not been determined. In this work, we studied the phylogenetic relationships of 40 recent enterovirus B isolates that belong to 11 different serotypes and 16 prototype enterovirus strains. A very obvious phylogeny conflict was observed in four genome regions analyzed. While for the VP1 genome region, the grouping of strains correlated with the serotype (Fig. 2), as widely reported previously (5, 32), for other genome regions, such grouping was not observed (Fig. 3 to 5). Moreover, grouping differed significantly between different genome regions, with only a few consistent groups including closely related strains. Similar results have been reported in a number of other studies (24, 33, 36).
On the basis of extensive work on recombination in poliovirus, the most feasible explanation for these observed discrepancies in the phylogenetic relationships of the strains studied seems to be ubiquitous recombination. Indeed, judging from other studies, such recombination was highly expected. What was unexpected, however, was the extent of recombination observed. Indeed, none of the 40 strains studied grouped with its prototype strain for the 3D region, and 39 of 40 did not group for the 5′ NTR. Similar observations was made for the 3D region in a recent study on recombination in circulating NPEVs (33). Thus, a vast majority of the strains studied differed from their prototypes, isolated in the 1950s, by at least two recombination events and were related to their prototype strains solely in the capsid region, which spans only one-third of the genome. However, these observations would not be as interesting without the reliable grouping of an overwhelming majority of the modern isolates, regardless of their serotypes, for the 5′ NTR and the 3D region. Therefore, it is possible to conclude that the 5′ NTRs and nonstructural regions of most of the modern enterovirus strains studied here are descendants of a common ancestor. For the 3D fragment, this common ancestor was more similar to the prototypes EV1 and EV30 and clearly distinct from other prototype enteroviruses. Obviously, the capsid and NSP genes of the strains studied followed clearly different evolutionary paths. The ubiquitous recombination seems to be so efficient that it enabled the 5′ NTR and NSP genome fragments to spread to different serotypes within the enterovirus B species. The marked differences in the 3D regions in strains isolated in the 1980s and 1990s suggest that such spreading of NSP genes to different serotypes could occur within a decade.
Our results suggest a novel model of enterovirus B genetics. At present, enterovirus serotypes merely represent a set of fairly discrete capsid genes that combine freely and frequently with many different variants of the 5′ NTR and NSP genes. This activity allows capsid, NTR, and NSP genes to evolve virtually independently. Thus, it seems that enterovirus B species (and, most likely, other enterovirus species) are an example of a gene symbiosis system, where genes, not species, are the elementary units of evolution. This notion, in turn, suggests that enterovirus B species, although comprised of 30 serotypes, should be considered a genetically uniform entity. Importantly, we have not detected among the strains studied any recombinants containing RNA sequences from viruses other than enterovirus B. Thus, the ability for frequent natural recombination could be an additional species criterion for enteroviruses.
A practical conclusion that follows is that when enteroviruses are identified only by serotype, information is obtained for one-third of the genome. The study of other genome regions could elucidate many important and poorly understood issues of enterovirus biology. The use of full-genome analysis for multiple isolates, involving sequencing or other techniques, does not seem feasible at present; however, such analysis should be the goal of typing approaches in the immediate future.
Acknowledgments
This work was supported in part by the Sigrid Juselius Foundation.
The strains sequenced were isolated as a part of the WHO Polio Eradication Initiative in the European Region. We express our acknowledgments to E. Nasirova (Republican Centre for Epidemiology and Hygiene, Baku, Azerbaijan), S. Sarkisyan (Republican Centre for Hygienic and Epidemic Surveillance, Yerevan, Armenia), S. Jabirov (Republican Centre for Immunoprophylaxis, Dushanbe, Tajikistan), T. Kutateladze (National Centre for Disease Control, Tbilisi, Georgia), K. Kasymbekova (Centre for Immunoprophylaxis, Bishkek, Kyrgyzstan), V. Gidirim (National Centre of Preventive Medicine, Kishinev, Moldavia), L. Yektova (Centre for State Sanitary Epidemiological Surveillance, Donetsk, Ukraine), I. Demchishina (Ukrainian Centre for State Sanitary Epidemiological Surveillance, Kiev, Ukraine), L. Kotlik (Central Laboratory of AIDS of Odessa, Odessa, Ukraine), G. Osipchuk (Republican Sanitary-Epidemiological Station, Tashkent, Uzbekistan), O. Utnitskaya (State Center of Sanitary-Epidemiological Surveillance, Ekaterinburg, Russian Federation), M. Bichourina (Pasteur Institute, St. Petersburg, Russian Federation), S. Kuribko (State Center of Sanitary-Epidemiological Surveillance, Moscow, Russian Federation), and E. Romanenko (State Center of Sanitary-Epidemiological Surveillance, Stavropol, Russian Federation) for providing the strains. Strains Kh1/94, Kh3/97, and Kh6/80 were obtained courtesy of V. I. Reznik (State Center of Sanitary-Epidemiological Surveillance in Khabarovsk Region, Khabarovsk, Russian Federation). We are grateful to Jussi Mantere for technical assistance with the DNA sequencer.
