Abstract
BMS-378806 is a recently discovered small-molecule human immunodeficiency virus type 1 (HIV-1) attachment inhibitor with good antiviral activity and pharmacokinetic properties. Here, we demonstrate that the compound targets viral entry by inhibiting the binding of the HIV-1 envelope gp120 protein to cellular CD4 receptors via a specific and competitive mechanism. BMS-378806 binds directly to gp120 at a stoichiometry of approximately 1:1, with a binding affinity similar to that of soluble CD4. The potential BMS-378806 target site was localized to a specific region within the CD4 binding pocket of gp120 by using HIV-1 gp120 variants carrying either compound-selected resistant substitutions or gp120-CD4 contact site mutations. Mapping of resistance substitutions to the HIV-1 envelope, and the lack of compound activity against a CD4-independent viral infection confirm the gp120-CD4 interactions as the target in infected cells. BMS-378806 therefore serves as a prototype for this new class of antiretroviral agents and validates gp120 as a viable target for small-molecule inhibitors.
Approximately 42 million individuals worldwide are now living with human immunodeficiency virus type 1 (HIV-1). Apart from the recently approved fusion inhibitor enfuvirtide, all of the presently approved drugs target either the HIV protease or reverse transcriptase (RT) enzymes (4). Combination therapies often reduce patient viremia to an undetectable level and significantly delay disease progression in most treated individuals (for a review, see reference 31). However, shortcomings, such as the emergence of drug-resistant HIV strains, pill burden, adverse side effects, and/or insufficient potencies, have resulted in 30 to 50% of therapeutic failures (36). Furthermore, drug-resistant HIV-1 variants have been detected in as many as 26% of newly infected, treatment-naive patients (9, 21, 37). Therefore, long-term suppression of HIV replication requires new therapies that are more patient friendly and effective against current resistant HIV-1 strains by targeting steps other than protease or RT in the HIV life cycle.
The HIV-1 entry process involves multiple stages and represents several sources of new targets, with the likelihood of providing novel classes of drugs (for reviews, see references 1, 8, 19, and 30). The first stage of HIV-1 entry (viral attachment) involves the binding of the viral envelope protein gp120 to the host cell receptor, CD4. This interaction triggers the conformational changes that facilitate the binding of gp120 to either coreceptor CCR5 or CXCR4. Subsequent structural adjustments enable the insertion of the viral gp41 fusion peptide into the host membrane and, finally, fusion of the viral and host membranes by a still unresolved mechanism (for reviews, see references 3 and 38). Therefore, viral attachment, coreceptor binding, and virus-cell fusion represent unique targets for drug development. Proof of concept has been obtained in the clinical trials for all three of these targets with an attachment inhibitor, PRO542 (a CD4-immunoglobulin G fusion protein) (11, 35), a CCR5 coreceptor antagonist, SC-351125 (SCH C) (J. Reynes, R. Rouzier, T. Kanouni, V. Baillat, B. Baroudy, A. Keung, C. Hogan, M. Markowitz, and M. Laughlin, 9th Conf. Retrovir. Opportunistic Infect., abstr. 1, 2002), and the fusion inhibitors enfuvirtide (T-20) (12, 13, 20) and T-1249 (G. D. Miralles, J. B. Lalezari, N. Bellos, G. Richmond, Y. Zhang, H. Murchison, R. Spence, C. Raskino, and R. A. DeMasi, 10th Conf. Retrovir. Opportunistic Infect., abstr. 14b, 2003).
The HIV envelope is required for viral entry and consists of exterior gp120 and transmembrane gp41 proteins. The crystal structure of the gp120 core, in complex with a two-domain fragment of CD4 and the Fab fragment of 17b, a gp120 monoclonal antibody, provides revealing insights into the process of HIV entry (17, 18). The gp120 core is composed of an inner and an outer domain, as well as a bridging sheet, which all contribute to CD4 binding. The conserved residues within the CD4 binding site of gp120 form a deep hydrophobic pocket and interact with the phenylalanine 43 (Phe43) residue of CD4. Hence, this deep depression has been referred to as the Phe43 cavity (17, 18).
BMS-378806 (Fig. 1) is a recently discovered small-molecule inhibitor with potent activity against various HIV-1 laboratory strains (R5−, X4−, and R5/X4−) and clinical isolates of the B subtype (20a). The compound inhibits HIV-1 infection early in the viral replication cycle and blocks the fusion of cells expressing the HIV-1 envelope with those displaying CD4 and coreceptors. Our initial results also show that BMS-378806 inhibits gp120-CD4 binding in an enzyme-linked immunosorbent assay (ELISA) and that the BMS-378806-selected resistant substitutions are localized to the HIV-1 envelope. The compound displays no inhibitory activity against HIV-1 RT, protease, and integrase, implying that BMS-378806 is inhibiting a new target. Moreover, the clinical potential of this compound has been suggested by its good pharmacokinetic and safety profile (20a).
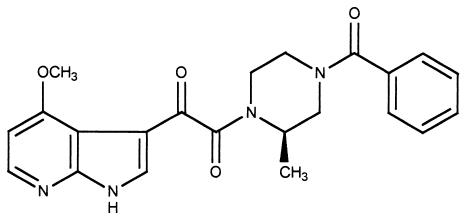
FIG. 1.
The structure of BMS-378806.
Here, we further characterize the interaction between BMS-378806 and the HIV-1 envelope by using a combination of biochemical and genetic approaches. The potential BMS-378806 binding site was mapped to a specific area within the CD4 binding pocket of the gp120 envelope protein. Our results demonstrate the existence of a functionally conserved region in the CD4 binding pocket of gp120 and validate this CD4 binding pocket as an anti-HIV target.
MATERIALS AND METHODS
Compounds.
BMS-378806 was synthesized at Bristol Myers Squibb. [3H]BMS-378806 was prepared at Vitrax by the tritiation of the corresponding dibromoderivative with T2 over Pd/C in N, N-dimethyl formamide. The final compound was purified by preparative high-performance liquid chromatography and had a specific activity of 38 Ci/mmol.
Proteins and antibodies.
D7324 (Aalto Bio Reagents Ltd., Dublin, Ireland) is an affinity-purified sheep polyclonal antibody to a 15-residue peptide from the conserved carboxy terminus of HIV-1LAI gp120. OKT4 is a CD4 monoclonal antibody and was purified from an OKT4-producing hybridoma cell line (ATCC CRL-8002). Soluble CD4 (sCD4) was obtained from ImmunoDiagnostics, Inc. HIV-1 gp120IIIB purified protein was purchased from Advanced Biotechnologies, Inc.
Cells and viruses.
Baby hamster kidney (BHK) cells, 293 embryonic kidney cells, NIH 3T3 mouse embryonic cells, and Cf2Th canine thymocyte cells were purchased from the American Type Culture Collection. The Cf2Th cell lines expressing CD4 or CD4/CCR5 were generated by transfecting pcDNA3.1 plasmids expressing human CD4 or/and CCR5 and confirmed by fluorescence-activated cell sorting analysis with the RPA-T4 anti-CD4 and the 2D7 anti-CCR5 monoclonal antibodies (BD Biosciences Pharmingen). pNL-ADA wild-type and pNL-ADA-S190R/N197S proviral DNA were constructed as described previously (14, 15). To prepare infectious HIV recombinant viruses, 293 cells were transfected with 10 μg of proviral DNA. Viral supernatants were harvested 72 h following transfection, clarified by centrifugation (200 × g for 10 min) and stored at −70°C.
