Abstract
To evaluate the role of Epstein-Barr Virus (EBV) nuclear antigen 3A (EBNA3A) in the continuous proliferation of EBV-infected primary B lymphocytes as lymphoblastoid cell lines (LCLs), we derived LCLs that are infected with a recombinant EBV genome that expresses EBNA3A fused to a 4-hydroxy-tamoxifen (4HT)-dependent mutant estrogen receptor hormone binding domain (EBNA3AHT). The LCLs grew similarly to wild-type LCLs in medium with 4HT despite a reduced level of EBNA3AHT fusion protein expression. In the absence of 4HT, EBNA3AHT moved from the nucleus to the cytoplasm and was degraded. EBNA3AHT-infected LCLs were unable to grow in medium without 4HT. The precise time to growth arrest varied inversely with cell density. Continued maintenance in medium without 4HT resulted in cell death, whereas readdition of 4HT restored cell growth. Expression of other EBNAs and LMP1, of CD23, and of c-myc was unaffected by EBNA3A inactivation. Wild-type EBNA3A expression from an oriP plasmid transfected into the LCLs protected the EBNA3AHT-infected LCLs from growth arrest and death in medium without 4HT, whereas EBNA3B or EBNA3C expression was unable to protect the LCLs from growth arrest and death. These experiments indicate that EBNA3A has a unique and critical role for the maintenance of LCL growth and ultimately survival. The EBNA3AHT-infected LCLs are also useful for genetic and biochemical analyses of the role of EBNA3A domains in LCL growth.
Epstein-Barr virus (EBV), a human gammaherpesvirus, can cause lymphocyte-proliferative diseases in immune-deficient people and is also etiologically associated with Burkitt's lymphoma, Hodgkin's disease, other B- and T-cell lymphomas, anaplastic nasopharyngeal carcinoma, and a small fraction of gastric carcinomas (for a review see reference 47). When EBV infects primary human B lymphocytes, they are efficiently transformed into continuously proliferating lymphoblastoid cell lines (LCLs) (19, 44). In LCLs EBV expresses six nuclear proteins (EBNA1, -2, -3A, -3B, -3C, and -LP), three integral membrane proteins (LMP1, -2A, and -2B), two small nonpolyadenylated RNAs (EBER1 and EBER2), and Bam A rightward transcripts (for a review see reference 30). EBNA1, -2, -3A, -3C, and -LP and LMP1 are necessary for LCL outgrowth, whereas the rest of the EBV genome is dispensable.
EBNA2 and EBNALP are expressed first in primary B-lymphocyte infection and coactivate transcription from cell and viral promoters (1, 17, 40). EBNA2 associates with the sequence-specific DNA binding protein RBP-Jκ/CBF-1/CSL and activates transcription from promoters containing RBP-Jκ binding sites (14, 18). EBNA2 activates the cell CD21, CD23, c-fgr, and c-myc promoters and the viral EBNA and LMP promoters and thereby has a crucial role in the conversion of primary human B lymphocytes into LCLs (1, 3, 8-10, 13, 16, 24, 31, 36, 37, 42, 43, 57-60, 64). EBNALP coactivates transcription with EBNA2 (1, 17, 40).
The role of EBNA3A, EBNA3B, and EBNA3C in LCL outgrowth and continued proliferation is only partially delineated. EBNA3A, EBNA3B, and EBNA3C are encoded by three related, tandem genes (for a review see reference 30). EBNA3A and EBNA3C are essential for EBV-mediated primary B-lymphocyte conversion to LCLs, whereas EBNA3B is dispensable (29, 52, 53). Like EBNA2, EBNA3A, EBNA3B, and EBNA3C stably associate with RBP-Jκ (22, 32, 39, 45, 48, 49, 56, 62). Altogether, EBNA2, EBNA3A, EBNA3B, and EBNA3C are associated with most of the cell's RBP-Jκ (8, 22, 39). This association potentially limits EBNA2 transcriptional up-regulation (8, 22). In transient-transfection assays, EBNA3A, EBNA3B, and EBNA3C reduce EBNA2 activation of the EBNA Cp promoter (6, 12, 34, 39, 45, 56). Furthermore, three- to fivefold overexpression of EBNA3A in an LCL disrupts EBNA2 association with RBP-Jκ, down-regulates c-myc, CD21, and CD23, and causes G0/G1 growth arrest (8). EBNA3C and to a lesser extent EBNA3A and EBNA3B can also coactivate the LMP1 promoter with EBNA2 (2, 35, 39, 63). EBNA3A, EBNA3B, and EBNA3C have EBV type common and evolutionarily conserved domains (12, 21, 50, 61). EBNA3A and EBNA3C have domains that can activate or repress transcription when fused to the Gal4 DNA binding domain in reporter assays with promoters that have multiple upstream Gal4 binding sites (4-6, 12, 39, 46). EBNA3A and -3C interact with cellular proteins that may mediate transcriptional activation or repression or affect cell growth (11, 15, 20, 26-28, 35, 41, 46, 51).
The experiments reported here focus on the role of EBNA3A in continued LCL proliferation. While recombinant genetic analyses placing a nonsense codon after codon 302 of the EBNA3A gene indicate that EBNA3A is critical for EBV-induced LCL outgrowth, two LCLs that have only a mutant EBNA3A were derived (29, 53). The growth properties of these LCLs were not characterized (29). An EBV-infected cell line derived from a leukemic patient also lacks EBNA3A, although the growth of these cells has not been shown to be dependent on EBV gene expression (33). Thus, EBNA3A may be required for initial LCL outgrowth but may not be required for continued LCL proliferation.
To investigate the role of EBNA3A in maintaining LCL growth, we have derived LCLs infected with an EBV recombinant genome that expresses a conditionally active EBNA3A. The EBNA3A open reading frame was fused in frame to a 4-hydroxy-tamoxifen (4HT)-dependent mutant estrogen receptor hormone binding domain (ERTM) (38) to create an open reading frame that encodes putative conditionally active EBNA3A EBNA3AHT. ERTM was preferred over ER because ERTM is not responsive to estrogen in serum or to phenol red dye in tissue culture medium and 4HT does not activate endogenous estrogen receptors (38). EBNA3AHT is anticipated to be docked, inactive, and possibly destabilized in the cytoplasm in the absence of 4HT added to cell culture medium and active in the cell nucleus in response to 4HT addition to the cell culture medium. We have also explored whether conditional EBNA3AHT expression in an LCL may be useful for genetic analysis of the role of EBNA3A in LCL growth.
MATERIALS AND METHODS
Cells.
BJAB is an EBV-negative B-lymphoma cell line. The HH514-16 subclone of the EBV-infected P3HR-1 (clone 16) Burkitt's lymphoma cell line (a gift from G. Miller) contains a type 2 EBV genome that has the EBNA2 open reading frame and part of the EBNALP open reading frame deleted, rendering it nontransforming. IB4 is a lymphoblastoid cell line transformed with type 1 EBV. Stable IB4 transfectants which express FLAG-tagged EBNA3A fused to the 4HT-dependent estrogen receptor hormone binding domain (IB4-fE3AHT) were made by cotransfecting PvuI-digested pcDNA3 (Invitrogen) and PvuI-digested pSG5-fE3AHT into IB4 cells, followed by selection with 0.8 mg of G418/ml. After identification, IB4-fE3AHT-expressing cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), l-glutamine, streptomycin, penicillin, and 0.4 mg of G418/ml. Primary human mononuclear cells were obtained from the blood of healthy adults or normal placentas by centrifugation over a cushion of Lymphoprep (Axis-Shield). Newly transformed EBNA3AHT LCLs were maintained in RPMI 1640 medium supplemented with 15% FBS, l-glutamine, streptomycin, penicillin, and 200 nM 4HT. Newly transformed control LCLs were maintained in the same medium, except for the absence of 4HT. All other cell lines were maintained in RPMI 1640 medium supplemented with 10% FBS, l-glutamine, streptomycin, and penicillin. Viable-cell numbers were determined by hemocytometer based on trypan blue exclusion.