REFERENCES
- 1.Andersson, P., K. Edman, and A. M. Lindberg. 2002. Molecular analysis of the echovirus 18 prototype: evidence of interserotypic recombination with echovirus 9. Virus Res. 85:71-83. [DOI] [PubMed] [Google Scholar]
- 2.Blomquist, S., A. L. Bruu, M. Stenvik, and T. Hovi. 2003. Characterization of a recombinant type 3/type 2 poliovirus isolated from a healthy vaccinee and containing a chimeric capsid protein VP1. J. Gen. Virol. 84:573-580. [DOI] [PubMed] [Google Scholar]
- 3.Boom, R., C. Sol, M. Salimans, C. Jansen, P. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cammack, N., A. Phillips, G. Dunn, V. Patel, and P. D. Minor. 1988. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology 167:507-514. [PubMed] [Google Scholar]
- 5.Caro, V., S. Guillot, F. Delpeyroux, and R. Crainic. 2001. Molecular strategy for ′serotyping' of human enteroviruses. J. Gen. Virol. 82:79-91. [DOI] [PubMed] [Google Scholar]
- 6.Chang, K. H., P. Auvinen, T. Hyypia, and G. Stanway. 1989. The nucleotide sequence of coxsackievirus A9: implications for receptor binding and enterovirus classification. J. Gen. Virol. 70:3269-3280. [DOI] [PubMed] [Google Scholar]
- 7.Chua, B. H., P. C. McMinn, S. K. Lam, and K. B. Chua. 2001. Comparison of the complete nucleotide sequence of echovirus 7 strain UMMC and the prototype (Wallace) strain demonstrates a significant genetic drift over time. J. Gen. Virol. 82:2629-2639. [DOI] [PubMed] [Google Scholar]
- 8.Cuervo, N., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, R. Crainic, and F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahllund, L., L. Nissinen, T. Pulli, V. Hyttinen, G. Stanway, and T. Hyypia. 1995. The genome of echovirus 11. Virus Res. 35:215-222. [DOI] [PubMed] [Google Scholar]
- 10.Dahourou, G., S. Guillot, O. Le Gall, and R. Crainic. 2002. Genetic recombination on wild-type poliovirus. J. Gen. Virol. 38:3103-3110. [DOI] [PubMed] [Google Scholar]
- 11.Duggal, R., and E. Wimmer. 1999. Genetic recombination of poliovirus in vitro and in vivo: temperature-dependent alteration of crossover sites. Virology 258:30-41. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 13.Furione, M., S. Guillot, D. Otelea, J. Balanant, A. Candrea, and R. Crainic. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology 196:199-208. [DOI] [PubMed] [Google Scholar]
- 14.Georgescu, M.-M., F. Delpeyroux, and R. Crainic. 1995. Tripartite organization of a natural type 2 vaccine/nonvaccine recombinant poliovirus. J. Gen. Virol. 76:2343-2348. [DOI] [PubMed] [Google Scholar]
- 15.Georgescu, M. M., F. Delpeyroux, M. Tardy-Panit, J. Balanant, M. Combiescu, A. A. Combiescu, S. Guillot, and R. Crainic. 1994. High diversity of poliovirus strains isolated from the central nervous system from patients with vaccine-associated paralytic poliomyelitis. J. Virol. 68:8089-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gratsch, T. E., and V. F. Righthand. 1994. Construction of a recombinant cDNA of echovirus 6 that established a persistent in vitro infection. Virology 201:341-348. [DOI] [PubMed] [Google Scholar]
- 17.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirst, G. K. 1962. Genetic recombination with Newcastle disease virus, polioviruses and influenza. Cold Spring Harbor Symp. Quant. Biol. 27:303-308. [DOI] [PubMed]
- 19.Hughes, P. J., C. North, P. D. Minor, and G. Stanway. 1989. The complete nucleotide sequence of coxsackievirus A21. J. Gen. Virol. 70:2943-2952. [DOI] [PubMed] [Google Scholar]
- 20.Iizuka, N., S. Kuge, and A. Nomoto. 1987. Complete nucleotide sequence of the genome of coxsackievirus B1. Virology 156:64-73. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins, O., J. D. Booth, P. D. Minor, and J. W. Almond. 1987. The complete nucleotide sequence of coxsackievirus B4 and its comparison to other members of the Picornaviridae. J. Gen. Virol. 68:1835-1848. [DOI] [PubMed] [Google Scholar]