Construction of wild-type and mutant envelope plasmids.
The pNL4-3 and codon-optimized pJRFL plasmids were obtained through the National Institutes of Health AIDS Reagent Repository. The pLAI plasmid was kindly provided by M. Emerman at the Fred Hutchinson Institute. The envelope genes were PCR amplified from these plasmids and cloned into the pSFV-SNA plasmid vector (28). The pSFV-SNA-gp120 DNA was linearized, and RNA was transcribed (Ambion). RNA transcripts were either electroporated (Bio-Rad) or transfected with DMRIE-C (Invitrogen) to BHK cells, and the gp120-containing media were collected and stored at −70°C.
Alternatively, the envelope genes amplified from these plasmids were cloned into a pTRE2 vector (BD Biosciences Pharmingen) (20a). The envelopes were mutagenized using the QuikChange site-directed mutagenesis kit (Stratagene). The numbering of the gp120 amino acid residues is based on the sequence of the prototypic HXBc2 strain of HIV-1, according to current convention (16). These envelope constructs were used either in cell fusion assays or for the generation of envelope proteins.
Purification of gp120 protein.
Wild-type gp120-containing supernatants from transfection of NIH 3T3 cells with gp120JRFL-pTRE plasmid were concentrated (Amicon ultrafiltration chamber lined with a YM30 membrane; Millipore, Bedford, Mass.). The concentrate was dialyzed twice against 20 mM Tris-HCl, pH 8.0, 0.15 M NaCl, 0.5 mM MgCl2, and 0.5 mM CaCl2 and absorbed onto 10 ml of lectin Sepharose 4B (Pharmacia) (preequilibrated with 10× volumes of buffer E, 20 mM Tris-HCl [pH 8.0], 0.5 mM MgCl2, and 0.5 mM CaCl2 at 4°C). After washing with buffer E containing 1 M NaCl, gp120 was eluted with 20 mM Tris-HCl (pH 8.0), 0.5 M α-methyl mannoside, 0.5 M α-methyl glucoside, and 0.1 mM EDTA, and the eluent was dialyzed twice against 25 mM Tris-HCl (pH 7.6) and then concentrated (Amicon). Protein was further purified over a HiTrap Q column (Pharmacia) by washing with buffer E and were then eluted with a 0 to 400 mM NaCl gradient. gp120-positive fractions were pooled, concentrated (Centricon YM30 Millipore filter) at 4°C, and stored at −70°C. Monomeric gp120 was purified further by application of the HiTrap eluent over Superdex 200 HR (10/30) with 20 mM Tris (pH 8.0) and 0.15 M NaCl. Monomeric gp120 fractions were verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stored at −70°C.
Cell-based fusion assay.
The details of a cell-based fusion assay were described previously (20a). Briefly, a HeLa target cell line that expresses CD4, CXCR4, and CCR5 was stably transduced with the pTRE-luciferase (luc) reporter plasmid (BD Biosciences Pharmingen). Upon activation by the pTet-off plasmid (expressing a transcriptional activator; BD Biosciences Pharmingen), the target cells would express the luciferase activity. The effector cells were prepared by transfecting 3 × 106 HeLa cells with 0.2 μg of the pTRE envelope construct and 1 μg of pTet-off by using Lipofectamine Plus (Invitrogen). The next day, the fusion process was initiated by mixing trypsinized effector and target cells in a 1:2 ratio and the mixtures were seeded in a 96-well plate at 5 ×104 cells/well followed by the addition of various concentrations of compound. Following an overnight incubation, the media were removed and the luciferase activity was measured by using a Steady-Glo luciferase assay system (Promega).
gp120-CD4 ELISA.
To measure gp120-CD4 binding, the wild-type or variant gp120 proteins were first captured onto a plate by D7324 antibody (24). CD4 binding was initiated by adding sCD4 to a gp120-coated plate (23). To determine the ability of BMS-378806 to compete with sCD4 for gp120 binding, the compound was added simultaneously with sCD4 and reactions were carried out in buffer C (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1% bovine serum albumin) for 2 h at room temperature. After washing with buffer B (20 mM Tris-HCl, 500 mM NaCl, 0.05% Tween 20 [pH 7.5]), the bound CD4 was detected with OKT4 antibody (0.36 μg/ml) and goat anti-mouse peroxidase conjugate (Bio-Rad). Bound antibody was detected with 3,3′,5,5′-tetramethylbenzidine chromogenic substrate for peroxidase (Pierce).
Measuring binding of [3H]BMS-378806 to gp120.
A scintillation proximity assay (SPA) was performed by first prebinding YSI protein A SPA beads (AP Biotech) with 0.36 μg of D7324 antibody overnight at 4°C. After washing with Tris-buffered saline, 30 mg of beads/ml was incubated with 60 μg of gp120/ml at room temperature for 1 h. Beads were then washed and resuspended at 30 mg/ml in phosphate-buffered saline. Aliquots (1 mg) were deposited into each well of a plate followed by various concentrations of tritiated compound. Plates were incubated for 4 h at 25°C and then analyzed on a TopCount NXT scintillation counter (Perkin-Elmer Life Sciences). Kd and Ki determinations were calculated from counts per minute bound by nonlinear least-squares analysis methods with GraphPad (San Diego, Calif.) Prism software (version 3.02).
Alternatively, Micro BioSpin 6 columns (Bio-Rad), capable of separating small ligands from large macromolecules, were used to measure the binding of [3H]BMS-378806 to gp120. Binding solution (30 μl) containing 25 mM Tris-HCl (pH 7.5), 125 mM NaCl, 5 to 200 nM gp120, and 100 nM [3H]BMS-378806 was allowed to equilibrate for 5 min, adsorbed to a Micro BioSpin 6 column, and centrifuged for 5 s. The eluent was collected and counted in a TopCount scintillation counter (Perkin-Elmer Life Sciences).
Additionally, a Flashplate (DuPontNEN) method was used to measure the binding of [3H]BMS-378806 to gp120. Supernatants containing gp120 variants were captured on a 96-well Flashplate as described for the gp120-CD4 binding ELISA. [3H]BMS-378806 (1 × 105 to 5 × 105 cpm) in buffer C was added to plates and kept at 4°C overnight. Supernatants were removed and plates were washed with cold buffer C, and an additional 100 μl of buffer C was added to the wells before reading in a TopCount scintillation counter. Signals from wells containing no gp120 were used as a background control.
Fluorescence determination of gp120-BMS-378806 binding.
Monomeric gp120JRFL or gp120IIIB (Advanced Biotechnologies, Inc.) were diluted in phosphate-buffered saline to the concentrations indicated, 200 μl of gp120 was added to a quartz cell at ambient temperature, and baseline spectra were recorded (excitation at 280 nm, emission at 343 nm, 10 nm/min; slit width, 6 cm) on a Perkin-Elmer LS50B luminescence spectrometer. Aliquots (0.5 or 1.0 μl) of stock BMS-378806 solutions were added with a 10-μl Hamilton syringe. Readings were made 1 min after sample mixing. Total volume changes in the experiments were less than 10% (vol/vol). The percent fluorescence reduction is defined as 100 × {[initial intensity − final intensity]/initial intensity} at 343 nm. The data were fit to a saturation-binding isotherm by a nonlinear least-squares fitting with GraphPad Prism software (version 3.02).