Plasmids and cosmid construction.
pSG5-flagEBNA3A, -EBNA3B, and -flagEBNA3C have been described previously (8, 35, 48, 49). The EBNA3A gene stop codon in pSG5-flagEBNA3A was replaced with a BamHI site by PCR. A BamHI-SalI fragment encoding the 4HT-responsive modified estrogen receptor hormone binding domain (HTER) from pBSKS+ERTM (a gift from T. Littlewood) was subcloned into the BamHI and XhoI sites of this modified pSG5-flagEBNA3A (pSG5-fE3AHT), resulting in in-frame fusion of FLAG-EBNA3A and HTER genes. Cosmid XF contains an XbaI (B95-8 bp 81853)-to-FseI (B95-8 bp 118079) fragment of B95-8 EBV DNA. A BssHII (B95-8 bp 94051)-to-StuI (B95-8 bp 95158) fragment of cosmid XF was replaced by a BssHII-EcoRV fragment of pSG5-fE3AHT which contains the coding sequence for the EBNA3A C terminus fused in frame to the HTER hormone binding domain (XF-EBNA3AHT). For creation of EBV recombinants, EcoRI A was used to marker rescue the P3HR1 EBV EBNA2 and EBNALP deletion mutation in P3HR-1 clone 16 cells. A cosmid that bridges between the mutated EBNA3A cosmid XF-EBNA3AHT and EcoRI A, pSECΔHpaI, has an HpaI-to-HpaI deletion (B95-8 bp 86020 to 101932) made with SalI E/C; the deleted segment contains EBNA3 genes. Lytic EBV replication was induced with pSVNaeI Z. To make the oriP plasmids expressing FLAG, enhanced green fluorescent protein (EGFP), FLAG-EBNA3A, EBNA3B, or FLAG-EBNA3C under the control of the simian virus 40 (SV40) promoter, SalI fragments containing SV40 promoter-driven FLAG, EGFP, FLAG-EBNA3A, EBNA3B, or FLAG-EBNA3C cassettes from pSG5-flag, -EGFP, -flagEBNA3A, -EBNA3B, or -flagEBNA3C, respectively, were subcloned into the SalI-digested pCEP4 vector (Invitrogen).
Generation of recombinant EBV.
Ten micrograms of linearized cosmid EcoRI A DNA, 40 μg of linearized cosmid XF-EBNA3AHT, and 20 μg of pSECΔHpaI were mixed with 40 μg of pSVNaeI Z and electroporated into 107 P3HR-1 clone 16 log-phase cells in 400 μl of complete medium in a cuvette (0.4-cm gap; Bio-Rad). Following 10 min at room temperature, the culture was pulsed with 200 V at 960 μF. Transfected P3HR-1 cells were cultured in 14 ml of complete medium for 5 days to allow EBV replication and release of virus into the culture medium. Lytic EBV infection was induced in B LCLs by transfection with 40 μg of pSVNaeI Z under similar conditions.
Primary B-lymphocyte infections.
Cell-free virus was prepared from transfected P3HR-1 or LCL cells by rapid freeze-thaw and centrifugation to remove cell debris. Primary human mononuclear cells were infected with virus at 37°C for 1 to 2 h, washed, and resuspended in complete medium containing 4HT. Aliquots of 200 μl were distributed into each well of 96-microwell plates (5 × 104 to 20 × 104 cells per well). Every 7 days after plating, 50% of the medium was replaced with fresh complete medium containing 4HT. LCLs were macroscopically visible 3 to 5 weeks after plating. LCL subclones singly infected with recombinant EBV were obtained from coinfected LCLs by limiting dilution. One or 10 LCL cells were cultured with 104 irradiated (8,800 cGy) IB4 cells in complete medium containing 4HT in 96-microwell plates. Every 7 days after plating, 50% of the medium was replaced with fresh complete medium containing 4HT. LCL subclones emerged at between 3 and 4 weeks after plating. Alternatively, recombinant EBV was segregated from the coinfecting P3HR-1 EBV by infection of primary B lymphocytes with cell-free virus from coinfected LCLs. Progeny LCLs were visible 3 to 5 weeks after plating.
PCR analyses.
Oligonucleotide primers for amplification of distinctive fragments from type 1 versus type 2 EBNA3A, -3B, or -3C have been described (50). Oligonucleotides 5′-E3AHT (ACATGTGTCAGGATGACGAG) and 3′-E3AHT (CTGAAGGGTCTAGAAGGATC) were used to amplify the junction between EBNA3A- and HTER-encoding DNA. Cell DNA was prepared for PCR, amplified for 40 cycles with annealing at 58°C, and analyzed by electrophoresis.
Western blot and immunofluorescence analyses.
Total cell proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted onto a nitrocellulose membrane (Bio-Rad), and reacted with EBV-immune human sera, rabbit anti-estrogen receptor α polyclonal antibody MC20 (Santa Cruz Biotechnology), sheep anti-EBNA3A polyclonal antibodies (Exalpha Biologicals), rabbit anti-c-myc polyclonal antibody N-262 (Santa Cruz Biotechnology), or mouse anti-LMP1 monoclonal antibody S12. Membranes were reacted with horseradish peroxidase-conjugated species-specific secondary antibodies (Santa Cruz Biotechnology) and developed with a chemiluminescence reagent (NEN). For immunofluorescence, cells were smeared on glass slides, air dried, fixed in 1:1 methanol-acetone at −20°C for 3 min, blocked with 20% goat serum (Gemini)-phosphate-buffered saline, incubated with rabbit anti-estrogen receptor α polyclonal antibodies, and then incubated with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (Jackson ImmunoResearch).
Fluorescence-activated cell sorter analyses for cell DNA and surface expression of CD21, CD23, and CD54.
Approximately 106 cells were fixed and stained with propidium iodide (Molecular Probes) or incubated live with phycoerythrin-conjugated CD21, CD23, or CD54 antibodies (Pharmingen) for 30 min and washed in phosphate-buffered saline supplemented with 2% FBS at 4°C. Cells were analyzed with a FACSCalibur (Becton Dickinson).
LCL growth.
LCL cells (2 × 105, 1 × 105, or 5 × 104) were cultured in 24-well plates in 1 ml of complete medium with or without 4HT. After 7 days, 1 ml of fresh medium was added; then every 3 or 4 days, 50% of the medium was replaced with fresh medium. Cells were counted every 3 or 4 days. In other experiments, 2 × 106 LCL cells were cultured in 25-cm2 culture flasks in 10 ml of complete medium (2 × 105 cells/ml) with or without 4HT. Every 3 or 4 days, the viable cell number was determined and cultures were diluted to 2 × 105 cells/ml with fresh medium to maintain optimum growth. Total cell numbers were calculated based on the expansion from the initial 2 × 106 cells.
Complementation experiments.
EBNA3AHT-infected LCLs (107) were transfected with 30 μg of oriP plasmid DNA expressing FLAG-EBNA3A, EBNA3B, FLAG-EBNA3C or a control oriP plasmid. In some experiments, 10 or 3 μg of plasmid DNA was used. For electroporation, LCLs were harvested during log-phase growth, washed once with complete medium, and resuspended in 400 μl of complete medium with DNA in a cuvette (0.4-cm gap; Bio-Rad). Following a 10-min incubation at 25°C, the culture was pulsed with 220 V at 960 μF. Transfected LCLs were cultured in 14 ml of LCL-conditioned medium with 4HT for 3 to 7 days. Cells were then washed, and 0.5 × 106 to 2 × 106 cells were cultured in 10 ml of complete medium with or without 4HT in a 25-cm2 culture flask. Every 3 to 8 days, cells were counted, cultures were split, and total cell numbers relative to those of the initial culture were calculated.
RESULTS
Reverse-genetic derivation of a recombinant EBV genome that expresses conditionally active EBNA3A.
To evaluate whether the EBNA3A open reading frame fused in frame with DNA encoding the 4HT-dependent mutant estrogen receptor hormone binding domain (HTER) would encode a stable fusion protein, a FLAG epitope-tagged wild-type EBNA3A fused through the EBNA3A C terminus to the N terminus of HTER (FLAG-EBNA3AHT) was expressed in B lymphocytes under the control of the SV40 promoter. When cells were grown in medium supplemented with 4HT, the fused open reading frames expressed a stable protein of ∼180 kDa, the size expected for a FLAG-EBNA3AHT fusion protein (data not shown). A single protein was readily detected on Western blots with FLAG-, EBNA3A-, and ER-specific antibodies, indicating that the N- and C-terminal parts of the fusion protein remain stably joined when the fusion protein is expressed in human B lymphocytes (data not shown). Without the addition of 4HT to cell culture medium, FLAG-EBNA3AHT was barely detectable, indicating that EBNA3AHT stability is dependent on 4HT (data not shown).
