Abstract
A satellite DNA sequence, As120a, specific to the A-genome chromosomes in the hexaploid oat, Avena sativa L., was isolated by subcloning a fragment with internal tandem repeats from a plasmid, pAs120, that had been obtained from an Avena strigosa (As genome) genomic library. Southern and in situ hybridization showed that sequences with homology to sequences within pAs120 were dispersed throughout the genome of diploid (A and C genomes), tetraploid (AC genomes), and hexaploid (ACD genomes) Avena species. In contrast, sequences homologous to As120a were found in two A-genome species (A. strigosa and Avena longiglumis) and in the hexaploid A. sativa whereas this sequence was little amplified in the tetraploid Avena murphyi and was absent in the remaining A- and C-genome diploid species. In situ hybridization of pAs120a to hexaploid oat species revealed the distribution of elements of the As120a repeated family over both arms of 14 of 42 chromosomes of this species. By using double in situ hybridization with pAs120a and a C genome-specific probe, three sets of 14 chromosomes were revealed corresponding to the A, C, and D genomes of the hexaploid species. Simultaneous in situ hybridizations with pAs120a and ribosomal probes were used to assign the SAT chromosomes of hexaploid species to their correct genomes. This work reports a sequence able to distinguish between the closely related A and D genomes of hexaploid oats. This sequence offers new opportunities to analyze the relationships of Avena species and to explore the possible evolution of various polyploid oat species.
Keywords: in situ hybridization/satellite DNA/genome evolution
The cultivated hexaploid oat Avena sativa L. belongs to a group of closely related species of the genus Avena. Species in this group share their genome(s) with one another, and it is to be presumed that some are the ancestors of others. An essential prerequisite for genome analysis in this genus was the discovery of the correct basic chromosome number, x = 7, and three ploidy levels, i.e., diploidy, tetraploidy, and hexaploidy. All diploid species contain either the AA or CC genomes, and these genomes are also present in the AACC tetraploid and AACCDD hexaploid species (1). The evolution of oat nuclear genomes was a complex process involving divergence at the diploid level from a common diploid ancestor and then convergence, followed by divergence, at the polyploid level (2).
Differences between the A genomes of diploid species were revealed by karyotype studies (1, 3), isozyme variations (4), and chromosome pairing in interspecific hybrids (2). Comparative karyotype studies between A. sativa and diploid Avena strigosa Schreb showed that the A. strigosa genome (As) matched very closely the putative A genome of hexaploid oat (1, 5). Molecular investigations based on genomic in situ hybridization (6, 7, 8) and on the physical position of 5S rDNA loci (9) suggested that both A and D genomes of A. sativa were highly homologous to A. strigosa. Attempts to distinguish between A and D genomes by genomic in situ hybridization by using DNA from different A-genome species as probe and different combinations of probe/blocker of diploid A-genome species (10) so far have failed. These observations together with the absence of DD genome diploid species supports the hypothesis that an A-genome diploid species could be the donor of both the A and D genomes of the hexaploid oat.
The C genome largely is differentiated from the A and D genomes. Chromosome C-banding studies indicated that the C-genome chromosomes contain large heterochromatic regions whereas the A- and D-genome chromosomes are mostly euchromatic (5, 11, 12). Different diploid species have been proposed as the putative donors of the C genome of hexaploids. Studies based on the number and morphology of satellite chromosomes pointed to Avena ventricosa Bal. ex Coss, or a combination of Avena canariensis Baum, Rajhathy et Sampson, and Avena eriantha Dur. (1). In situ hybridization using A. eriantha showed the presence of this chromatin in the C genome of hexaploid species (6–8). Although the identification of diploid species in the evolution of the hexaploids is still controversial, evidences favor the involvement of the tetraploid oats Avena maroccana Gdgr. (formerly described as Avena magna) and Avena murphyi Ladiz. as the putative AC-donors. These evidences are based on chromosome pairing (13) and domestication of the tetraploid species by crossing them with cultivated oats (14). Although A. strigosa DNA has been able to paint A-genome chromosomes of AACC species (7, 8), chromosome pairing between AA diploid and AACC tetraploid species was insufficient to support the proposition that the As genome was present in tetraploid species (15). These different viewpoints seem to suggest that various diploid species might be involved in the evolution of each A and C genomes.