Virus replication and drug susceptibility assays.
ADA-S190R/N197S virus was propagated for up to 1 month to allow gradual viral adaptation in the CD4-negative Cf2Th/CCR5 cell line as previously described (15). Replication kinetics of the ADA mutant virus was monitored by a p24 ELISA following infection at a multiplicity of infection of 0.01. To test drug susceptibility, cells were infected with ADA-S190R/N197S at an multiplicity of infection of 0.01 and incubated in the presence of BMS-378806 for 5 days. Virus yields were quantitated by a p24 ELISA. Results from at least two independent experiments were used to calculate the 50% effective concentrations (EC50).
RESULTS
Competitive inhibition of gp120-CD4 binding by BMS-378806.
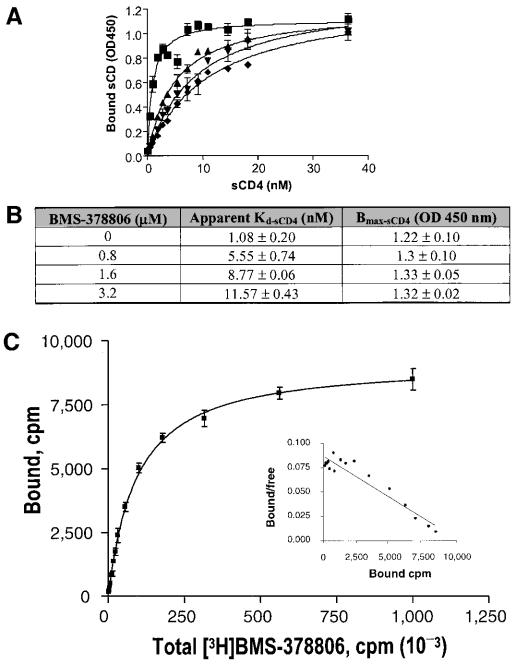
Results from our initial studies suggested that BMS-378806 blocks the interaction between viral gp120 and cellular CD4 receptors by an ELISA method (20a). However, it was not clear as to the mechanism by which BMS-378806 exerts its activity. To fully characterize the nature of BMS-378806 inhibition, a saturation binding experiment was performed, also using an ELISA method, to measure the apparent dissociation equilibrium constant (Kd) and the maximal binding capacity (Bmax) of sCD4 to gp120 under increasing amounts of the compound. Results showed concomitant increases in the apparent Kd values of sCD4 and the compound concentrations used (Fig. 2A), while the maximal level of sCD4 bound (Bmax) remained constant (Fig. 2A and B). These data imply that the higher the levels of BMS-378806, the more sCD4 it will take to reach the same level of sCD4 occupancy on gp120. Occupancy of gp120 by BMS-378806 can be completely overcome by the presence of high concentrations of sCD4. Therefore, the nature of the BMS-378806 inhibition to the binding of sCD4 to gp120 is in agreement with the classical mechanism of competitive inhibition (22, 29).
FIG. 2.
Competitive inhibition of gp120-CD4 binding by BMS-378806 and binding affinity of BMS-378806 to gp120. (A) Saturation binding curve of BMS-378806. The levels of sCD4 bound to gp120 in the absence or presence of various concentrations of BMS-378806 and in escalating concentrations of sCD4 were measured by using the gp120-CD4 binding ELISA. BMS-378806 concentrations: ▪, 0 μM; ▴, 0.8 μM; ▾, 1.6 μM; ⧫, 3.2 μM. Data were analyzed according to the one-site binding model with GraphPad Prism. OD450, optical density at 450 nm. (B) Apparent binding constants for sCD4 in the absence and presence of BMS-378806. The Kd and Bmax values were derived by using nonlinear regression analysis with GraphPad Prism. Values were the means ± standard errors of the means, representing two independent experiments. (C) Binding affinity of BMS-378806 to gp120 by SPA. [3H]BMS-378806 binding to the gp120 protein was measured by employing gp120-coated SPA beads. The amount of gp120-bound [3H]BMS-378806 was determined by using a Packard TopCount scintillation counter. The results from Scatchard analysis of binding isotherm are shown in the insert. The x axis represents bound (corrected), and the y axis indicates bound/free.
Binding affinity of BMS-378806 to gp120.
To further elucidate how BMS-378806 interferes with the binding of the gp120 protein to CD4, the binding of [3H]BMS-378806 to gp120JRFL was measured by an SPA (10, 27). Results showed that BMS-378806 binds directly to gp120, with a Kd of 21.1 ± 1.9 nM as estimated by a nonlinear least-squares curve fitting analysis (Fig. 2C). The binding isotherm is consistent with that of a single site on the target protein by Scatchard analysis (7, 33). By comparison, the binding of sCD4 to gp120JRFL or gp120BH10 gave Kd values of 28 ± 3.5 nM (40) or 22 ± 6 nM (25), respectively.
In confirming the specific binding of BMS-378806 to gp120, Biacore experiments showed that a strong resonance signal was seen with the gp120JRFL-coated biosensor chip upon compound binding but not with the sCD4-coated surface (data not shown). These results, as well as those reported elsewhere with a Micro Biospin column filtration method (20a), demonstrate that BMS-378806 binds directly and selectively to gp120.
Reversibility of BMS-378806 binding and Ki determination.
For BMS-378806 to have clinical application, a property to be avoided is the chemical reactivity. To address this concern, an initial study showed that the preincubation and subsequent removal of BMS-378806 from the gp120-coated plates resulted in negligible inhibitory effects on the sCD4 binding to gp120 and gave an early indication that the binding of BMS-378806 to gp120 was reversible (data not shown).
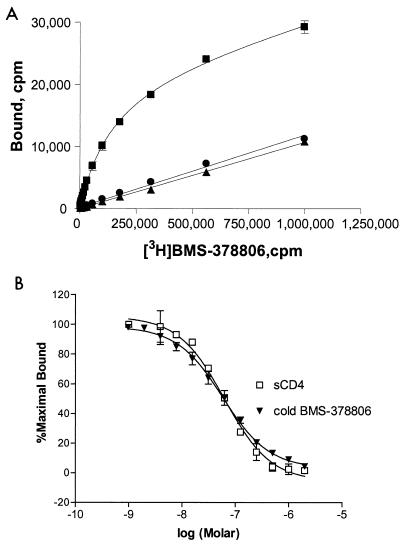
This observation was strengthened by the results showing that unlabeled BMS-378806 was able to displace the [3H]BMS-378806 bound to gp120JRFL by using an SPA method (Fig. 3A). Briefly, after mixing all assay components, signals were measured after 4 h to ensure equilibrium binding between [3H]BMS-378806 and gp120 (Fig. 3A). Unlabeled BMS-378806, at a concentration in 2,857-fold excess (60 μM) of the Kd, was then added to the labeled compound-protein complex and incubated for an additional 15 h. Binding of [3H]BMS-378806 was reduced (Fig. 3A) to the same level as control samples lacking gp120 (Fig. 3A). This reduction in the [3H]BMS-378806 signal was not due to label decay, since in a separate control experiment, the signal detected from the labeled compound bound to the beads remained constant over 1 week (data not shown). Thus, the displacement of [3H]BMS-378806 by unlabeled BMS-378806 implied that the specific binding of BMS-378806 to gp120 is reversible.
