Abstract
Treatment with antimetabolites results in chemically induced low nucleoside triphosphate pools and cell cycle arrest in exponentially growing cells. Since steady-state levels of hepatitis C virus (HCV) replicon RNA were shown to be dependent on exponential growth of Huh-7 cells, the effects of antimetabolites for several nucleoside biosynthesis pathways on cell growth and HCV RNA levels were investigated. A specific anti-HCV replicon effect was defined as (i) minimal interference with the exponential cell growth, (ii) minimal reduction in cellular host RNA levels, and (iii) reduction of the HCV RNA copy number per cell compared to that of the untreated control. While most antimetabolites caused a cytostatic effect on cell growth, only inhibitors of the de novo pyrimidine ribonucleoside biosynthesis mimicked observations seen in confluent replicon cells, i.e., cytostasis combined with a sharp decrease in replicon copy number per cell. These results suggest that high levels of CTP and UTP are critical parameters for maintaining the steady-state level replication of HCV replicon in Huh-7 cells.
Despite the availability of infectious cDNA clones of the hepatitis C virus (HCV), efficient in vitro replication has not been observed (3). After transfection of subgenomic HCV RNA replicons that also express the neomycin phosphotransferase gene selection marker, HCV replication has been reported previously in the human hepatoma cell line Huh-7 (2, 21). Such HCV replicon-harboring cell lines could be cultivated for more than a year without signs of cytopathogenicity (26). High levels of HCV RNAs can be maintained in cells passaged under continuous selection with G418. In addition, high-level replication was reflected in the observed adaptations of the HCV replicon to the host cell (20).
A tight coupling of the amount of intracellular HCV RNA and cell growth was observed. High levels of viral RNA were found in exponentially growing cells, but this was followed by a sharp decline in RNA levels when cells reached a confluent state (26, 29). This suggests that cellular factors required for RNA replication and/or translation vary in abundance and become limited in resting cells. Several proteins have been suggested, but none of them has as yet been positively identified to be directly responsible (9, 15, 16, 19, 25, 30). However, there are no reports in which the availability of the cellular nucleoside triphosphate pools were linked to the loss of steady-state level replication of HCV replicon RNA in confluent cells. It is well-known that confluent cells mainly depend on salvage nucleoside biosynthetic pathways, resulting in lowered concentrations of endogenous nucleoside pools. As most antimetabolite agents of the de novo nucleoside biosynthesis are known to chemically deplete nucleoside pools and induce a concentration-dependent cytostasis, evaluating these compounds against HCV replicon can shed light on the relationship between nucleoside pools and replicon dynamics. However, before doing so, a definition of specificity of antiviral effect in HCV replicon cells is required.
Specificity of the antiviral effect in the HCV replicon system.
Currently, alpha interferon (IFN-α) and ribavirin (Fig. 1) are the only drugs approved for treatment of HCV infection. Apart from these two agents, several other compounds have been reported to exert specific antiviral activity against HCV (6, 31; S. S. Carroll, R. L. LaFemina, D. L. Hall, A. L. Himmelberger, L. C. Kuo, M. MacCoss, D. B. Olsen, C. A. Rutkowski, J. E. Tomassini, H. Ann, B. Bhat, N. Bhat, P. D. Cook, A. B. Eldrup, C. J. Guinosso, M. Prhavc, and T. Prakash, 25 July 2002, U.S. Patent Office; J. P. Sommadossi and P. LaColla, 6 Dec. 2002).
FIG. 1.
Chemical structures of various nucleosides and antimetabolites.
In order to demonstrate the ability of the system to respond to specific inhibition, a series of control experiments were performed. IFN-α-2a, ribavirin, 2′-C-CH3-C (Fig. 1), and 2′-C-CH3-A (Fig. 1) were tested over a range of concentrations for their ability to reduce the HCV RNA levels in exponentially growing replicon cells after 4 days of exposure as described previously (29). At 100 IU/ml, IFN-α-2a had a minimal effect on the rRNA levels, and after correcting for this toxicity, a specific antiviral effect of 1.36 ± 0.37 log10 reduction in HCV RNA was observed (Table 1). As previously reported, IFN-α-2a showed a corrected 90% effective concentration (EC90) of 4.5 IU/ml after 96 h of incubation (29). Similar experiments and calculations were performed for 2′-C-CH3-C (EC90, 10.4 μM), ribavirin (EC90, ∼100 μM), and for 2′-C-CH3-A (EC90, 1.4 μM) (Table 1).
TABLE 1.