In addition to polyploid, intergenomic translocations are considered to be a significant force in oat evolution. Cytological and molecular evidence has shown that the C and the A/D genomes underwent structural rearrangements after they were incorporated into polyploid species (6–9, 16, 17). However, the inability to distinguish between the A and D genomes makes the identification of the A and D chromosomes involved in intergenomic translocations impossible. A detailed restriction fragment length polymorphism linkage map for the hexaploid oat containing 38 linkage groups has been established (17), but, so far, the assignment of the linkage groups to specific chromosomes has not been accomplished. Joining molecular cytogenetics with genetic chromosome analysis appears to be the best approach for developing an oat cytogenetic map and assigning the linkage groups.
The aim of this paper was to present a comprehensive strategy for genome painting of hexaploid species by using different categories of DNA probes. A repeated sequence from A. strigosa was isolated, sequenced, and located on oat chromosomes. This sequence was used in combination with either 18S-5.8S-26S or 5S rDNA probes (18, 19) and a C-genome specific sequence previously isolated (20) for identification of both individual and C-genome chromosomes in in situ hybridization experiments.
MATERIALS AND METHODS
Plant Materials.
The oat species used were as follows: (i) AA genome species, A. strigosa (PI 258729) from the John Innes Center (Norwich, U.K.), Avena damascena (CAV 0258) and A. canariensis (CAV 3873) from the Plant Research Centre (Ottawa), and Avena longiglumis Dur. (BGRC 052993) from the Institüt für Pflanzenban und Pflanzenzücthung (Braunschweig, Germany); (ii) CC genome species, A. eriantha (CAV 0063) and Avena clauda Dur. (CAV 0001) from the Plant Research Centre; (iii) AACC genome species, A. maroccana (CAV 4388) and A. murphyi (Cc 7120) from the Plant Research Centre and the Welsh Plant Breeding Station (Aberystwith, UK), respectively; and (iv) AACCDD genome species, Avena byzantina C. Koch cv. ‘Kanota’ from the University of Osaka, Japan Avena sterilis L. (PI 411958) from the National Small Grain Collection (Beltsville, MD), and A. sativa, including cultivars ‘Previsión’ and ‘Pandora’ from the National Institute of Seeds (Madrid, Spain) and cultivar ‘Extra Klock’ from the Nordic Gene Bank (Lund, Sweden). Total DNA was isolated from young leaves by using standard techniques (21).
Cloning, Screening, and Sequencing of Repeated Sequences.
A. strigosa genomic DNA was digested partially with NdeII and was cloned into the BamHI site of pUC19. The ligation mixture was used to transform Escherichia coli strain XL1. A total of 719 positive colonies were plated on a grid and were transferred onto positively charged nylon membranes. Two replicates of each membrane were screened with [32P]dCTP-labeled genomic DNA probes of A. strigosa and A. eriantha by a standard colony hybridization procedure (22). Sixty-nine clones that strongly hybridized to A. strigosa DNA but not to A. eriantha DNA were selected. Plasmids were extracted and subjected to a second screening by Southern blot analysis and hybridization with the genomic probes. Twenty-one plasmids were selected as probable carriers of A-genome specific repetitive sequences. The pAs120 clone was selected for further analysis by Southern and in situ hybridization. A NdeII fragment, pAs120a, was subcloned in plasmid pUC19. The DNA sequences of pAs120 and pAs120a were determined for both strands by using an Applied Biosystems Automated DNA sequencer (model 377). Sequence similarity searches at the European Molecular Biology Laboratory database were performed.
Southern Blotting.
Genomic DNA from 11 oat species was restricted, was electrophoresed in 1.5% agarose gels, and was transferred onto nylon membranes. Hybridization probes were labeled with Digoxigenin-11dUTP by a PCR (23). Hybridization, posthybridization washes, and chemiluminescence detection were performed as described (24). Membranes were exposed to Hyperfilm-ECL (Amersham) for 1–3 h.
Fluorescent in Situ Hybridization (FISH).