FIG. 3.
Reversibility of BMS-378806-gp120 binding and Ki determination. (A) Competition of the gp120-bound [3H]BMS-378806 by cold compound. Binding of [3H]BMS-378806 to gp120JRFL (monomer) was performed with the gp120-bound SPA. After reaching equilibrium (at 4 h) (squares), specific binding was reversed to background levels (circles) by the addition of 60 μM unlabeled BMS-378806 (2,857-fold excess of Kd, at 15 h). Background binding was similar to that of control SPA beads lacking bound gp120 (triangles). (B) Ki determination for BMS-378806 and sCD4. The Ki values were determined by adding 106 cpm of [3H]BMS-378806 to SPA reaction mixtures containing various concentrations of unlabeled BMS-378806 or sCD4. The data reported here were expressed as percentages of the maximal counts per minute bound and represented the averages of at least four sets of replicates.
An SPA-based competition assay was next performed to determine Ki values. Both BMS-378806 and sCD4 readily displaced [3H]BMS-378806 bound to gp120JRFL, and monophasic competition curves were obtained (Fig. 3B). The Ki values obtained for BMS-378806 (24.9 ± 0.8 nM) and sCD4 (24.5 ± 2.5 nM) were comparable. Together, the results from these studies indicate that BMS-378806 binds reversibly to the envelope protein and inhibits gp120-CD4 binding competitively with a binding affinity similar to that of sCD4.
Binding stoichiometry of BMS-378806 to envelope gp120.
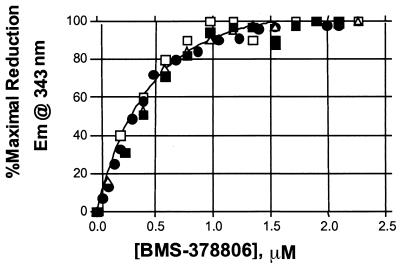
The binding stoichiometry was determined by estimating the molar ratios of gp120 and compound required to achieve a saturable fluorescence reduction upon complex formation. A total of 6 tryptophans are present in the gp120 core protein (17, 18). The positioning of 4 tryptophans at or near the CD4 binding pocket of gp120 prompted the speculation that BMS-378806 might interact with some of these residues. By monitoring the changes in tryptophan fluorescence emission (2, 5) from the gp120IIIB protein upon titrating increasing concentrations of BMS-378806 or an inactive analog, we were able to show that BMS-378806 binding to gp120 specifically induced a reduction in gp120 fluorescence (data not shown).
The binding stoichiometry for the BMS-378806-gp120 complex was then determined by titrating the compound into a solution containing 1.0 μM monomeric gp120JRFL protein. Under these conditions, a concentration-dependent, saturable fluorescence reduction was observed at an approximately 1:1 ratio of BMS-378806 to gp120JRFL (Fig. 4). The data imply that one molecule of BMS-378806 interacts with a single monomeric gp120 protein.
FIG. 4.
Binding stoichiometry of BMS-378806 to gp120 protein. gp120JRFL (1.0 μM) was titrated with BMS-378806, and the observed percent reduction in fluorescence from each of four independent experiments was plotted first as a function of BMS-378806 concentration (data not shown). Each of these data sets was then normalized to the percent maximal fluorescence reduction observed for each experiment in order to analyze the four sets of data together. The individual, normalized data sets are depicted as open squares, closed squares, closed diamonds, and open triangles. The curve represents the best fit to a saturation binding isotherm determined by using GraphPad Prism.
Lack of activity against infection by a CD4-independent HIV-1 variant.
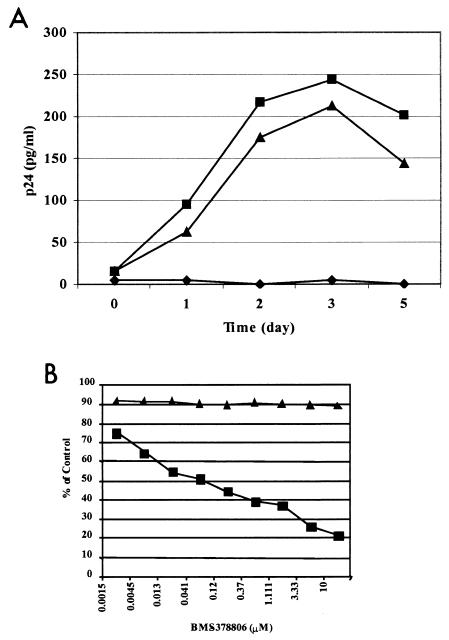
To determine whether the anti-HIV-1 activity of BMS-378806 is mediated exclusively via interference with gp120-CD4 binding, the effect of the compound was evaluated against a CD4-independent strain of HIV-1. A mutant virus (ADA S190R/N197S), shown previously to replicate in a CD4-negative Cf2Th CCR5+ cell line (14, 15), exhibited comparable replication rates measured for up to 5 days in both CD4-negative Cf2Th CCR5+ and Cf2Th CD4+ CCR5+ cells. This result indicated that the productive replication of this mutant HIV-1 does not require CD4, although the virus may prefer the host cells expressing CD4 (Fig. 5A). Subsequent drug susceptibility testing showed that BMS-378806 inhibited the replication of the mutant virus in Cf2Th CD4+ CCR5+ cells with an EC50 of 61 nM (Fig. 5B). However, a concentration of the compound of up to 10 μM failed to show inhibitory activity against the mutant virus in CD4-negative Cf2Th CCR5 cells (Fig. 5B). The lack of inhibitory activity in the context of a CD4-independent viral infection supports the previous conclusion that BMS-378806 inhibits viral infection primarily through interference with the interaction between gp120 and CD4.
FIG. 5.
Lack of inhibition against a CD4-independent HIV-1 variant. (A) Replication kinetics of the ADA-S190R/N197S variant in Cf2Th cell lines. Virus titers in the supernatants of Cf2Th-CD4+ CCR5+ (square), CD4-negative Cf2Th-CCR5+ (triangle), and Cf2Th-CD4− CCR5− (diamond) cell cultures following infection with the recombinant virus ADA-S190R/N197S were monitored with a p24 ELISA. (B) Effect of BMS-378806 on HIV infection of CD4-negative and CD4-positive cells. The antiviral activity of BMS-378806 in Cf2Th-CD4+ CCR5+ (square) and CD4-negative Cf2Th-CCR5+ (triangle) cell cultures was determined 3 days following viral infection with ADA-S190R/N197S. The results are presented as percentages of inhibition relative to their respective untreated infected cell cultures. Results from a representative experiment are shown.
Targeting a distinct region within the CD4 binding pocket of gp120.