Antiviral and cytotoxicity effects of antimetabolites on HCV replicon-transfected Huh-7 cells
| Compound | log10 RNA reductiona at 100 μM for: |
Corrected HCV RNA |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HCV | rRNA | log10 reductiona at 100 μM | log10 reduction at 10 μM | EC90, μM | |||||||
| IMPDH inhibitors (EC 1.1.1.205) | |||||||||||
| Mizoribine | 0.29 ± 0.74 | 0.21 ± 0.50 | 0.08 ± 0.82 | −0.14 ± 0.12 | >100 | ||||||
| Tiazofurin | 0.86 ± 0.27 | 0.99 ± 0.35 | −0.13 ± 0.37 | 0.04 ± 0.10 | >100 | ||||||
| Mycophenolic acid | 1.15 ± 0.43 | 1.09 ± 0.28 | 0.07 ± 0.47 | 0.22 ± 0.01 | >100 | ||||||
| C2-MAD | 1.09 ± 0.21 | 1.00 ± 0.15 | 0.08 ± 0.24 | 0.36 ± 0.21 | >100 | ||||||
| Ribonucleotide reductase inhibitors (EC 1.17.4.1; EC 1.17.4.2) | |||||||||||
| Guanazole | 0.25 ± 0.11 | 0.07 ± 0.03 | 0.32 ± 0.08 | 0.05 ± 0.08 | >100 | ||||||
| Hydroxyurea | 0.17 ± 0.08 | 0.25 ± 0.20 | −0.08 ± 0.16 | 0.06 ± 0.04 | >100 | ||||||
| Tezacytabine | 1.59 ± 0.08 | 1.78 ± 0.69 | −0.19 ± 0.49 | 0.63 ± 0.07 | >100 | ||||||
| Deferoxamine | 1.00 ± 0.06 | 0.92 ± 0.08 | 0.08 ± 0.03 | 0.17 ± 0.11 | >100 | ||||||
| CTP synthase inhibitors (E.C. 6.3.4.2.) | |||||||||||
| CP-C | 1.97 ± 0.38 | 0.91 ± 0.13 | 1.06 ± 0.26 | 0.64 ± 0.10 | 25 | ||||||
| CPE-C | 2.47 ± 0.33 | 1.21 ± 0.16 | 1.26 ± 0.51 | 1.43 ± 0.01 | 2.5 | ||||||
| 3-DU | 1.41 ± 0.09 | 0.48 ± 0.11 | 0.94 ± 0.20 | 0.13 ± 0.10 | ∼100 | ||||||
| dFdC | 1.87 ± 0.16 | 0.59 ± 0.05 | 1.29 ± 0.11 | 1.32 ± 0.08 | Too toxic | ||||||
| OMPDC inhibitors (EC 4.1.1.23) | |||||||||||
| 6-Azauridine | 0.25 ± 0.09 | 0.61 ± 0.18 | −0.36 ± 0.16 | 0.12 ± 0.05 | >100 | ||||||
| 2-Thio-6-azauridine | 0.16 ± 0.04 | −0.02 ± 0.12 | 0.19 ± 0.09 | 0.12 ± 0.10 | >100 | ||||||
| PZF | 1.88 ± 0.05 | 0.42 ± 0.03 | 1.46 ± 0.08 | 1.16 ± 0.21 | 3.80 | ||||||
| ATC inhibitors (EC 2.1.3.2) | |||||||||||
| PALA | 1.77 ± 0.02 | 0.48 ± 0.02 | 1.30 ± 0.05 | 1.18 ± 0.11 | 7.60 | ||||||
| Dihydroorotate dehydrogenase inhibitors (E.C. 1.3.3.1) | |||||||||||
| Brequinar (NSC-368390) | −0.05 ± 0.05 | 0.29 ± 0.01 | −0.34 ± 0.04 | −0.17 ± 0.09 | >100 | ||||||
| Dichloroallyl lawsone (NSC-126771) | 1.27 ± 0.02 | 2.13 ± 0.15 | −0.86 ± 0.17 | −0.52 ± 0.01 | >100 | ||||||
| Thymidylate synthase inhibitors (E.C. 2.1.1.45) | |||||||||||
| 2′-Deoxy-5-fluorouridine | 0.76 ± 0.06 | 0.73 ± 0.35 | 0.04 ± 0.25 | 0.23 ± 0.05 | >100 | ||||||
| Methotrexate | 0.18 ± 0.01 | 0.07 ± 0.10 | 0.11 ± 0.09 | 0.15 ± 0.01 | >100 | ||||||
| Controls | |||||||||||
| IFN-α-2a | 1.57 ± 0.26 | 0.21 ± 0.21 | 1.36 ± 0.37 | NAb | 4.5 IU/ml | ||||||
| Ribavirin | 1.96 ± 0.28 | 0.91 ± 0.12 | 1.05 ± 0.29 | 0.16 ± 0.10 | ∼100 | ||||||
| 2′-C-CH3-A | 2.32 ± 0.11 | 2.96 ± 0.08 | −0.64 ± 0.18 | 2.05 | 1.4 | ||||||
| 2′-C-CH3-C | 2.20 ± 0.52 | −0.02 ± 0.05 | 2.21 ± 0.47 | 1.0 | 10.4 | ||||||
IFN tested at 100 IU/ml; dFdC tested initially at 50 μM; a meaningful EC90 value could not be attributed because of extreme toxicity.
NA, not applicable.