Chromosome preparation, probe labeling and FISH were as described (16). Five DNA probes were used for FISH analysis: (i) Clone pTa71, containing a 9-kilobase EcoRI fragment including the 18S-5.8S-26S rDNA gene and spacer sequences from Triticum aestivum (18); (ii) clone pTa794, including a 410-bp 5S rDNA gene and intergeneric spacer isolated from T. aestivum (19); (iii and iv) clones pAs 120 and pAs 120a isolated from A. strigosa (this work); and (v) clone pAm1, a satellite DNA specific to the oat C genome containing an insert of 464 bp, isolated from A. murphyi (20).
RESULTS
Molecular Structure and Chromosomal Organization of the As120 Repeat Sequence.
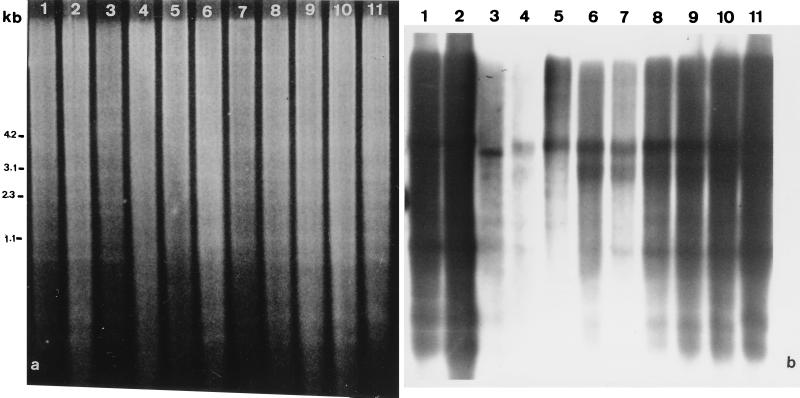
In Southern hybridization experiments, sequences hybridizing with pAs120, a plasmid containing DNA isolated from the AsAs diploid oat A. strigosa, were detected in all oat species tested (Fig. 1). The hybridization intensity profiles after genomic DNA digestion with EcoRV showed high levels of hybridization in the diploid species A. strigosa (AsAs) and A. longiglumis (AlAl), the tetraploid species A. murphyi, and the hexaploid species studied. Moderate levels were observed in the diploid A genome species A. canariensis (AcAc) and A. damascena (AdAd), diploid C genome species A. eriantha and A. clauda, and the tetraploid species A. maroccana. A smear pattern of hybridization with several distinct bands was observed, indicating that some sequences with homology to the probe were present in several discrete configurations whereas others were interspersed with unrelated sequences. The A. murphyi and A. clauda (data not shown) patterns showed a different distribution of several bands relative to the patterns of the other species.
Figure 1.
Southern blot analysis of the DNAs of diploid, tetraploid, and hexaploid oats. The species, in lanes: 1, A. strigosa; 2, A. longiglumis; 3, A. canariensis; 4, A. damascena; 5, A. eriantha; 6, A. murphyi; 7, A. maroccana; 8 and 10, respectively, A. sativa cvs. Prevision and Extra Klock; 9, A. byzantina cv. Kanota; 11, A. sterilis. (a) EtdBr-stained 1.5% agarose gel after separation of DNA samples cut with EcoRV showing similar amounts of plant DNA in each lane. (b) Autoradiogram after hybridization to pAs120 probe.