To determine the BMS-378806 binding region within the gp120 protein, the binding activity of the compound was investigated by using drug-resistant gp120 variants and CD4 contact site envelope mutants. The BMS-378806-selected substitutions were identified previously as I595F, M475I, M434I/V, K655E, R350K, S440R, V68A, D185N, and M426L, all residing in the HIV-1 envelope (20a). To confirm the roles these changes have in resistance development, the substitutions were introduced into either HIV-1LAI or HIV-1NL4-3 envelope backgrounds and the resulting gp120 variants were examined in both envelope-mediated cell fusion and gp120-CD4 binding ELISAs. Individually, the changes in M426, M434, or M475 residues were all shown to exhibit significant decreases in susceptibility to BMS-378806 with both assays and in either virus background, indicating that they are the key resistant substitutions to BMS-378806. However, these variants maintained near normal levels of binding affinity to sCD4 (Table 1). The M434V and M434T substitutions also significantly affected the drug susceptibility of HIVLAI or HIVNL4-3 envelopes. Since both M426L and M475I substitutions reside at or near the CD4 contact residues of gp120 (18), it is highly probable that BMS-378806 targets the CD4 binding pocket of gp120. In contrast, substitutions V68A, S185N, R350K, S440R, I595F, and K655E only conferred low levels of or no resistance. V68A and I595F are the secondary changes that enhanced the resistance of other substitutions as evidenced by results from double-mutant studies (V68A/M426L, V68A/M434I, V68A/M475I, or M434I/I595F), whereas R350K appeared to temper the resistance of M475I (Table 1). Interestingly, the cell fusion data also showed that M426L, M434I, and M475I substitutions conferred greater resistance levels in the HIV-1NL4-3 envelope than the HIV-1LAI background, suggesting that viral background may influence the resistance phenotype (32). Data accumulated to date indicated that most of the BMS-378806-selected substitutions listed above play important roles in resistance development.
TABLE 1.
Effect of selective amino acid changes on BMS-378806 efficacy in both cell fusion and gp120/CD4 binding studies
| Envelope | Substitution(s)b | Cell fusion
|
gp120-CD4 binding
|
CD4 affinity | Closest CD4 contact point(s)c | ||
|---|---|---|---|---|---|---|---|
| EC50 (nM) | Reduction (fold)a | IC50 (μM) | Reduction (fold)a | ||||
| HIV-1LAI | None (wild type) | 8.8 | 1 | 0.33 | 1 | 1 | NAd |
| V68A | 40 | 5 | 0.31 | 1 | 0.8 | NA | |
| M426L | 1,588 | 180 | >30 | >91 | 1 | M426 | |
| M434I | 83 | 9 | 9.17 | 28 | 0.8 | V430 | |
| M434V | 140 | 16 | 22.30 | 67 | 0.9 | V430 | |
| S440R | 11.5 | 1 | 0.67 | 2 | 0.6 | V430 | |
| M475I | 1,491 | 169 | 27.51 | 83 | 1.2 | D474 | |
| V68A/M426L | 7,917 | 900 | >30 | >91 | 0.8 | NA/M426 | |
| V68A/M434I | 4,392 | 499 | NA | NA | 1.2 | NA/V430 | |
| V68/M475I | 4,038 | 459 | NA | NA | 1 | NA/D474 | |
| M434I/I595F | 3,297 | 374 | NA | NA | NA | V430/NA | |
| I595F | 45 | 5 | NA | NA | NA | NA | |
| K655E | 18 | 2 | NA | NA | NA | NA | |
| HIV-1NL4-3 | None (wild type) | 8.6 | 1 | 0.31 | 1 | 1 | NA |
| D185N | 13 | 2 | 0.21 | 1 | 1 | NA | |
| R350K | 3 | 1 | 0.27 | 1 | 1.1 | S365 | |
| M426L | 31,154 | 3,623 | >30 | >97 | 0.8 | M426 | |
| M434T | >1,000 | >116 | >30 | >97 | 0.7 | V430 | |
| M475I | 14,978 | 1,742 | >30 | >97 | 0.7 | D474 | |
| R350K/M475I | 882 | 103 | 3.95 | 13 | 0.9 | S365/D474 | |
Reduction in susceptibility.
The gp120-CD4 binding ELISA was used to determine both the inhibitory activity of BMS-378806 against sCD4 binding and the binding affinity of sCD4 to the respective gp120 variants. Individual substitutions that contribute to significant reduction in BMS-378806 susceptibilities are in bold.
CD4 contact points were referenced from reference 18.
NA, not available.
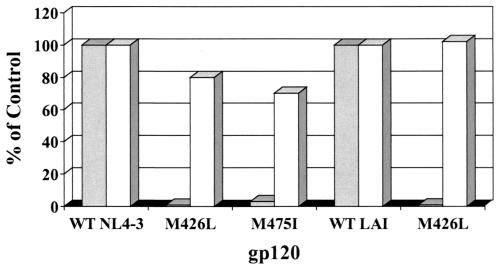
We next determined the binding activity of [3H]BMS-378806 to HIV-1NL4-3 and HIV-1LAI gp120 proteins harboring either the M426L or M475I major substitution. These two changes severely affected compound binding as determined by a FlashPlate assay, resulting in ∼17- to 100-fold binding decreases compared to that of the wild-type gp120 (Fig. 6). In contrast, sCD4 bound equally well to both wild-type and resistant gp120 proteins (Fig. 6). Similar results were obtained with the HIV-1JRFL strain carrying two aforementioned substitutions (data not shown). Importantly, these collective results imply that the compound may target the CD4 binding pocket of gp120 and share binding residues with sCD4.
FIG. 6.
Binding of BMS-378806 to gp120-resistant variants is significantly reduced. The y axis denotes the percentage of BMS-378806 binding to variant gp120 proteins versus the envelope protein of corresponding wild-type HIV-1 strain (control). The x axis represents the various gp120 proteins tested. The data from the [3H]BMS-378806 binding experiment are shown in gray, and those from sCD4 are depicted in white.
To further study the potential binding locations of BMS-378806 in the gp120-sCD4 binding cavity, several CD4 contact site mutants (D368R, E370R, I371F, W427V, and D457R) (18) were generated. Among them, D368R, E370R, W427V, and D457R were previously reported to be defective in gp120-CD4 binding (26). Affinity of these gp120 variant proteins to sCD4 was determined by the gp120-CD4 binding ELISA, while affinity to the compound was assessed by the Micro BioSpin binding assay with [3H]BMS-378806. As expected, all five mutations significantly reduced gp120 binding affinity for CD4 (Table 2). However, only W427V completely negated sCD4 and BMS-378806 binding, while the D368R, E370R, E371F, and D457R mutations had modest or no effects on BMS-378806 binding. This result implies that W427 plays a role in mediating compound binding to gp120. Furthermore, the S375W change, with the tryptophan residue filling in the CD4-Phe43 binding pocket, was recently shown to cause gp120 to assume a conformation closer to the CD4-bound state (39). Here, the S375W protein variant was also shown to completely lose the ability to bind to BMS-378806 (Table 2), indicating that the compound binding site may be situated in this CD4-Phe43 binding pocket of gp120.
TABLE 2.