To derive an EBV genome that expresses EBNA3AHT instead of wild-type EBNA3A, DNA encoding the EBNA3A C terminus was replaced with that encoding the C terminus of FLAG-EBNA3AHT in a 40-kbp type 1 EBV DNA cosmid clone. The recombinant XF-EBNA3AHT cosmid DNA was transfected into P3HR-1 cells, which are infected with a replication-competent and transformation-incompetent type 2 EBV, together with EBV EcoRI A DNA, which can recombine with the P3HR-1 EBV and restore transformation competence, plasmid SECΔHpaI, which can recombine with EcoR1 A and XF-EBNA3A to facilitate the incorporation of XF-EBNA3A, and pSVNaeZ, which induces lytic EBV replication (7, 52-54). The resultant virus was used to infect primary human B lymphocytes. Infected cells were plated in complete medium supplemented with 4HT in microwells at the anticipated end point dilution of recombinant transforming virus so as to enable the clonal outgrowth of infected LCLs. The LCLs were screened by PCR using primers which distinguish between the P3HR-1 type 2 and XF-EBNA3AHT type 1 EBV DNAs. Approximately 9% (4 of 46) of the LCLs were positive for type 1 EBNA3A (data not shown). All four LCLs were also infected with parental P3HR-1 EBV, which is produced in vast excess and has type 2 EBNA3A DNA.
To segregate the type 1 EBNA3AHT-expressing recombinant EBV genome from the coinfecting P3HR-1 EBV genome, LCL 41, which had recombinant EBV and P3HR-1 EBV at a molar ratio of 1:1, was subcloned by limiting dilution in irradiated IB4 cells in complete medium supplemented with 4HT. Of 46 subclones, 19 contained only recombinant type 1 EBNA3A. The other LCLs all contained both type 1 and type 2 EBNA3A (data not shown). Only 7 of the 19 LCL subclones could be expanded continuously in complete medium with 4HT. Subclones 41-3 and 41-13 were used for further analyses. Other LCLs infected with putative EBNA3AHT-expressing recombinant virus were obtained by inducing lytic-virus replication in the coinfected LCL 41 cells and infecting primary human B lymphocytes with the resultant virus. An LCL that was coinfected with recombinant virus and P3HR-1 EBV and that was permissive for virus replication after transfection with pSVNAEZ was obtained. Infection of primary B lymphocytes with the resultant virus resulted in 182 progeny LCLs, of which 13 were infected with only type 1 EBNA3A DNA, 127 contained only type 2 EBNA3A DNA, and 42 contained both type 1 and type 2 DNAs (data not shown). Five of the 13 LCLs that had only type 1 EBNA3A were successfully expanded. Three of these, LCLs 83, 124, and 163, were used for further analysis. Since wild-type recombinant virus-infected LCLs can usually be continuously expanded in culture, successful continuous expansion of only 7 of 19 LCL 41 subclones and of only 5 of 13 other LCLs infected with EBNA3AHT-expressing recombinant EBV may indicate that LCLs infected with EBNA3AHT-expressing recombinant EBV are relatively deficient in outgrowth.
LCLs 41-3, 41-13, 83, 124, and 163, which were infected with putative type 1 EBNA3AHT-expressing recombinant EBV, were reanalyzed by PCR for the presence of type 1 EBNA3AHT and surrounding DNA (Fig. 1). These LCLs contain type 1 EBNA3A and EBNA3B DNA, lack type 2 EBNA3A and EBNA3B DNA, and have only type 2 EBNA3C DNA, indicating that homologous recombination between the EBNA3B and EBNA3C primer sites had occurred (Fig. 1B to D). All five LCLs contain the EBNA3AHT fusion DNA, as determined by PCR with primers from EBNA3A and HTER (Fig. 1E). Thus, each of the five LCLs infected with EBNA3AHT expressing recombinant EBV has only type 1 EBNA3A and EBNA3B and type 2 EBNA3C.
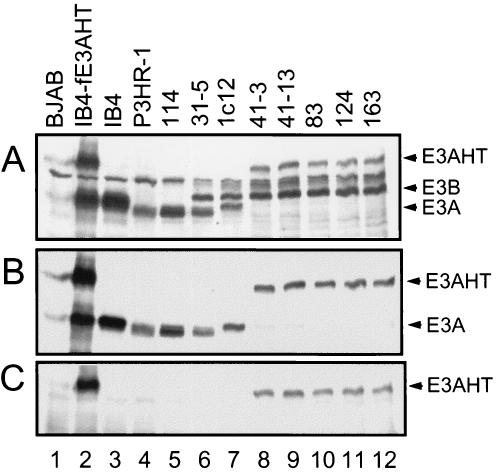
FIG. 1.
PCR analyses of the EBNA3A, EBNA3B, and EBNA3C genotypes of LCLs 41-3, 41-13, 83, 124, and 163, which were infected with recombinant EBV genomes that express EBNA3A fused in frame to EBNA3AHT. (A) Schematic representation of the EBV genome EBNA3 region within the targeting construct and location of the PCR primers. (B to D) Specific primer pairs distinguish between EBV type 1 and type 2 EBNA3A, -3B, and -3C genes in the EBV type 1-infected IB4 LCL and the EBV type 2-infected P3HR-1 Burkitt tumor cell (50). The EBNA3A (E3A) primers result in the amplification of 237- or 276-bp fragments from type 2 (T2) or type 1 (T1) DNA, respectively. The EBNA3B (E3B) primers result in amplification of 218- or 183-bp fragments from type 2 or type 1 DNA, respectively. The EBNA3C (E3C) primers result in the amplification of 179- or 158-bp fragments from type 2 or type 1 DNA, respectively. (E) Use of primers for amplification of the junction between EBNA3A and HTER genes (E3AHT primers). Lane 1, control amplification with primers only; lanes 2 and 3, amplifications with DNA from EBV type 2- and type 1-infected cells; lanes 4 to 8, amplifications of EBNA3AHT-infected LCL genomic DNA; lane 9, amplification of DNA from a stable IB4 cell line transfected with and expressing a FLAG-tagged EBNA3AHT fusion protein (IB4-fE3AHT).
Western blot analyses using EBV-immune human serum confirmed that the five EBNA3AHT-infected LCLs do not express either wild-type 1 or 2 EBNA3A, both of which are ∼130 kDa, and express only a larger EBNA3AHT, ∼180 kDa, which is also specifically recognized by estrogen receptor α and EBNA3A antibodies (Fig. 2 and data not shown). When these five LCLs were grown in the presence of 4HT, EBNA3AHT expression was ∼25% of that of the wild-type EBNA3A protein in control LCLs, as established by comparative twofold dilution and Western blotting (data not shown). Note that in Fig. 2A the amounts of EBNA3AHT in the 41-3, 41-13, 83, 124, and 163 LCLs are substantially less than the amount of EBNA3A in the 1c12 LCL; the 1c12 LCL lane is also underloaded, as is evident from the smaller amount of EBNA3B in the 1c12 LCL than in the EBNA3AHT-infected LCLs. The lower level of EBNA3AHT in the LCLs infected with recombinant EBV expressing EBNA3AHT than of EBNA3A in LCLs infected with wild-type EBV expressing wild-type EBNA3A is likely due to a destabilizing effect of HTER fusion with EBNA3A.
FIG. 2.
EBNA3 protein expression in LCLs infected with EBNA3AHT-expressing EBV recombinants. Immunoblots were incubated with EBV-immune human serum (A), sheep anti-EBNA3A polyclonal antibodies (B), or rabbit anti-estrogen receptor α polyclonal antibodies (C). Lane 1, lysates from EBV-negative BJAB human B lymphoblasts as a negative control; lanes 2 to 4, lysates from IB4 LCLs stably transfected with a FLAG-tagged EBNA3AHT (IB4-fE3AHT), IB4 cells, and P3HR-1 cells, respectively; lanes 5 and 6, lysates from LCLs 114 and 31-5, with type 2 wild-type EBNA3A-infected LCLs; lane 7, type 1 wild-type EBNA3A-infected LCL 1c12; lanes 8 to 12, five EBNA3AHT-infected LCLs. All LCLs, except for 114, expressed type 1 EBNA3B (E3B). All five EBNA3AHT-infected LCLs expressed a large EBNA3AHT (E3AHT) fusion protein and did not express wild-type 1 or 2 EBNA3A (E3A). Type 2 EBNA3C (E3C) is just below a background control band and was faintly detected by this EBV-immune human serum.