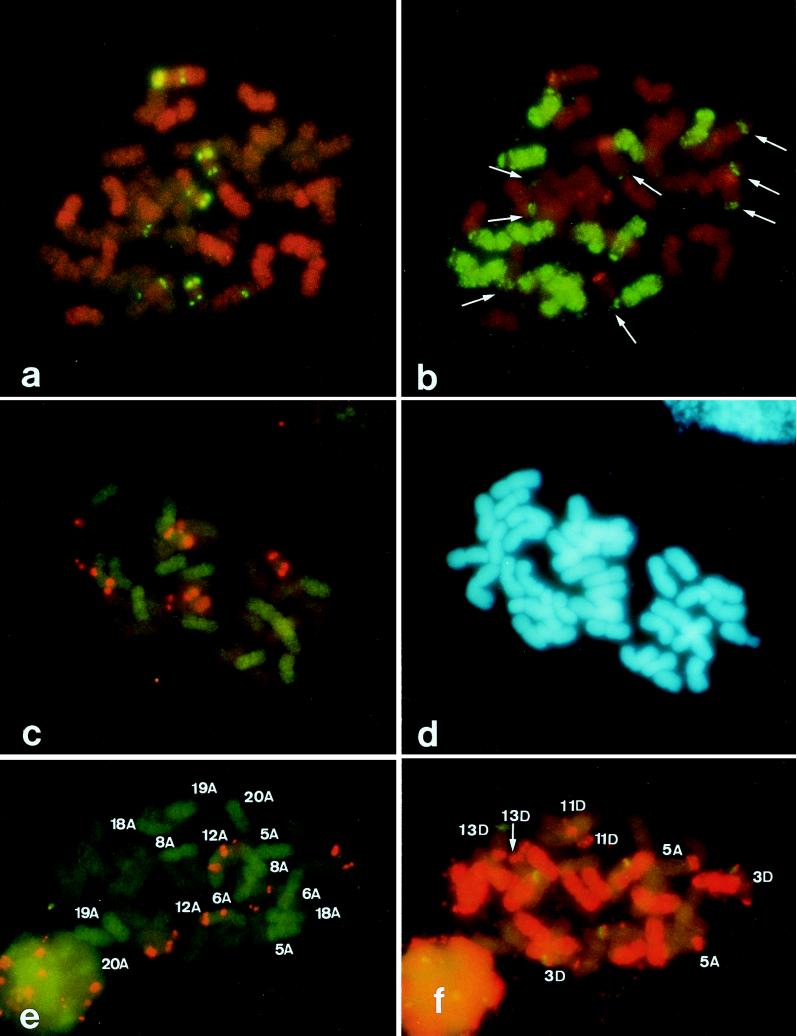
When pAs120 was hybridized to A. sativa mitotic metaphase plates, two chromosome patterns were identified (Fig. 5a). Fourteen chromosomes appeared highly hybridized, showing a dispersed distribution of the sequences across the chromosomes, whereas the remaining 28 chromosomes showed a much less intense signal. Of these 28 chromosomes, 14 were identified further as belonging to the C genome after hybridization with probe pAm1 (Fig. 5b). Taken together, all of these results suggested that the sequence cloned in pAs120 might be detecting repetitive sequences differently organized or of different composition in each oat genome. To test this hypothesis, the nucleotide sequence of the insert DNA of the pAs120 clone was determined (Fig. 2). The insert was 670 bp in length and had a 56% AT content. No relationships with known sequences at the nucleotide level were detected in the European Molecular Biology Laboratory database. Comparisons of the As120 sequence with itself by Harr plot graphic program (dnasis, Hitachi, Tokyo) revealed that the pAs120 insert was composed of two elements. The first consisted of four direct repeats arranged in tandem extending from the 1-nt to the 389-nt position. No internal subrepeats were found in the second element, which extended from the 390-nt to the 670-nt. To determine the genomic organization of the tandemly repeated element, the pAs120 insert was excised from the plasmid, was digested with NdeII, and was fractionated by agarose-gel electrophoresis. The 389-bp fragment was subcloned into pUC19, and the resulting clone, pAs120a, was characterized further.
Figure 5.
FISH of metaphase plates of A. sativa cvs. Extra Klock (a and b) and Prevision (c, d, e, and f). (a) Simultaneous visualization of hybridization sites of digoxigenin-labeled pTa794 (green) and rhodamine-labeled pAs120 (orange). (b) The same cell as in a, shown after in situ hybridization with digoxigenin-labeled pAm1 (green) and rhodamine-labeled pTa71 (red). (c) Double FISH of digoxigenin-labeled pAs120 a (green) and rhodamine-labeled pTa 794 (orange). (d) The same cell as in c, counterstained with 4′,6-diamidino-2-phenylindole. (e) Double FISH of digoxigenin-labeled pAs120a (green) and rhodamine-labeled pTa794 (orange). (f) The same cell as in e, shown after in situ hybridization with rhodamine-labeled pAm1 (orange) and digoxigenin-labeled pTa71 (green). In e, the numbers indicate the A-genome chromosomes. In f, the numbers indicate the A-C and D-C translocated chromosomes.
Figure 2.