Binding activity of gp120 protein variants to BMS-378806 or sCD4
| gp120 mutation | Criterion | Relative sCD4 affinity to gp120HXB2a | Relative sCD4 affinity to gp120JRFLb | Relative [3H]BMS-378806 affinity to gp120JRFLc | Direct or closest CD4 contact pointd |
|---|---|---|---|---|---|
| None (wild type) | 1 | 1 | 1 | ||
| D368R | Important for CD4 binding | <0.004 | <0.01 | 0.97 | Yes |
| E370R | Important for CD4 binding | <0.003 | <0.01 | 0.14 | Yes |
| I371F | Important for CD4 binding | Not reported | 0.01 | 0.54 | Yes |
| W427V | Important for CD4 binding | <0.006 | <0.01 | <0.01 | Yes |
| D457R | Important for CD4 binding | 0.15 | Not done | 1.16 | Yes |
| S375W | Phe43 cavity | Not reported | 1.66 | 0.02 | I371 |
Relative affinity of CD4 to the gp120HXB2 variants was reported in reference 26.
Relative sCD4 affinities to gp120JRFL variants were determined with the gp120-CD4 binding ELISA.
Relative binding affinity of BMS-378806 to gp120JRFL variants. The Micro BioSpin column assay was employed to measure the quantity of [3H]BMS-378806 bound to gp120JRFL. Substitutions that significantly impacted both BMS-378806 and sCD4 binding to gp120 are in bold.
CD4 contact point information was referenced from reference 18.
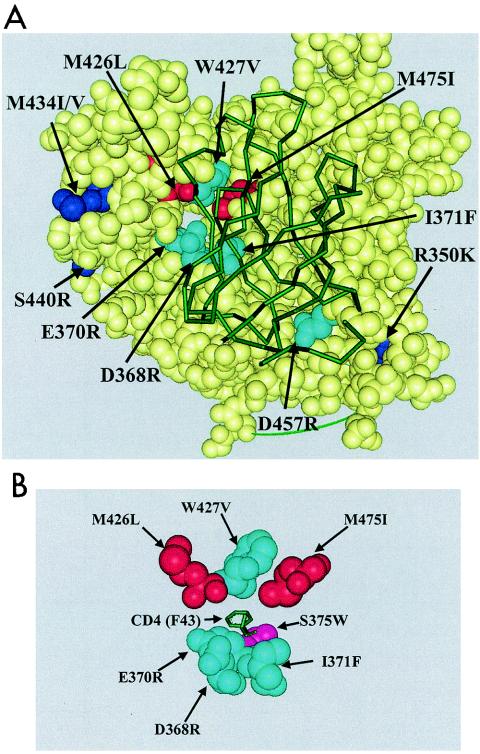
The mechanisms by which these CD4 contact site mutations affect compound binding may include conformational alterations, steric hindrance, or involvement in the actual binding to BMS-378806. However, it is intriguing that the W427 residue, judged important for compound binding by these approaches, is geographically clustered with the M426 and M475 resides, which were shown to be relevant to compound binding via resistance studies. Together, the data suggest that this compound may interact with a select subset of gp120 residues, including M426, W427, and M475. Most importantly, these data localized the BMS-378806 binding to a site deep within the CD4-Phe43 pocket of gp120 (18) (Fig. 7A). The spatial relationships between these residues, S375W (located at the base of the CD4-Phe43 pocket) and CD4 Phe43 residue are shown in Fig. 7B. The S375W residue lies just below the Phe43 residue of CD4, at the base of the CD4 binding pocket of gp120.
FIG. 7.
Locations of BMS-378806-selected resistant substitutions and CD4 contact site mutations on gp120. (A) A space-filled model with atom spheres sized to the van der Waals radii was prepared with WebLab Viewerpro 4.0 (Accelrys, Inc., San Diego, Calif.). Yellow spheres, gp120 residues; red spheres, BMS-378806-selected substitutions located in the CD4 binding pocket (M426L and M475I); dark blue spheres, BMS-378806-selected substitutions (R350K, M434I/V, and S440R); turquoise spheres, CD4 contact site mutations (D368R, E370R, I371F, W427V, and D457R). The sCD4 protein is represented by a green diagram overlapping gp120. (B) Placement of the S375W residue. A simplified model shows only the relative locations of the CD4 contact site mutations (in turquoise) and the BMS-378806-selected substitutions (in red). The S375W residue (in pink) lies just below the Phe43 (F43) residue of CD4 (in green), at the base of the CD4 binding pocket.
DISCUSSION
Previous results from time-of-addition, cell fusion, and resistance studies suggested that the HIV-1 envelope is the likely target for BMS-378806 (20a). Here, we demonstrate that BMS-378806 inhibits the interaction between gp120 and sCD4 via a competitive mechanism and that this inhibition is the result of a specific compound binding to gp120 and not to CD4 (Fig. 2). This interaction is reversible (Fig. 3), and the compound binds to the gp120 monomer with a stoichiometry of approximately 1:1 (Fig. 4). BMS-378806-directed inhibition of viral replication appears to be mediated exclusively through the inhibition of gp120-CD4 interactions, as it was shown to be inactive during CD4-independent infection (Fig. 5).
Results from direct binding assays (Fig. 2C) of BMS-378806 to gp120JRFL generated a Kd of 21 nM, which is similar to published reports of CD4 binding to gp120JRFL (Kd of 28 nM) (40) and gp120BH10 (Kd of 22 nM) (25). Interestingly, data from binding competition assays (Fig. 3) indicated a Ki value of 24.9 nM for BMS-378806 binding to gp120JRFL, which was similar to that of sCD4 (24.5 nM). The comparable values imply that BMS-378806 interacts with gp120 with a binding affinity similar to that of sCD4. However, the Ki obtained is ∼10-fold higher than the potency (EC50) of BMS-378806 against the HIV-1JRFL infection. This difference is possibly due to varying degrees of sensitivities between two dissimilar assays. In addition, a monomeric form of soluble gp120 was used in the in vitro binding studies, whereas a trimeric gp120-gp41 structure is believed to be an integral part of the viral envelope in the anti-HIV infection assays. This structural disparity is also likely to contribute to the observed differences in Ki values and EC50s, as the binding of an inhibitor onto a gp120 subunit of a virion-associated envelope protein could now have an impact on its neighboring gp120 as well as the gp41 subunits in envelope trimers. The mode of gp120-inhibitor interaction with regard to virion particle-associated HIV-1 gp120 is currently under investigation.
Data from biochemical experiments indicated that BMS-378806 binding to gp120 is competitive with sCD4 (Fig. 2A and B), implying that BMS-378806 (molecular mass, 406 Da) may share some of the same binding determinants with sCD4 (molecular mass, 40,000 Da). However, it is puzzling how the binding of a small molecule to the gp120 cavity could possibly result in the exclusion of the much larger CD4 protein. One plausible explanation is that conformational (allosteric) changes may occur in gp120 upon compound binding and prevent the subsequent binding to CD4 receptors. Another possibility is that compound binding occupies a crucial structural determinant required for productive CD4 docking. Understanding the detailed interaction between BMS-378806 and gp120 will ultimately require an X-ray crystallographic study of the compound-gp120 complex, which will be a formidable task. To date, the only available crystal structure of gp120 was obtained with a gp120 core complexed with sCD4 and 17b antibody. Unfortunately, the exclusion of sCD4 by BMS-378806 complicates the cocrystallization work. Therefore, an initial determination of the BMS-378806 target site was attempted by using a combination of genetic and biochemical approaches.