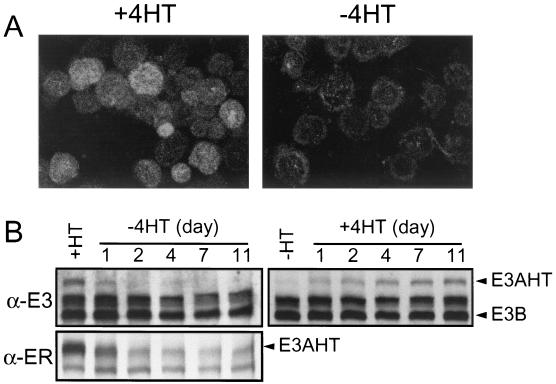
EBNA3AHT localized to cell nuclei when cells were grown in medium supplemented with 4HT and relocalized to the cytoplasm when cells were grown in complete medium without 4HT (Fig. 3A and data not shown). Background cytoplasmic fluorescence in control cells that lack EBNA3AHT was minimal. EBNA3AHT decreased more than 50% within 1 day of incubation of EBNA3AHT-infected LCLs in medium without 4HT and was barely detectable after 2 days in culture medium without 4HT (Fig. 3B). When cells were transferred to medium with 4HT, EBNA3AHT reaccumulated over ∼4 days to ∼50% of stable expression levels and reached stable expression levels between 7 and 11 days (Fig. 3B). Overall, these data indicate that EBNA3AHT stability and nuclear localization are dependent on 4HT. In summary, EBNA3AHT levels in recombinant EBV-infected LCLs in the presence of 4HT are only ∼25% of EBNA3A levels in wild-type LCLs and growth in medium without 4HT results in at least a 75% additional decrease in EBNA3AHT levels as well as dislocation of EBNA3AHT from the nucleus to the cytoplasm. In the absence of 4HT, EBNA3AHT is dislocated to the cytoplasm and expressed at levels that are less than 10% of the EBNA3A levels in wild-type LCLs. Thus, EBNA3AHT-infected LCLs maintained in the absence of 4HT are nearly completely EBNA3A deficient.
FIG. 3.
Regulation of EBNA3AHT fusion protein by 4HT. (A) Immunofluorescent staining of stable IB4 transfectants expressing a FLAG-tagged EBNA3AHT fusion protein (IB4-fE3AHT) with an anti-estrogen receptor α antibody and a fluorescein isothiocyanate-conjugated secondary antibody. IB4-fE3AHT cells cultured in the presence (left) or absence (right) of 4HT for 3 days were dried onto glass slides, fixed, and stained. (B) Immunoblot of EBNA3AHT expression in lysates of LCL 41-3 infected with EBV expressing recombinant EBNA3AHT. The LCL had been maintained in complete medium for 7 days with or without 4HT and then switched for the indicated number of days to complete medium without (left) or with (right) 4HT, respectively. Protein lysates from cells at each time point were resolved by sodium dodecyl sulfate-6% polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and probed with an EBV-immune human serum which recognizes EBNA3s (α-E3) or with an anti-estrogen receptor α antibody (α-ER).
Growth of EBNA3AHT-expressing LCLs is 4HT dependent.
The growth of the EBNA3AHT-expressing LCLs in complete medium with or without 4HT was assessed. Cells were seeded at 0.5 × 105 to 2 × 105 cells/ml in complete medium with or without 4HT, and the live-cell number was determined every several days for 3 weeks. Cultures were fed weekly with an equal volume of fresh medium as is appropriate for LCLs unless they are in maximum-log-phase growth, wherein they double every 24 to 48 h. LCLs reach saturation at ∼0.8 × 106 to 1 × 106 cells/ml. When seeded at 2 × 105 cells/ml, EBNA3AHT- and wild-type EBNA3A-expressing cells grew at similar rates for 6 to 8 days in medium with or without 4HT (Fig. 4 and data not shown). Thereafter, the growth of EBNA3AHT-expressing cells slowed in medium without 4HT, relative to the growth of EBNA3AHT-expressing cells in medium with 4HT or to that of wild-type EBNA3A-expressing cells (Fig. 4 and data not shown). EBNA3A- and EBNA3AHT-expressing, 4HT-treated cells continued to grow to more than 20 × 105 total cells (106 cells/ml) and subsequently decreased (Fig. 4), because the cells were then at saturation density. When these cells were fed more frequently, they continued in log phase indefinitely (data not shown). In contrast, EBNA3AHT-expressing cells never grew beyond 15 × 105 total cells (∼8 × 105 cells/ml) in medium without 4HT. When these cells were fed more frequently, they stopped growing sooner. These data indicate that the growth of cells infected with EBNA3AHT-expressing recombinant EBV becomes EBNA3A dependent within 6 to 9 days after 4HT is withdrawn, even when the initial cell density is sufficient for rapid entry into log-phase growth. Although EBNA3AHT is inactivated and degraded within 2 days without 4HT, the effect of EBNA3AHT inactivation on cell growth is not evident for several days thereafter.
FIG. 4.
Comparison of growth rates of EBNA3AHT-infected LCLs in the presence and absence of 4HT. Five LCLs infected with EBV expressing recombinant EBNA3AHT (E3AHT LCLs) and two control LCLs were cultured with (open circles) or without (solid circles) 4HT in 24-well plates as described in Materials and Methods. Total cell numbers in each well were plotted at each time point. ND, not done. Each growth curve experiment was done twice with similar results.
The onset of the growth defect that accompanies EBNA3AHT inactivation was more rapid when cells were cultured at 0.5 × 105 to 1 × 105 cells/ml. EBNA3AHT- and wild-type EBNA3A-expressing LCLs were similar in their delayed growth when plated at 0.5 × 105 to 1 × 105 cells/ml of complete medium. After a 3- to 6-day delay at 105 cells/ml or a 6- to 9-day delay at 0.5 × 105 cells/ml, during which the cells conditioned the medium, wild-type EBNA3A- and EBNA3AHT-expressing cells in medium with 4HT grew at the same rate as cells plated at 2 × 105 cells/ml. In marked contrast, EBNA3AHT-expressing cells plated at 0.5 × 105 or 1 × 105 cells/ml of complete medium without 4HT failed to grow or grew poorly. EBNA3AHT-expressing-cell growth was minimal or stopped after 6 to 12 days in medium without 4HT, and cell numbers slowly decreased thereafter (Fig. 4 and data not shown). Thus, the role of EBNA3A in cell growth is evident within 6 to 8 days, when cells are plated at 1 × 105 or 0.5 × 105 cells/ml.
The effect of 4HT withdrawal on the cell cycle distribution of EBNA3AHT-expressing LCLs over the ensuing 20 days after plating at 105 cells/ml in medium without 4HT was assessed by propidium iodide staining and fluorescence-activated cell sorter analysis (Fig. 5 and data not shown). In the presence of 4HT ∼73% of the cells were in G0/G1, 7% were in S, and 13% were in G2/M, with 7% hypodiploid at days 7, 14, and 20 after plating. In contrast, in the absence of 4HT, the percentage in G0/G1 fell from 74 to 66%, the percentage in S fell from 6 to 3%, and the percentage in G2/M fell from 10 to 7%, while the percentages of hypodiploid cells increased to 10, 13, and 24% at days 7, 14, and 20, respectively, after plating. Thus, EBNA3A inactivation resulted in 50% fewer cells in S or G2/M and three times as many hypodiploid cells at day 20. Thus, EBNA3A inactivation results in decreased cell cycle entry and increased apoptosis.
FIG. 5.
EBNA3AHT inactivation results in a progressive decrease of cells in S or G2/M and an increase in hypodiploid cells. EBNA3AHT-infected LCL 41-13 cells were plated at 105 cells/ml in the presence (+4HT) or absence (−4HT) of 4HT. Cells were fed every 3 or 4 days. At 7, 14, and 20 days after plating, cells were fixed, stained with propidium iodide, and analyzed by FACSCalibur.
To further examine the role of EBNA3A in cell growth at optimum cell density, which is between 2 × 105 and 6 × 105 cells/ml, EBNA3AHT-expressing cells were seeded into medium at 2 × 105 cells/ml with or without 4HT. The cell density was readjusted every several days to 2 × 105 cells/ml of partially new medium with or without 4HT. All five EBNA3AHT-expressing LCLs grew exponentially in medium with 4HT, but the same cells gradually stopped growing after 6 to 12 days in medium without 4HT. In medium without 4HT, the viable-cell count began to decrease at 16 to 20 days (Fig. 6A). The growth of wild-type EBNA3A-expressing control LCLs was similar to that of the EBNA3AHT-expressing LCLs growing with 4HT and was unaffected by the presence or absence of 4HT (Fig. 6A). These experiments further indicate that EBNA3A has a critical role in LCL growth.