- 22.Kirkegaard, K., and D. Baltimore. 1986. The mechanism of RNA recombination in poliovirus. Cell 47:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klump, W. M., I. Bergmann, B. C. Muller, D. Ameis, and R. Kandolf. 1990. Complete nucleotide sequence of infectious coxsackievirus B3 cDNA:two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J. Virol. 64:1573-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopecka, H., B. Brown, and M. Pallanch. 1995. Genotypic variation in coxsackievirus B5 isolates from three different outbreaks in the United States. Virus Res. 38:125-136. [DOI] [PubMed] [Google Scholar]
- 25.Ledinko, N. 1963. Genetic recombination with poliovirus type 1: studies of crosses between a normal horse-serum resistant mutant and several guanidine-resistant mutants of the same strain. Virology 20:107-119. [DOI] [PubMed] [Google Scholar]
- 26.Lindberg, A. M., S. Johansson, and A. Andersson. 1999. Echovirus 5: infectious transcripts and complete nucleotide sequence from uncloned cDNA. Virus Res. 59:75-87. [DOI] [PubMed] [Google Scholar]
- 27.Lipskaya, G. I., A. R. Muzychenko, O. K. Kutitova, S. V. Maslova, M. Equestre, S. G. Drozdov, R. Perez Bercoff, and V. I. Agol. 1991. Frequent isolation of intertypic poliovirus recombinants with serotype 2 specificity from vaccine-associated polio cases. J. Med. Virol. 35:290-296. [DOI] [PubMed] [Google Scholar]
- 28.Lukashev, A. N., V. A. Lashkevich, G. A. Koroleva, J. Ilonen, G. G. Karganova, V. I. Reznik, and A. E. Hinkkanen. 2003. Molecular epidemiology of enteroviruses causing uveitis and multisystem hemorrhagic disease of infants. Virology 307:45-53. [DOI] [PubMed] [Google Scholar]
- 29.Macadam, A. J., C. Arnold, J. Howlett, A. John, S. Marsden, F. Taffs, P. Reeve, N. Hamada, K. Wareham, J. Almond, N. Cammack, and P. D. Minor. 1989. Reversion of the attenuated and temperature-sensitive phenotypes of the Sabin type 3 stain of poliovirus in vaccinees. Virology 172:408-414. [DOI] [PubMed] [Google Scholar]
- 30.Martino, T. A., R. Tellier, M. Petric, D. M. Irwin, A. Afshar, and P. P. Liu. 1999. The complete consensus sequence of coxsackievirus B6 and generation of infectious clones by long RT-PCR. Virus Res. 64:77-86. [DOI] [PubMed] [Google Scholar]
- 31.Minor, P. D., A. John, M. Ferguson, and J. P. Icenogle. 1986. Antigenic and molecular evolution of the vaccine strain of type 3 poliovirus during the period of excretion by primary vaccinee. J. Gen. Virol. 67:693-706. [DOI] [PubMed] [Google Scholar]
- 32.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oprisan, G., M. Combiescu, S. Guillot, V. Caro, A. Combiescu, F. Delpeyroux, and R. Crainic. 2002. Natural genetic recombination between co-circulating heterotypic enteroviruses. J. Gen. Virol. 83:2193-2200. [DOI] [PubMed] [Google Scholar]
- 34.Pallansch, M. A., and R. P. Roos. 2001. Enteroviruses: polioviruses, coxsackieviruses, echoviruses and newer enteroviruses, p. 723-775. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 35.Ryan, M. D., O. Jenkins, P. J. Hughes, A. Brown, N. J. Knowles, D. Booth, P. D. Minor, and J. W. Almond. 1990. The complete nucleotide sequence of enterovirus type 70: relationships with other members of the Picornaviridae. J. Gen. Virol. 71:2291-2299. [DOI] [PubMed] [Google Scholar]
- 36.Santti, J., H. Harvala, L. Kinnunen, and T. Hyypia. 2000. Molecular epidemiology and evolution of coxsackievirus A9. J. Gen. Virol. 81:1-12. [DOI] [PubMed] [Google Scholar]
- 37.Santti, J., T. Hyypia, L. Kinnunen, and M. Salminen. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J., T. Gibson, F. Plewniak, F. Jeanmougin, and D. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, G., G. Wilsden, N. J. Knowles, and J. W. McCauley. 1993. Complete nucleotide sequence of a coxsackie B5 virus and its relationship to swine vesicular disease virus. J. Gen. Virol. 74:845-853. [DOI] [PubMed] [Google Scholar]
- 40.Zimmermann, H., H. J. Eggers, and B. Nelsen-Salz. 1996. Molecular cloning and sequence determination of the complete genome of the virulent echovirus 9 strain Barty. Virus Genes 12:149-154. [DOI] [PubMed] [Google Scholar]