Previously generated HIV-1 envelope variants harboring the BMS-378806-selected substitutions provide a set of unique tools for mapping the compound-gp120 interaction sites. These substitutions (V68A, S185N, R350K, M426L, M434I/V, S440R, M475I, I595F, and K655E) are mapped to the entire gp160 envelope (20a). This result is not surprising, since gp120 protein functions largely through conformational changes and a conformation usually consists of discontinuous regions of gp120. Nonetheless, among the BMS-378806-selected gp120 changes, M426L, M434I, and M475I emerged as the major resistance substitutions, based on the magnitude of effects on compound susceptibility in cell fusion and gp120-CD4 binding assays (Table 1). The M434I substitution, located outside the CD4 binding pocket, is likely to confer a drug resistance phenotype by inducing gp120 conformational changes. Importantly, both M426L and M475I changes, situated near each other, are located at or near the CD4 contact residues in the gp120 structure model (18). The results suggested that BMS-378806 might interact with residues 426 and 475 of gp120.
To further define the binding mechanism of the compound, we postulated that if BMS-378806 targets a critical CD4 contact site on gp120, the mutations at this particular site should abolish the binding of both BMS-378806 and sCD4 to gp120. This proposal was tested with the gp120-CD4 contact site information published previously (18) as well as the gp120 mutations known to be defective in CD4 binding (26). Our data (Table 2) showed that of the five gp120-CD4 contact site mutations studied (D368R, E370R, I371F, W427V, and D457R) that are defective in CD4 binding, only W427V was completely devoid of binding to BMS-378806 (<1%) while E370R retained 14% of the activity. Therefore, the W427V mutation eliminated the ability of gp120 to bind to both the compound and sCD4. Importantly, in the gp120 structural models (17, 18), the W427V change and the major BMS-378806-selected substitutions, M426L and M475I, are clustered at one distinct region of the CD4 binding pocket, with the E370R residue residing in close proximity (Fig. 7). In other words, these three geographically proximal mutations of gp120 are each capable of negating the binding of gp120 to BMS-378806. The simplest explanation of this finding is that these three gp120 residues are involved in compound interactions.
Next, the determination of whether BMS-378806 shares the same cavity as the Phe43 residue of sCD4 was carried out. A recent report (39) indicated that the S375W change, located near the binding site of the Phe43 residue of CD4, allows the gp120 protein to assume a conformation closer to the CD4-bound form. We found that the gp120 variant carrying this mutation, with tryptophan filling the Phe43 cavity, was also greatly defective in BMS-378806 binding (Table 2). These data suggested that the compound may bind to the same cavity as the Phe43 residue of sCD4. The S375 residue is also proximal to residues M426, W427, and M475, judging from the published gp120 structural models (Fig. 7B). However, we caution that an accurate understanding of the BMS-378806 binding site requires structural information of a pre-CD4-bound form of gp120, which is currently unavailable. Furthermore, the interaction of various gp120 loops with the compound also needs to be addressed.
HIV envelope proteins are highly variable and flexible, allowing viruses to effectively evade the host immune response. The challenges facing those who are in search of a vaccine highlight the degree of difficulty in locating conserved epitopes on the HIV-1 envelope. The gp120 crystal structures provide the first insight into the structural orientation of sCD4 when bound to the gp120 core protein and reveal the presence of a conserved hydrophobic pocket within gp120 which serves as a contact point with sCD4 (17, 18). Numerous attempts to design CD4 mimicking peptides as anti-HIV agents have yielded disappointing clinical results (6, 34). Only recently has PRO542 demonstrated antiviral efficacy in phase I/II trial, presumably due to its improved protein stability and the enhanced affinity to gp120 (11, 35). However, this large protein precludes the development of an oral formulation. Nonetheless, its positive clinical result provides a proof of concept for HIV attachment inhibitors and validates the CD4 binding pocket on HIV-1 as a viable drug target. BMS-378806, which targets this same pocket, and can be delivered orally, represents a prototype for a new class of HIV-1 inhibitors.
In summary, BMS-378806 is a representative of a new class of HIV-1 attachment inhibitors whose mechanism of action is to selectively bind to the viral envelope protein and inhibit binding to cellular CD4 receptors, thereby inhibiting HIV-1 infection. This compound may also be useful as a tool to probe the structure of gp120 and to help further the understanding of the HIV entry process.
REFERENCES
- 1.Blair, W. S., P. F. Lin, N. A. Meanwell, and P. B. Wallace. 2000. HIV-1 entry-an expanding portal for drug discovery. Drug Discov. Today 5:183-194. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, I. D., and R. A. Dwek. 1984. Biological spectroscopy. Benjamin/Cummings, Menlo Park, Calif.
- 3.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, O. J., and A. S. Fauci. 2001. Current strategies in the treatment of HIV infection. Adv. Intern. Med. 46:207-246. [PubMed] [Google Scholar]
- 5.Copeland, R. A. 2000. A practical introduction to structure, mechanism, and data analysis, p. 104. In V. C. H. Wiley (ed.), Enzymes, 2nd ed. John Wiley & Sons, New York, N.Y.
- 6.Daar, E. S., X. L. Li, T. Moudgil, and D. D. Ho. 1990. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc. Natl. Acad. Sci. USA 87:6574-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahlquist, F. 1978. The meaning of Scatchard and Hill plots. Methods Enzymol. 48:270-299. [DOI] [PubMed] [Google Scholar]
- 8.D'Souza, M. P., J. S. Cairns, and S. F. Plaeger. 2000. Current evidence and future directions for targeting HIV entry. JAMA 284:215-222. [DOI] [PubMed] [Google Scholar]
- 9.Grant, R. M., F. M. Hecht, M. Warmerdam, L. Liu, T. Liegler, C. J. Petropoulos, N. S. Hellmann, M. Chesney, M. P. Busch, and J. O. Kahn. 2002. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA 288:181-188. [DOI] [PubMed] [Google Scholar]
- 10.Hood, C. M., V. A. Kelly, M. I. Bird, and C. J. Britten. 1998. Measurement of a(1-3)fucosyltransferase activity using scintillation proximity assay. Anal. Biochem. 255:8-12. [DOI] [PubMed] [Google Scholar]
- 11.Jacobson, J. M., I. Lowy, C. V. Fletcher, T. J. O'Neill, D. N. Tran, T. J. Ketas, A. Trkola, M. E. Klotman, P. J. Maddon, W. C. Olson, and R. J. Israel. 2000. Single-dose safety, pharmacology, and antiviral activity of the human immunodeficiency virus (HIV) type 1 entry inhibitor PRO 542 in HIV-infected adults. J. Infect. Dis. 182:326-329. [DOI] [PubMed] [Google Scholar]