FIG. 6.
Growth and survival of LCL cells infected with EBV expressing recombinant EBNA3AHT are dependent on 4HT. (A) LCLs infected with EBV expressing recombinant EBNA3AHT (E3AHT LCLs) and wild-type control LCLs were cultured at 2 × 105 cells/ml in 10 ml of complete medium with (open circles) or without (solid circles) 4HT in 25-cm2 culture flasks. Every 3 to 4 days, cultures were split as described in Materials and Methods. Cells were counted at the charted intervals, and total cell numbers derived from the initial cultures were calculated and plotted at each time point. (B) LCLs infected with EBV expressing recombinant EBNA3AHT and control LCLs were cultured in the absence of 4HT for 7 days. Cells were then cultured with (open circles) or without (solid circles) 4HT as in panel A, and total cell numbers were plotted at each time point. Each experiment was done twice with similar results.
To determine whether cells that survive under conditions of EBNA3AHT inactivation can be restored to growth by EBNA3AHT activation, EBNA3AHT-expressing LCLs were cultured in the absence of 4HT for 7 days and then resuspended at 2 × 105 cells/ml of complete medium with or without 4HT. After 7 days in medium without 4HT, the cells remained growth arrested for 9 days in the presence or absence of 4HT. Thereafter, the EBNA3AHT-expressing cells which were in medium with 4HT grew in log phase at a rate similar to that for wild-type EBNA3A-expressing LCLs, whereas cells in medium without 4HT died at a substantial rate. These experiments indicate that EBNA3AHT reactivation can restore growth to cells that have stopped growing as a consequence of EBNA3AHT inactivation. Since cell growth in new medium at 2 × 105 cells/ml is delayed for 2 to 3 days to condition the medium (Fig. 5) and since complete reactivation of EBNA3AHT levels requires at least 7 days (Fig. 3), both factors probably contribute to the 9- to 12-day delay in cell growth after plating in medium with 4HT.
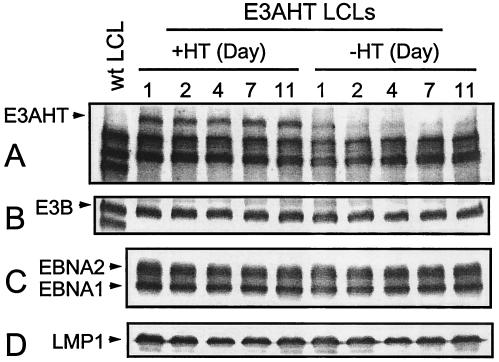
EBNA3AHT inactivation does not affect expression of other EBNAs or LMP1.
Since EBNA3A can affect the EBV Cp EBNA or the LMP1 promoter, EBNA3AHT activation or inactivation could alter EBNALP, EBNA2, EBNA3C, EBNA1, or LMP1 expression and thereby affect LCL growth and survival. We therefore compared EBNA and LMP1 expression in EBNA3AHT-expressing LCLs that had been growing for 11 days in the presence and absence of 4HT with EBNA expression in wild-type EBNA3A-expressing LCLs (Fig. 7 and data not shown). In the presence and in the absence of 4HT, EBNA3AHT-expressing LCLs expressed EBNA1, -2, and -3B and LMP1 at levels which were indistinguishable from those expressed by control LCLs, even though EBNA3AHT was barely detectable in the EBNA3AHT-expressing LCLs by 2 days after 4HT withdrawal. Since all EBNA mRNAs in LCLs are splice products of the transcripts from the same Cp or Wp EBNA promoters and since EBNA1, EBNA2, EBNA3B, and LMP1 protein levels do not change over 11 days of EBNA3A inactivation, EBNA3AHT inactivation does not directly affect EBNA or LMP1 expression.
FIG. 7.
EBNA3AHT inactivation has minimal effect on latent EBV gene expression. LCL 41-13 cells infected with EBV expressing recombinant EBNA3AHT were washed and cultured with (+4HT) or without (−4HT) 4HT for indicated numbers of days. Protein lysates were prepared from EBNA3AHT-infected LCL cells (E3AHT LCLs) or from control LCL 1c12 cells (wt LCL), and Western blot analyses used EBV-immune human serum to detect EBNA3AHT (A), EBNA3B (B), and EBNA2 and EBNA1 (C) and used the S12 monoclonal antibody to detect LMP1 (D).
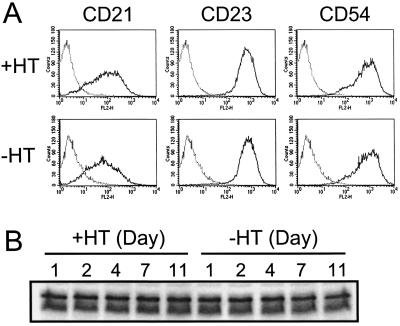
EBNA3AHT inactivation does not affect CD23 or c-myc expression but may have a small affect on CD21 expression.
Since EBNA3A associates with RBP-Jκ and can affect EBNA2-mediated cell gene transcription, we investigated the effect of EBNA3AHT inactivation on levels of CD21, CD23, and c-myc expression, which are EBNA2 regulated. EBNA3AHT-expressing LCLs were cultured with or without 4HT for 11 days. Levels of cell surface expression of CD21 and CD23 on EBNA3AHT-expressing LCLs cultured with or without 4HT for 7 days were compared by flow cytometry (Fig. 8). Mean CD21 expression decreased slightly but reproducibly in the 41-3 and 41-13 subclones, whereas expression of CD23 and a putative control surface marker, CD54, did not change. Expression of CD21 after 14 days of culture was similarly decreased to that after 7 days of culture, and CD23 expression remained unchanged (data not shown). There were also small changes in LCL 83 and 124 CD21 RNA, but not in LCL 163 RNA, after 4 days of growth in medium without 4HT. Like expression of CD23, expression of c-myc was unaffected by EBNA3AHT activation or inactivation; levels of c-myc expression in EBNA3AHT-expressing LCLs cultured with or without 4HT for 11 days were not different (Fig. 8B). Thus, EBNA3A does not affect c-myc or CD23 expression but may have a small role in CD21 expression.
FIG. 8.
Inactivation of EBNA3AHT in LCLs infected with EBV expressing recombinant EBNA3AHT does not substantially alter CD23 or c-myc expression but may slightly reduce CD21 expression. (A) LCL 41-13 cells infected with EBV expressing recombinant EBNA3AHT were washed and cultured with (+HT) or without (−HT) 4HT for 7 days. Cells were incubated with phycoerythrin-conjugated CD21 or CD23 monoclonal antibodies or with putative control CD54 monoclonal antibodies (solid lines) or without antibodies (dotted lines) and analyzed by flow cytometry. (B) EBNA3AHT-infected LCL 41-13 cells were cultured with or without 4HT for the indicated numbers of days. Protein lysates were prepared from cells at each time point, and Western blot analysis was performed with an anti-c-myc antibody.
Wild-type EBNA3A expression can enable the growth of EBNA3AHT-infected LCLs in medium without 4HT.