Nucleotide sequence of a 670-bp fragment cloned in pAs120, which contains a tandem repetitive segment (dark lines) and a interspersed repetitive segment (light lines). The nucleotide sequence data are stored in the European Molecular Biology Laboratory, GenBank, and DNA Data Base in Japan nucleotide sequence databases under the accession no. AJ001922.
Molecular Structure and Chromosomal Organization of the As120a Repeat Sequence.
The consensus sequence of the four monomers cloned in plasmid pAs120a was 114 bp long with an AT content of 62% (Fig. 3). No relevant internal direct repeats were detected, though an imperfect inverted repeat of ≈32 bp was found in the consensus sequence. The nucleotide sequences of the inverted repeats shared 75% homology with each other, which may allow them to form hairpin structures. The four repeated units were not identical to each other and showed between 57.8 and 81.6% homology to the consensus sequence. The number of base deletions, 60, was higher than the number of substitutions–additions, 40, which appeared to be spread randomly over the whole sequence.
Figure 3.
Sequence of a 389-bp fragment cloned in pAs120a that includes four closely related monomers. The consensus sequence has been written on the first line. Nucleotides common to the consensus are indicated by hyphens, sequences differences are denoted by the relevant nucleotide, deletions are indicated by dots, and insertions are written under the sequence. The two imperfect inverse repeats are overlined.
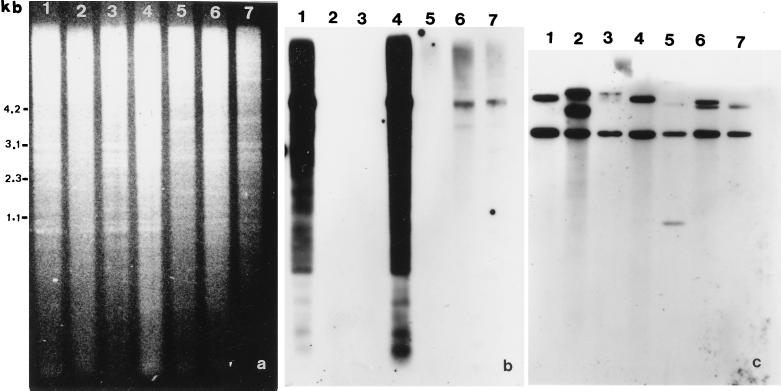
The organization of the As120a sequence in oat species was studied by digesting genomic DNA with MunI (Fig. 4). Substantial hybridization occurred with both A. strigosa (As genome) and A. longiglumis (Al genome) DNA. pAs120a hybridized to a minimum extent to A. sativa (ACD genomes) and A. murphyi (AC genomes) DNA. The hybridization pattern of A. maroccana (data not shown) was similar to that of A. murphyi. No hybridization signal was detected with either A. canariensis (Ac genome), A. damascena (Ad genome), or A. eriantha (Cp genome) (Fig. 4b). Differences in hybridization signal with pAs120a could not be attributed to differences in the quantity of DNA loaded in each gel lane, as shown by both the ethidium bromide gel (Fig. 4a) and the autoradiogram obtained after reprobing the blot with the ribosomal DNA probe pTa71 (Fig. 4c).
Figure 4.
Southern blot analysis of the MunI-digested genomic DNA of diploid and polyploid oats. The species, in lanes: 1, A. strigosa; 2, A. damascena; 3, A. canariensis; 4, A. longiglumis; 5, A. eriantha; 6, A. murphyi; 7, A. sativa cv. Extra Klock. (a) EtdBr-stained 1.5% agarose gel of DNA samples. (b) Autoradiogram of a after transfer and hybridization with pAs120a probe. (c) Autoradiogram showing rehybridization of blot in b with pTa71 probe.