- 12.Kilby, J., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 13.Kilby, J. M., J. P. Lalezari, J. J. Eron, M. Carlson, C. Cohen, R. C. Arduino, J. C. Goodgame, J. E. Gallant, P. Volberding, R. L. Murphy, F. Valentine, M. S. Saag, E. L. Nelson, P. R. Sista, and A. Dusek. 2002. The safety, plasma pharmacokinetics, and antiviral activity of subcutaneous enfuvirtide (T-20), a peptide inhibitor of gp41-mediated virus fusion, in HIV-infected adults. AIDS Res. Hum. Retrovir. 18:685-693. [DOI] [PubMed] [Google Scholar]
- 14.Kolchinsky, P., E. Kiprilov, P. Bartley, R. Rubinstein, and J. Sodroski. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolchinsky, P., T. Mirzabekov, M. Farzan, E. Kiprilov, M. Cayabyb, L. J. Mooney, H. Choe, and J. Sodroski. 1999. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J. Virol. 73:8120-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korber, B., F. Foley, C. Kuiken, S. Pillai, and J. Sodroski. 1998. Numbering positions in HIV relative to HXBc2, p. III-102-III-103. In B. Korber, C. Kuiken, B. Foley, B. H. Hahn, F. E. McCutchan, J. W. Mellors, and J. Sodroski (ed.), Human retroviruses and AIDS. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 17.Kwong, P. D., R. Wyatt, S. Mcajeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 18.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaBranche, C. C., G. Galasso, J. P. Moore, D. Bolognesi, M. S. Hirsch, and S. M. Hammer. 2001. HIV fusion and its inhibition. Antivir. Res. 50:95-115. [DOI] [PubMed] [Google Scholar]
- 20.Lalezeri, J. P., K. Henry, M. O'Hearn, J. S. G. Montaner, P. Piliero, B. Trottier, S. Walmsley, C. Cohen, D. R. Kuritzkes, J. J. Eron, J. Chung, R. DeMasi, L. Donatacci, C. Drobnes, J. Delehanty, M. Salgo, and the TORO 1 Study Group. 2003. Enfurvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 348:2175-2185. [DOI] [PubMed] [Google Scholar]
- 20a.Lin, P. F., W. Blair, T. Wang, T. Spicer, Q. Guo, N. Zhou, Y. F. Gong, H.-G. H. Wang, R. Rose, G. Yamanaka, B. Robinson, C. B. Li, R. Fridell, C. Deminie, G. Demers, Z. Yang, L. Zadjura, N. Meanwell, and R. Colonno. A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 21.Little, S. J., S. Holte, J.-P. Routy, E. Daar, M. Markowitz, A. C. Collier, R. A. Koup, J. W. Mellors, E. Connick, B. Conway, M. Kilby, L. Wang, J. M. Whitcomb, N. S. Hellmann, and D. D. Richman. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347:385-394. [DOI] [PubMed] [Google Scholar]
- 22.Masuda, Y., T. Sugo, T. Kikuchi, A. Kawata, M. Satoh, Y. Fujisawa, Y. Itoh, M. Wakimasu, and T. Ohtaki. 1996. Receptor binding and antagonist properties of a novel endothelin receptor antagonist, TAK-044 [cyclo[D-alpha-aspartyl-3-[(4-phenylpiperazin-1-yl) carbonyl]-L-alanyl-L-alpha-aspartyl-D-2-(2-thienyl) glycyl-L-leucyl-D-tryptophyl]disodium salt], in human endothelin A and endothelin B receptors. J. Pharmacol. Exp. Ther. 279:675-685. [PubMed] [Google Scholar]
- 23.Moore, J. P. 1990. Simple methods for monitoring HIV-1 and HIV-2 gp120 binding to soluble CD4 by enzyme-linked immunosorbent assay: HIV-2 has a 25-fold lower affinity than HIV-1 for soluble CD4. AIDS 4:297-305. [DOI] [PubMed] [Google Scholar]
- 24.Moore, J. P., and R. F. Jarrett. 1988. Sensitive ELISA for the gp120 and gp160 surface glycoproteins of HIV-1. AIDS Res. Hum. Retrovir. 4:369-379. [DOI] [PubMed] [Google Scholar]
- 25.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olshevsky, U., E. Helseth, C. Furman, J. Li, W. Haseltine, and J. Sodroski. 1990. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J. Virol. 64:5701-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel, S., A. Harris, G. O'Beirne, N. D. Cook, and C. W. Taylor. 1996. Kinetic analysis of inositol trisphosphate binding to pure inositol trisphosphate receptors using scintillation proximity assay. Biochem. Biophys. Res. Commun. 221:821-825. [DOI] [PubMed] [Google Scholar]
- 28.Paul, N. L., M. Marsh, J. A. McKeating, T. F. Schulz, P. Liljestrom, H. Garoff, and R. A. Weiss. 1993. Expression of HIV-1 envelope glycoproteins by Semliki Forest virus vectors. AIDS Res. Hum. Retrovir. 9:963-970. [DOI] [PubMed] [Google Scholar]
- 29.Pedersen, J. B., and W. E. Lindup. 1994. Interpretation and analysis of receptor binding experiments which yield non-linear Scatchard plots and binding constants dependent upon receptor concentration. Biochem. Pharmacol. 47:179-185. [DOI] [PubMed] [Google Scholar]
- 30.Pierson, T. C., and R. W. Doms. 2003. HIV-1 entry inhibitors: new targets, novel therapies. Immunol. Lett. 85:113-118. [DOI] [PubMed] [Google Scholar]
- 31.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 32.Rose, R. E., Y. F. Gong, J. A. Greytok, C. M. Bechtold, B. J. Terry, B. S. Robinson, M. Alam, R. J. Colonno, and P. F. Lin. 1996. Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors. Proc. Natl. Acad. Sci. USA 93:1648-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scatchard, G. 1949. The attractions of proteins for small molecules and ions. Ann. N. Y. Acad. Sci. 51:660-672. [Google Scholar]
- 34.Schacker, T., R. W. Coombs, A. C. Collier, J. E. Zeh, I. Fox, J. Alam, K. Nelson, E. Eggert, and L. Corey. 1994. The effects of high-dose recombinant soluble CD4 on human immunodeficiency virus type 1 viremia. J. Infect. Dis. 169:37-40. [DOI] [PubMed] [Google Scholar]
- 35.Shearer, W. T., R. J. Israel, S. Starr, C. V. Fletcher, D. Wara, M. Rathore, J. Church, J. DeVille, T. Fenton, B. Graham, P. Samson, S. Staprans, J. McNamara, J. Moye, P. J. Maddon, and W. C. Olson. 2000. Recombinant CD4-IgG2 in human immunodeficiency virus type 1-infected children: phase 1/2 study. J. Infect. Dis. 182:1774-1779. [DOI] [PubMed] [Google Scholar]
- 36.Volberding, P. 1999. Advances in the medical management of patients with HIV-1 infection: an overview. AIDS 13:S1-S9. [PubMed] [Google Scholar]
- 37.Wegner, S. A., S. K. Brodine, J. R. Mascola, S. A. Tasker, R. A. Shaffer, M. J. Starkey, A. Barile, G. J. Martin, N. Aronson, W. W. Emmons, K. Stephan, S. Bloor, J. Vingerhoets, K. Hertogs, and B. Larder. 2000. Prevalence of genotypic and phenotypic resistance to anti-retroviral drugs in a cohort of therapy-naive HIV-1 infected US military personnel. AIDS 14:1009-1015. [DOI] [PubMed] [Google Scholar]
- 38.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 39.Xiang, S. H., P. D. Kwong, R. Gupta, C. D. Rizzuto, D. J. Casper, R. Wyatt, L. Wang, W. A. Hendrickson, M. L. Doyle, and J. Sodroski. 2002. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 76:9888-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, W., A. P. Godillot, R. Wyatt, J. Sodroski, and I. Chaiken. 2001. Antibody 17b binding at the coreceptor site weakens the kinetics of the interaction of envelope glycoprotein gp120 with CD4. Biochemistry 40:1662-1670. [DOI] [PubMed] [Google Scholar]