Having established that EBNA3AHT-infected LCLs are conditionally dependent on 4HT for proliferation, we evaluated whether these cells are formally dependent on wild-type EBNA3A for their growth since that would enable them to be used for genetic analyses of the components of EBNA3A necessary for cell growth. To evaluate whether heterologous EBNA3A expression can complement EBNA3AHT inactivation and enable EBNA3AHT LCL growth in medium without 4HT, LCLs were transfected with an oriP plasmid expressing wild-type FLAG-tagged EBNA3A or with an oriP plasmid control. The LCL transfection efficiency at day 3 with a control oriP plasmid that expresses EGFP in the place of FLAG-tagged EBNA3A was 20 to 40%, as estimated from the number of EGFP-positive cells. The efficiency of transfection as estimated from the number of EGFP-positive cells after transfection with an oriP plasmid that expresses EGFP fused in frame with the N terminus of EBNA3A was ∼30%. Having established that transfection of LCLs with oriP-based expression vectors resulted in expression of EBNA3A in a substantial fraction of the transfected cells, EBNA3AHT-infected LCLs were transfected with a vector control or FLAG-tagged wild-type EBNA3A oriP expression plasmid, cultured in medium with 4HT for 7 days, and transferred to fresh medium with or without 4HT. After seven additional days, control vector-transfected cells continued to grow in the presence of 4HT but died in the absence of 4HT (Fig. 9). In contrast, wild-type-EBNA3A-transfected cells grew almost as well in the absence of 4HT as in the presence of 4HT (Fig. 9A). Over 21 days in culture without any selection other than 4HT withdrawal, wild-type EBNA3A expression stabilized at a higher level in cells growing in the absence of 4HT than in cells growing in the presence of 4HT, further evidence that wild-type EBNA3A expression particularly enhances the proliferation of EBNA3AHT-infected LCLs in the absence of 4HT (Fig. 9). The level of wild-type EBNA3A in the EBNA3AHT-infected LCLs growing in the absence of 4HT at day 21 was slightly higher than the level of EBNA3A in wild-type LCLs or in EBNA3AHT-infected LCLs growing in the presence of 4HT, even when EBNA3AHT is included (Fig. 9 and data not shown). Similar growth-complementing effects of wild-type EBNA3A expression were demonstrated in the three other EBNA3AHT-infected LCLs grown in medium without 4HT (data not shown). Experiments done with an oriP plasmid that expresses EGFP fused in frame with wild-type EBNA3A resulted in at least 50% EGFP-positive cells at day 21 or 29 after transfection and plating in medium without 4HT (data not shown). Thus, by day 21 or 29 most or all of the EBNA3AHT-infected cells that were maintained in the absence of 4HT had selected for expression of wild-type EBNA3A. As expected, transfection of the four EBNA3AHT-infected LCLs with the vector control plasmid did not enable cell growth in medium without 4HT (Fig. 9 and data not shown). These data indicate that the growth defect of EBNA3AHT-infected LCLs under conditions of EBNA3AHT inactivation can be transcomplemented by expression of wild-type EBNA3A; the growth effect of wild-type EBNA3A is evident within 2 weeks of transfection. Thus, transfection of EBNA3AHT-infected LCLs with a wild-type EBNA3A expression plasmid and transfer to medium without 4HT constitute a rapid and robust assay for wild-type EBNA3A's role in LCL proliferation and survival.
FIG. 9.
Expression of wild-type EBNA3A enables the continuous proliferation of LCLs infected with EBV expressing recombinant EBNA3AHT in medium without 4HT. (A) EBNA3AHT-infected LCL 41-3 cells were transfected with 30 μg of the oriP plasmid expressing FLAG-EBNA3A (oriP-fE3A) or a control oriP plasmid expressing EGFP (oriP-cont) and cultured in conditioned medium with 4HT for 7 days. Then, cells were washed and resuspended at 2 × 106 cells/10 ml of complete medium with (+4HT) or without (−4HT) 4HT in 25-cm2 culture flasks (day 0). Every 7 days, cells were counted and all cultures were split. The total cell numbers derived from the initial cultures were calculated and plotted at each time point. Protein lysates were prepared from these cells, and Western blot analyses (B) were performed using EBV-immune human serum. The data are representative of five similar experiments.
Wild-type EBNA3A expression enables the growth of EBNA3AHT-infected LCLs in medium without 4HT, but EBNA3B or EBNA3C do not.
To assess whether additional EBNA3B or EBNA3C expression can substitute for EBNA3A expression in sustaining LCL proliferation, EBNA3B, EBNA3C, and EBNA3A were compared for their ability to complement the growth defect caused by EBNA3AHT inactivation (Fig. 10). Transfection of EBNA3AHT-infected LCLs with 3, 10, or 30 μg of wild-type EBNA3A expression plasmid resulted in dose-dependent increases in EBNA3A expression, as revealed by Western blotting done 3 days after transfection. All doses had similar positive effects and enabled continued LCL proliferation in medium without 4HT. In multiple experiments, transfection with similar amounts of EBNA3B and EBNA3C expression plasmids did not enable LCL growth in medium without 4HT, despite levels of FLAG-tagged-EBNA3B or -EBNA3C expression with 30 μg of expression vector that were comparable to levels of FLAG-tagged-EBNA3A expression with 10 μg of the expression vector (Fig. 10 and data not shown). In some experiments, transfection of EBNA3AHT-infected LCLs with an EBNA3C expression plasmid enabled a transient positive partial effect on EBNA3AHT-infected LCL growth in medium without 4HT. Overall, these experiments indicate that EBNA3B or EBNA3C overexpression cannot substitute for EBNA3A expression in maintaining LCL growth. Thus, EBNA3A has unique effects that are critical for LCL cell growth.
FIG. 10.
EBNA3B or EBNA3C overexpression cannot substitute for EBNA3A to complement the growth deficiency of LCL cells infected with EBV expressing recombinant EBNA3AHT evident following transfer to complete medium without 4HT. (A) EBNA3AHT-infected LCL 41-13 cells were transfected with the indicated amount of the oriP plasmid expressing FLAG-EBNA3A (fE3A), EBNA3B (E3B), or FLAG-EBNA3C (fE3C) or with a control oriP plasmid (Cont) and cultured in conditioned medium with 4HT for 3 days. The cells were washed, and 5 × 105 cells were cultured in 10 ml of complete medium with (+4HT) or without (−4HT) 4HT in 25-cm2 culture flasks (day 0). Every 5 to 7 days, cells were counted and cultures were fed with similar amounts of fresh medium. Total numbers of cells derived from the initial cultures were calculated and plotted at each time point. (B) Protein lysates from these cells 3 days after transfection were Western blotted with EBV-immune human serum to detect EBNA3A, EBNA3B, and EBNA3C. The data are representative of five similar experiments. Arrowheads indicate fE3A, E3B, or fE3C.
DISCUSSION
These experiments indicate that EBNA3A has a critical role in maintaining the proliferation of EBV-infected primary B lymphocytes as LCLs. The transformation-defective P3HR-1 genome was used as the genetic background for isolation of a recombinant EBV genome which homologously incorporated a transfected DNA fragment that restored exons encoding part of EBNALP and all of EBNA2 and which is therefore able to transform human B lymphocytes into LCLs. LCL 41 was identified as a cell line infected with an EBV genome that had also homologously recombined with a second transfected EBV DNA fragment, which had genomic EBNA3A DNA fused in frame to 4HT-dependent estrogen receptor hormone binding domain DNA (38). This LCL was coinfected with the parental P3HR-1 virus. Subclones which had only the recombinant EBNA3AHT EBV genome were isolated in complete medium supplemented with 4HT. Multiple other cell lines were derived by sequential infection and growth transformation of primary human B lymphocytes with virus from the original dually infected LCL 41. Many of these LCLs grew in medium supplemented with 4HT and were infected with only an EBNA3AHT-expressing recombinant EBV genome.
Incubation of LCLs infected with EBV expressing recombinant EBNA3AHT in medium without 4HT caused EBNA3AHT to localize to the cytoplasm and to be degraded. EBNA3AHT inactivation in LCLs infected with EBV expressing recombinant EBNA3AHT, including two subclones of the original cell line and three independently infected LCLs, resulted in inhibition of cell growth and ultimately in decreased cell survival. Following EBNA3AHT inactivation, the LCL cells continued to proliferate without EBNA3A for several days but then stopped growing and slowly died. c-myc levels remained high despite the absence of EBNA3A. These data indicate that EBNA3A is an essential modulator of one or more biochemical processes, which can remain functionally adequate for more than a complete cell cycle before the putative process(es) deteriorates below a level that is consistent with cell growth and cell survival.
This delayed effect of EBNA3AHT inactivation on cell growth may be due to an effect on a promoter of a cell gene important for growth since the protein product of that gene could have a long half-life or a substantial reservoir. The growth of LCLs is dependent on secreted and nonsecreted growth factors, including interleukin-6 (IL-6) and tumor necrosis factor; IL-6 transcription can be dependent on regulation through an upstream RBP-Jκ site (25, 55). The onset of the growth defect following EBNA3AHT inactivation in LCLs infected with EBV expressing recombinant EBNA3AHT is dependent on cell concentration, as would be expected for a growth factor effect. However, LCL growth is in general cell concentration dependent, and plating at lower cell density may indirectly exacerbate the effects of EBNA3AHT inactivation. Our attempts to prevent the growth-inhibitory effects of EBNA3AHT inactivation by frequent feeding with LCL-conditioned medium or by mixing these recombinant EBV-infected LCLs with irradiated wild-type LCLs have so far failed to complement and provide sustained LCL growth following EBNA3AHT inactivation.