To determine the chromosomal distribution of this repeated family, pAs120a was used as a probe by in situ hybridization. The nomenclature used in previously reported karyotype analyses (1, 5, 9) was followed for chromosome designation. Moreover, the identification of chromosomes carrying ribosomal genes or belonging to the C genome were achieved by using ribosomal (5) and pAm1 (9) clones, respectively, both by simultaneous FISH experiments and by reprobing the same metaphase plates. The digoxigenin-labeled insert of pAs120a hybridized strongly and evenly over the entire length of all A. strigosa chromosomes, but it failed to hybridize with A. murphyi and A. maroccana metaphases (data not shown). When digoxigenin-labeled pAs120a was hybridized with A. sativa metaphases, only 14 chromosomes were hybridized (Fig. 5c). When digoxigenin-labeled pAs120a and rhodamine-labeled pAm1 were hybridized with A. sativa metaphases, three sets of chromosomes, each of 14 chromosomes, clearly were identified (Fig. 5 e and f). The hybridization pattern of one set was like that of the diploid A. strigosa and was assigned to the A-genome (green in Fig. 5 c and e). A second chromosome set hybridized with the pAm1 insert, thus identifying the C-genome chromosomes (red in Fig. 5f). The remaining chromosomes were unhybridized with either probe and were assigned to the D genome. The A-genome chromosomes could be further identified as follows: One satellite chromosome pair was identified as 12A by the presence of complementary sequences to pTa71 and pTa794. The presence/absence of terminal sequences homologous to pAm1 (9) and karyotype morphology enabled the remaining A chromosomes to be identified tentatively (Fig. 5e). Five chromosome pairs showed absence of Am1 sequences, i. e. 6A, 8A, 18A, 19A, and 20A. The remaining one, 5A, showed hybridization (at a site of a C genome translocation) with pAm1.
Regarding the D-genome chromosomes, three chromosome pairs were identified through hybridization with ribosomal and pAm1 probes (Fig. 5f). Two of these three pairs were satellite chromosomes, and they were identified as 3D and 13D. Moreover, the 3D chromosome pair contained 5S sequences on both chromosome arms in a way similar to that of 12A. The additional D-genome chromosome was identified as 11D because it showed Am1 sequences (at a site of a C genome translocation) on the long chromosome arm. Therefore, two satellite chromosomes of A. sativa were assigned to the D genome, and one was assigned to the A genome. Consequently, one A-genome chromosome pair, 5A, and three D-genome chromosome pairs, 3D, 11D, and 13D, were identified tentatively based on translocations with C-genome chromosomes in the hexaploid genome.
DISCUSSION
The present study describes the characterization of a cloned repetitive DNA fragment, isolated from the A. strigosa genome, in 11 oat species. Nucleotide sequence determination showed that the fragment was composed of a tandemly repeated DNA sequence adjacent to an unrelated sequence. The two elements of the clone pAs120 revealed repetitive families that appear to have become differently organized during the evolution of the oat species analyzed. These differences have permitted discrimination between otherwise closely related A genomes of the diploid species and between the A and D genome of the hexaploids. When the repeated sequence insert in pAs120a was used as probe to restriction enzyme digested oat DNA, no hybridization bands were observed on the lanes carrying genomic DNA of the A genome diploid taxa A. damascena and A. canariensis or the C diploid taxa A. eriantha and A. clauda (Fig. 4). However, clone pAs120 hybridized, although at varying levels, with DNA of all of the species analyzed. This indicates that the tandemly repeated sequence As120a was amplified in the genome of a progenitor of A. strigosa and A. longiglumis subsequent to the differentiation of these species from the ancestral progenitor of A genome species. These results in general agree with the speciation model for the A genome diploid species proposed by Thomas (2) based mainly on chromosome pairing. Both the length of the repeats and the existence of four monomers in the insert of pAs 120a (Fig. 3) seem to indicate that the As120a sequence could be classified as a satellite DNA sequence. Satellite DNA has been shown to be preferentially located in constitutive heterochromatin (25). However, it has been pointed out (26) that only large clusters of tandem repeats stain as C-bands. The distribution of sequences homologous to As120a across the chromosomes of A. strigosa (data not shown) and the A genome of A. sativa (Fig. 5 c and e) suggests that this is the case, given the terminal location of C-bands in the chromosomes of these species (3, 5, 7). The detection of imperfect subrepeats of 32 bp would indicate that the As120a family has evolved from duplications of shorter units. Moreover, the high number of nucleotide substitutions, deletions, and insertions found reflect the dynamic and rapid change of this tandemly repeated DNA sequence that is not as homogenized as other plant satellite sequences (16, 27, 28). A comparison with the Am1 satellite C-genome specific sequence, which showed a 92% homology among its monomers (16), confirms the independent evolution rates of each repetitive family.
Because pAs120a only hybridized with 14 chromosomes in the AACCDD hexaploid species, we postulated that this satellite DNA was specific for one of the genomes of cultivated oats. The genome identity was revealed partially by reprobing with pAm1, which hybridizes with the 14 chromosomes of the C-genome. The two probes recognized different chromosomes. This result demonstrate that, if the As120a sequence is genome specific, it is specific for either the A or the D genome. It has been demonstrated that the D genome is related closely to the A genome in terms of repetitive sequences (6–8). Chromosome identification and the pattern of in situ hybridization in diploid and polyploid species by using ribosomal DNA sequences suggested that some A genome diploid species should be the donor of the D genome of hexaploid species (9). These observations confounded the assignment of the genome marked by the pAs120a probe. Because A. strigosa has been considered the A genome donor of the hexaploid species (1, 2) and the As120a sequence was isolated from that species, the chromosome set of the hexaploid species carrying sequences complementary to As120a was designated the A genome of the hexaploids. After hybridization with pAs120a, Southern blots of A. maroccana and A. murphyi showed a faint fragment with a similar size of that of A. strigosa (Fig. 4b). This indicates a close genetic and phylogenetic relationship among these species. However, the lack of in situ hybridization of pAs120a with the tetraploid species chromosomes suggests that a great genomic reorganization of As120a sequences took place in the present-day AACC tetraploids. This reorganization could have originated a reduced concentration of As120a sequences at each genomic location, undetectable by in situ hybridization. Whether these observations invalidate the involvement of A. strigosa in the origin of the AACC tetraploids needs further investigation. Analysis is required of the hybridization pattern of the probe to other tetraploids. Alternatively, some other diploid species, such as A. damascena or A. canariensis, or even one not yet described, could be involved in the origin of these tetraploids. Meiotic behavior of triploid and tetraploid hybrids supports this supposition (2).
To further identify individual chromosomes, ribosomal and A- and C-specific genome probes were used in simultaneous FISH. In hexaploids oats, the two chromosome pairs carrying both 18S-5.6S-26S and 5S rDNA loci belong one each to the A and D genomes. This corroborates their previous assignments by C-banding analysis (5). In contrast, the third chromosome pair, which carries a 18S-5.6S-26S rDNA locus, belongs to the D genome. Similarly, other chromosome pairs classified as belonging to the A genome can now be identified precisely as D-genome chromosomes. The potential of painting particular genomes might be of special use for identifying the chromosomes involved in aneuploid series and the reassignment of chromosome designations in diploid and polyploid oats. Moreover, discrimination between genomes could be a powerful tool for following introgression in experiments involving interspecific crosses. Phylogenetic relationships also could be established more accurately.
Acknowledgments
The authors express gratitude to Dr. M. Leggett for critical reading of the manuscript and fruitful discussions. This study was supported by Dirección General de Investigación Científica y Técnica of Spain (PB92-0173, PB95-0329, and HB1996-0035).
ABBREVIATION
- FISH
fluorescent in situ hybridization
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the European Molecular Biology Laboratory, GenBank, and DNA Data Base (Japan) databases (accession no. AJ001922).