The EBNA3AHT fusion protein is somewhat destabilized by the HTER domain, and levels in the presence of 4HT are only ∼25% of wild-type EBNA3A levels. Despite the lower EBNA3AHT levels, EBNA3AHT in the presence of 4HT has seemingly wild-type effects on cell growth. Thus, either wild-type EBNA3A levels are in excess of levels required for wild-type effects or the EBNA3AHT fusion is a “relative gain of function” that enables less protein to function at wild-type levels. The observation that 10-fold-different amounts of wild-type EBNA3A expression vector and substantially different amounts of EBNA3A protein can transcomplement LCL proliferation when EBNA3AHT-infected recombinant LCLs are grown in the absence of 4HT indicates that LCL proliferation can be sustained by a range of wild-type EBNA3A expression levels in the absence of a potentially confounding effect of the EBNA3AHT fusion protein. Interestingly, in multiple experiments, wild-type EBNA3A expression levels were maintained over many weeks in culture at nearly wild-type levels only in the setting of 4HT withdrawal, consistent with intrinsic selection for optimal EBNA3A expression levels. Indeed, EBNA3A, under conditions of forced more-than-threefold overexpression in an LCL, causes growth arrest by binding to RBP-Jk, by dissociating EBNA2 from RBP-Jκ, and by down-regulating c-myc (8). In contrast to growth arrest induced by EBNA3AHT inactivation, which is associated with continued c-myc expression and apoptosis, growth arrest induced by EBNA3A overexpression is associated with substantially inhibited c-myc expression and is not followed by apoptosis, unless c-myc is activated (8). Thus, one function of EBNA3A may be to protect against the proapoptotic effects of c-myc expression (23).
At normal EBNA3A levels in LCLs, EBNA3A extensively associates with the cellular transcription factor RBP-Jκ and is presumed to modulate RBP-Jκ-mediated transcription (6, 8, 12, 22, 34, 49, 62). EBNA3A has a weak repressive effect on a multimerized Cp promoter when expressed in the absence of other EBNAs (6). EBNA3A domains that repress or activate transcription when fused to a Gal4 DNA binding domain have been identified; however, EBNA3A association with RBP-Jκ may inhibit binding to cognate DNA (5, 6, 56). The most profound transcriptional effects of EBNA3A have been evident when it is coexpressed with EBNA2 in transient-transfection assays in the presence of an EBNA2-dependent reporter. In such assays, EBNA3A expression strongly represses EBNA2 activation of the EBV Cp EBNA promoter and slightly coactivates expression from the EBV LMP1 promoter with EBNA2 (34, 35, 56, 62). Effects on cell or viral gene expression at physiologically appropriate levels have not been investigated. If EBNA3A's role in transcriptional regulation is to “fine tune” the effects of EBNA2, studies of EBNA3A would be particularly dependent on physiologic concentrations of the other EBNA proteins. In this regard, EBNA3AHT-infected recombinant LCLs are a potentially useful system for studying the putative transcriptional effects of EBNA3A. The data presented here indicate that EBNA3AHT inactivation in LCLs does not affect viral EBNA or LMP1 expression or cellular CD23 or c-myc expression, although CD21 expression may be slightly down-regulated by EBNA3AHT inactivation. Studies are in progress to identify the putative effects of EBNA3A on cell gene transcription through transcriptional profiling of EBNA3AHT-infected LCLs that have been switched to medium with or without 4HT.
An important aspect of these experiments is that they demonstrate for the first time that EBNA3A has a role in LCL growth that is unequivocally distinct from that of EBNA3B and EBNA3C. Expression of EBNA3A at several levels prevented growth arrest following EBNA3AHT inactivation, whereas similar levels of additional EBNA3B or EBNA3C had minimal or no effect. Thus, EBNA3A has a unique and important role in LCL growth and survival. The ability to complement EBNA3AHT inactivation in these LCLs with oriP-based expression vectors provides a new genetic system to rapidly assess the requirement for specific EBNA3A domains and amino acids in LCL proliferation and survival. This should facilitate the identification of those biochemical interactions that are critical for the EBNA3A role(s) and may identify EBNA3A domains that promote B-cell proliferation by unexpected mechanisms.
Acknowledgments
We thank Bo Zhao, Ellen Cahir-McFarland, and Ken Izumi for helpful discussions and suggestions.
This research was supported by grants from the National Cancer Institute of the USPHS (CA47006 and CA87661). E.J. received support from grant 1 K08 AI49943-02 from the National Institute of Allergy and Infectious Diseases of the USPHS.
REFERENCES
- 1.Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. [DOI] [PubMed] [Google Scholar]
- 2.Allday, M. J., and P. J. Farrell. 1994. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J. Virol. 68:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aman, P., M. Rowe, C. Kai, J. Finke, L. Rymo, E. Klein, and G. Klein. 1990. Effect of the EBNA-2 gene on the surface antigen phenotype of transfected EBV-negative B-lymphoma lines. Int. J. Cancer 45:77-82. [DOI] [PubMed] [Google Scholar]
- 4.Bain, M., R. J. Watson, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J. Virol. 70:2481-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourillot, P. Y., L. Waltzer, A. Sergeant, and E. Manet. 1998. Transcriptional repression by the Epstein-Barr virus EBNA3A protein tethered to DNA does not require RBP-Jκ. J. Gen. Virol. 79:363-370. [DOI] [PubMed] [Google Scholar]
- 6.Cludts, I., and P. J. Farrell. 1998. Multiple functions within the Epstein-Barr virus EBNA-3A protein. J. Virol. 72:1862-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, J. I., F. Wang, and E. Kieff. 1991. Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J. Virol. 65:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper, A., E. Johannsen, S. Maruo, E. Cahir-McFarland, D. Illanes, D. Davidson, and E. Kieff. 2003. EBNA3A association with RBP-Jκ down-regulates c-myc and Epstein-Barr virus-transformed lymphoblast growth. J. Virol. 77:999-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordier, M., A. Calender, M. Billaud, U. Zimber, G. Rousselet, O. Pavlish, J. Banchereau, T. Tursz, G. Bornkamm, and G. M. Lenoir. 1990. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces expression of B-cell activation molecules CD21 and CD23. J. Virol. 64:1002-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordier-Bussat, M., M. Billaud, A. Calender, and G. M. Lenoir. 1993. Epstein-Barr virus (EBV) nuclear-antigen-2-induced up-regulation of CD21 and CD23 molecules is dependent on a permissive cellular context. Int. J. Cancer 53:153-160. [DOI] [PubMed] [Google Scholar]
- 11.Cotter, M. A., II, and E. S. Robertson. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol. Cell. Biol. 20:5722-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalbies-Tran, R., E. Stigger-Rosser, T. Dotson, and C. E. Sample. 2001. Amino acids of Epstein-Barr virus nuclear antigen 3A essential for repression of Jκ-mediated transcription and their evolutionary conservation. J. Virol. 75:90-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuentes-Panana, E. M., R. Peng, G. Brewer, J. Tan, and P. D. Ling. 2000. Regulation of the Epstein-Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway. J. Virol. 74:8166-8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossman, S. R., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grundhoff, A. T., E. Kremmer, O. Tureci, A. Glieden, C. Gindorf, J. Atz, N. Mueller-Lantzsch, W. H. Schubach, and F. A. Grasser. 1999. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 274:19136-19144. [DOI] [PubMed] [Google Scholar]
- 16.Gutkind, J. S., D. C. Link, S. Katamine, P. Lacal, T. Miki, T. J. Ley, and K. C. Robbins. 1991. A novel c-fgr exon utilized in Epstein-Barr virus-infected B lymphocytes but not in normal monocytes. Mol. Cell. Biol. 11:1500-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harada, S., and E. Kieff. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 71:6611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 19.Henle, W., V. Diehl, G. Kohn, H. zur Hausen, and G. Henle. 1967. Herpes-type virus and chromosome marker in normal leukocytes after growth with irradiated Burkitt cells. Science 157:1064-1065. [DOI] [PubMed] [Google Scholar]
- 20.Hickabottom, M., G. A. Parker, P. Freemont, T. Crook, and M. J. Allday. 2002. Two nonconsensus sites in the Epstein-Barr virus oncoprotein EBNA3A cooperate to bind the co-repressor carboxyl-terminal-binding protein (CtBP). J. Biol. Chem. 277:47197-47204. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, H., Y. G. Cho, and F. Wang. 2000. Structural, functional, and genetic comparisons of Epstein-Barr virus nuclear antigen 3A, 3B, and 3C homologues encoded by the rhesus lymphocryptovirus. J. Virol. 74:5921-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johannsen, E., C. L. Miller, S. R. Grossman, and E. Kieff. 1996. EBNA-2 and EBNA-3C extensively and mutually exclusively associate with RBPJκ in Epstein-Barr virus-transformed B lymphocytes. J. Virol. 70:4179-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juin, P., A. Hunt, T. Littlewood, B. Griffiths, L. B. Swigart, S. Korsmeyer, and G. Evan. 2002. c-Myc functionally cooperates with Bax to induce apoptosis. Mol. Cell. Biol. 22:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaiser, C., G. Laux, D. Eick, N. Jochner, G. W. Bornkamm, and B. Kempkes. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73:4481-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kannabiran, C., X. Zeng, and L. D. Vales. 1997. The mammalian transcriptional repressor RBP (CBF1) regulates interleukin-6 gene expression. Mol. Cell. Biol. 17:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kashuba, E., V. Kashuba, K. Pokrovskaja, G. Klein, and L. Szekely. 2000. Epstein-Barr virus encoded nuclear protein EBNA-3 binds XAP-2, a protein associated with hepatitis B virus X antigen. Oncogene 19:1801-1806. [DOI] [PubMed] [Google Scholar]
- 27.Kashuba, E., V. Kashuba, T. Sandalova, G. Klein, and L. Szekely. 28 August 2002, posting date. Epstein-Barr virus encoded nuclear protein EBNA-3 binds a novel human uridine kinase/uracil phosphoribosyltransferase. BMC Cell Biol. 3:23. [Online.] http://www.biomedcentral.com/1471-2121/3/23. [DOI] [PMC free article] [PubMed]
- 28.Kashuba, E., K. Pokrovskaja, G. Klein, and L. Szekely. 1999. Epstein-Barr virus-encoded nuclear protein EBNA-3 interacts with the epsilon-subunit of the T-complex protein 1 chaperonin complex. J. Hum. Virol. 2:33-37. [PubMed] [Google Scholar]
- 29.Kempkes, B., D. Pich, R. Zeidler, B. Sugden, and W. Hammerschmidt. 1995. Immortalization of human B lymphocytes by a plasmid containing 71 kilobase pairs of Epstein-Barr virus DNA. J. Virol. 69:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kieff, E., and A. Rickinson. 2001. Epstein-Barr virus, p. 2511-2574. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 31.Knutson, J. C. 1990. The level of c-fgr RNA is increased by EBNA-2, an Epstein-Barr virus gene required for B-cell immortalization. J. Virol. 64:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krauer, K. G., N. Kienzle, D. B. Young, and T. B. Sculley. 1996. Epstein-Barr nuclear antigen-3 and -4 interact with RBP-2N, a major isoform of RBP-Jκ in B lymphocytes. Virology 226:346-353. [DOI] [PubMed] [Google Scholar]
- 33.Lee, W., Y. H. Hwang, S. K. Lee, C. Subramanian, and E. S. Robertson. 2001. An Epstein-Barr virus isolated from a lymphoblastoid cell line has a 16-kilobase-pair deletion which includes gp350 and the Epstein-Barr virus nuclear antigen 3A. J. Virol. 75:8556-8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Roux, A., B. Kerdiles, D. Walls, J. F. Dedieu, and M. Perricaudet. 1994. The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology 205:596-602. [DOI] [PubMed] [Google Scholar]
- 35.Lin, J., E. Johannsen, E. Robertson, and E. Kieff. 2002. Epstein-Barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA-2. J. Virol. 76:232-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling, P. D., J. J. Hsieh, I. K. Ruf, D. R. Rawlins, and S. D. Hayward. 1994. EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1. J. Virol. 68:5375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ling, P. D., D. R. Rawlins, and S. D. Hayward. 1993. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc. Natl. Acad. Sci. USA 90:9237-9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall, D., and C. Sample. 1995. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J. Virol. 69:3624-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nitsche, F., A. Bell, and A. Rickinson. 1997. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J. Virol. 71:6619-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orre, R. S., M. A. Cotter II, C. Subramanian, and E. S. Robertson. 2000. Prothymosin alpha functions as a cellular oncoprotein by inducing transformation of rodent fibroblasts in vitro. J. Biol. Chem. 276:1794-1799. [DOI] [PubMed] [Google Scholar]
- 42.Patel, M., S. J. Leevers, and P. M. Brickell. 1990. Regulation of c-fgr proto-oncogene expression in Epstein-Barr virus infected B-cell lines. Int. J. Cancer 45:342-346. [DOI] [PubMed] [Google Scholar]
- 43.Peng, R., J. Tan, and P. D. Ling. 2000. Conserved regions in the Epstein-Barr virus leader protein define distinct domains required for nuclear localization and transcriptional cooperation with EBNA2. J. Virol. 74:9953-9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pope, J. H., M. K. Horne, and W. Scott. 1968. Transformation of foetal human keukocytes in vitro by filtrates of a human leukaemic cell line containing herpes-like virus. Int. J. Cancer 3:857-866. [DOI] [PubMed] [Google Scholar]
- 45.Radkov, S. A., M. Bain, P. J. Farrell, M. West, M. Rowe, and M. J. Allday. 1997. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J. Virol. 71:8552-8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radkov, S. A., R. Touitou, A. Brehm, M. Rowe, M. West, T. Kouzarides, and M. J. Allday. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rickinson, A., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2628. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 48.Robertson, E. S., S. Grossman, E. Johannsen, C. Miller, J. Lin, B. Tomkinson, and E. Kieff. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J. Virol. 69:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robertson, E. S., J. Lin, and E. Kieff. 1996. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJκ. J. Virol. 70:3068-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sample, J., L. Young, B. Martin, T. Chatman, E. Kieff, and A. Rickinson. 1990. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J. Virol. 64:4084-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian, C., M. A. Cotter II, and E. S. Robertson. 2001. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat. Med. 7:350-355. [DOI] [PubMed] [Google Scholar]
- 52.Tomkinson, B., and E. Kieff. 1992. Use of second-site homologous recombination to demonstrate that Epstein-Barr virus nuclear protein 3B is not important for lymphocyte infection or growth transformation in vitro. J. Virol. 66:2893-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomkinson, B., E. Robertson, R. Yalamanchili, R. Longnecker, and E. Kieff. 1993. Epstein-Barr virus recombinants from overlapping cosmid fragments. J. Virol. 67:7298-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vales, L. D., and E. M. Friedl. 2002. Binding of C/EBP and RBP (CBF1) to overlapping sites regulates interleukin-6 gene expression. J. Biol. Chem. 277:42438-42446. [DOI] [PubMed] [Google Scholar]
- 56.Waltzer, L., M. Perricaudet, A. Sergeant, and E. Manet. 1996. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-Jκ-EBNA2-activated transcription by inhibiting the binding of RBP-Jκ to DNA. J. Virol. 70:5909-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, F., C. Gregory, C. Sample, M. Rowe, D. Liebowitz, R. Murray, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 64:2309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, F., C. D. Gregory, M. Rowe, A. B. Rickinson, D. Wang, M. Birkenbach, H. Kikutani, T. Kishimoto, and E. Kieff. 1987. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc. Natl. Acad. Sci. USA 84:3452-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, F., H. Kikutani, S. F. Tsang, T. Kishimoto, and E. Kieff. 1991. Epstein-Barr virus nuclear protein 2 transactivates a cis-acting CD23 DNA element. J. Virol. 65:4101-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, F., S. F. Tsang, M. G. Kurilla, J. I. Cohen, and E. Kieff. 1990. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J. Virol. 64:3407-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao, B., R. Dalbies-Tran, H. Jiang, I. K. Ruf, J. T. Sample, F. Wang, and C. E. Sample. 2003. Transcriptional regulatory properties of Epstein-Barr virus nuclear antigen 3C are conserved in simian lymphocryptoviruses. J. Virol. 77:5639-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao, B., D. R. Marshall, and C. E. Sample. 1996. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J. Virol. 70:4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an spi-1/Spi-B binding site. J. Virol. 74:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zimber-Strobl, U., K. O. Suentzenich, G. Laux, D. Eick, M. Cordier, A. Calender, M. Billaud, G. M. Lenoir, and G. W. Bornkamm. 1991. Epstein-Barr virus nuclear antigen 2 activates transcription of the terminal protein gene. J. Virol. 65:415-423. [DOI] [PMC free article] [PubMed] [Google Scholar]