References
- 1. Rajhathy T, Thomas H. Cytogenetics of Oats. Ottawa: Miscellaneous Publication of the Genetics Society of Canada; 1974. , No. 2, pp. 1–90. [Google Scholar]
- 2.Thomas H. In: Oat Science and Technology. Marshall H G, Sorrells M E, editors. Madison, WI: American Society of Agronomy and Crop Science Society of America; 1992. pp. 473–507. [Google Scholar]
- 3.Fominaya A, Vega C, Ferrer E. Genome. 1988;30:627–632. [Google Scholar]
- 4.Sánchez de la Hoz P, Fominaya A. Theor Appl Genet. 1989;77:735–741. doi: 10.1007/BF00261252. [DOI] [PubMed] [Google Scholar]
- 5.Linares C, Vega C, Ferrer E, Fominaya A. Theor Appl Genet. 1992;83:650–654. doi: 10.1007/BF00226911. [DOI] [PubMed] [Google Scholar]
- 6.Chen Q, Armstrong K. Genome. 1994;37:607–612. doi: 10.1139/g94-086. [DOI] [PubMed] [Google Scholar]
- 7.Jellen E N, Gill B S, Cox T S. Genome. 1994;37:613–618. doi: 10.1139/g94-087. [DOI] [PubMed] [Google Scholar]
- 8.Leggett J M, Markhand G S. In: Kew Chromosome Conference IV. Brandham P E, Bennett M D, editors. Kew, U.K.: Royal Botanic Gardens; 1995. pp. 133–139. [Google Scholar]
- 9.Linares C, Gónzalez J, Ferrer E, Fominaya A. Genome. 1996;39:535–542. doi: 10.1139/g96-068. [DOI] [PubMed] [Google Scholar]
- 10.Leggett J M, Markhand G S. Aberystwyth Cell Genetics Group 7th Annual Conference. Aberystwyth, U.K.: Institute of Grassland and Environmental Research; 1997. p. 45. [Google Scholar]
- 11.Fominaya A, Vega C, Ferrer E. Genome. 1988;30:633–638. [Google Scholar]
- 12.Jellen E N, Phillips R L, Rines H W. Genome. 1993;36:1129–1137. doi: 10.1139/g93-151. [DOI] [PubMed] [Google Scholar]
- 13.Ladizinsky G, Zohari D. Euphytica. 1971;20:380–395. [Google Scholar]
- 14.Ladizinsky G. Theor Appl Genet. 1995;91:639–646. doi: 10.1007/BF00223291. [DOI] [PubMed] [Google Scholar]
- 15.Ladizinsky G. Evolution. 1969;23:676–684. doi: 10.1111/j.1558-5646.1969.tb03549.x. [DOI] [PubMed] [Google Scholar]
- 16.Fominaya A, Hueros G, Loarce Y, Ferrer E. Genome. 1995;38:548–557. doi: 10.1139/g95-071. [DOI] [PubMed] [Google Scholar]
- 17.O’Donoughue L S, Kianian S F, Rayapati P J, Penner G A, Sorrells M E, Tanskley S D, Phillips R L, Rines H W, Lee M, Fedak G, et al. Genome. 1995;38:368–380. doi: 10.1139/g95-048. [DOI] [PubMed] [Google Scholar]
- 18.Gerlach W L, Bedbrook J R. Nucleic Acids Res. 1979;7:1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlach W L, Dier T A. Nucleic Acids Res. 1980;8:4851–4865. doi: 10.1093/nar/8.21.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solano R, Hueros G, Fominaya A, Ferrer E. Theor Appl Genet. 1992;83:602–607. doi: 10.1007/BF00226904. [DOI] [PubMed] [Google Scholar]
- 21.Sharp P J, Desai S, Gale M D. Theor Appl Genet. 1988;76:691–699. doi: 10.1007/BF00303514. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. In: Molecular Cloning: A Laboratory Manual. Nolam C, editor. Plainview, New York: Cold Spring Harbor Lab. Press; 1989. pp. 1.90–1.95. [Google Scholar]
- 23.Hoisington D, Khairallal M, González de León D. Laboratory Protocols: CIMMYT Applied Molecular Genetics Laboratory. México D. F., Mexico: Centro Internacional de Mejoramiento de Maiz y Trigo; 1994. pp. 28–33. [Google Scholar]
- 24.Loarce Y, Hueros G, Ferrer E. Theor Appl Genet. 1996;93:1112–1118. doi: 10.1007/BF00230133. [DOI] [PubMed] [Google Scholar]
- 25.Smyth D R. Chromosoma. 1991;100:355–359. [Google Scholar]
- 26.Kunze B, Weichenhan D, Virks P, Traut W, Winking H. Cytogenet Cell Genet. 1996;73:86–91. doi: 10.1159/000134314. [DOI] [PubMed] [Google Scholar]
- 27.Lapitan N L V. Genome. 1992;35:171–181. [Google Scholar]
- 28.Ingham L D, Hanna W W, Baier J W, Hannah L C. Mol Gen Genet. 1993;238:350–356. doi: 10.1007/BF00291993. [DOI] [PubMed] [Google Scholar]