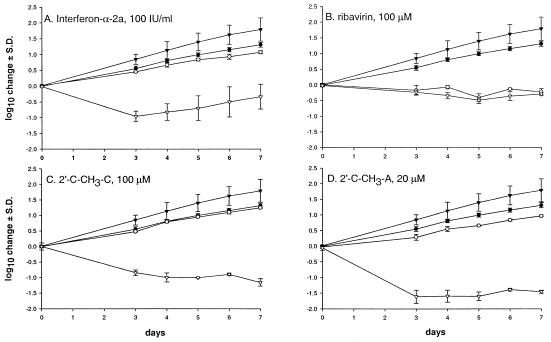
However, the EC90 determined on day 4 is a single static efficacy measurement that does not account for the influence of cell growth dynamics on HCV RNA replication, i.e., the compound-related changes in the obligate requirement for logarithmic cell growth. Therefore, experiments were conducted to monitor HCV RNA levels and the cell growth dynamics over a 7-day period. A total of 104 cells per well were seeded in a 24-well plate, and at the end of the incubation step, cells were counted using the trypan blue exclusion method, and replicon RNA was quantified as previously described (29). IFN-α-2a-treated cells grew significantly slower (day 7, 1.07 ± 0.06 log10 increase from day 0) than the untreated cell controls (1.31 ± 0.08 log10 increase from day 0; P = 0.003) (Fig. 2A). In addition, a significant drop in HCV RNA levels was observed over the 7-day period (control, 1.79 ± 0.4; IFN-α-2a, −0.53 ± 0.4; P = 0.0005). The rebound in replicon RNA from day 4 onward was also noted previously by Cheney et al. (7).
FIG. 2.
Dynamics of cell growth and HCV RNA levels after exposure to control anti-HCV compounds. HCV replicon cells were seeded at approximately 104 cells per well in a 24-well plate. Over a 7-day period, cells were counted daily, and rRNA and HCV RNA were quantified by quantitative reverse transcription-PCR. (A) IFN-α-2a at 100 IU/ml. (B) Ribavirin at 100 μM. (C) 2′-C-CH3-C at 100 μM. (D) 2′-C-CH3-A at 20 μM. •, cell proliferation in the absence of the compound; ○, cell proliferation in the presence of the compound; ▾, HCV RNA levels in untreated cells; ▿, HCV RNA levels in the presence of the compound. The points shown are averages ± standard deviations (S.D.) (error bars) for three independent experiments.
Ribavirin tested at 100 μM completely inhibited cell proliferation (0.22 ± 0.1 log10 reduction in cell growth at day 7 compared to day 0 or 1.53 log10 reduction compared to the no-treatment control on day 7) (Fig. 2B). Although ribavirin reduced HCV RNA by 2.08 log10 on day 7 compared to untreated controls, the ratio of HCV RNA copy number per cell in treatment versus no-treatment controls changed only marginally, suggesting that the antiviral activity is not specific.
2′-C-CH3-C (Fig. 2C) was tested at 100 μM and found to be very effective in reducing the HCV RNA levels with no apparent effects on the dynamics of the cell proliferation. The in vitro toxicity profile of 2′-C-CH3-A precluded antiviral testing at 100 μM; however, at 20 μM this compound reduced the HCV levels significantly while minimally affecting cell proliferation (Fig. 2D) (Table 1). Thus, determination of a specific antiviral effect on the HCV RNA replicon depends on at least some, if not a combination of all, of the following conditions: (i) no effect on exponential cell growth, (ii) no or limited reduction of cellular host RNA levels, and (iii) significant reduction of HCV RNA copy number per cell compared to that of the untreated controls.
Antiviral effect of antimetabolites.
One of the most important questions in testing antimetabolites for antireplicon (or antiviral) activity is whether induction of the cytostatic and/or cytotoxic condition can be separated from specific antiviral activity, since “dead cells don't make virus.” There are many reports in which specific antiviral activity has been ascribed to antimetabolites (1, 11, 22, 23, 28), but to date, no systematic study has been conducted to determine their effects on HCV subgenomic replicon. In addition, the antiviral action of antimetabolites such as ribavirin remains controversial (17, 18, 29).
The results of our experiments are summarized in Table 1. Although several of these antimetabolites significantly lowered HCV RNA levels, a similar inhibitory effect was exerted on rRNA levels. After correcting for cellular toxicity, the majority of these antimetabolites had no specific antiviral activity against HCV (EC90 > 100 μM).
However, certain compounds that inhibited enzymes responsible for the de novo biosynthesis of UTP and CTP, such as aspartate transcarbamoylase (ATC; EC 2.1.3.2), orotidine 5′-monophosphate decarboxylase (OMPDC; EC 4.1.1.23), and CTP synthase (CTPS; EC 6.3.4.2) showed modest antiviral activity. EC90 values (at 96 h of incubation) were determined for these compounds (Fig. 1) as follows: cyclopentylcytosine (CP-C), 25 μM; cyclopentenylcytosine (CPE-C), 2.5 μM; 3-deazaurine (3-DU), ∼100 μM; pyrazofurin (PZF; NSC-143095), 3.8 μM; N-(phosphonoacetyl)-l-aspartate (PALA; NSC-224131), 7.6 μM; and 2′-deoxy-2′,2′-difluorocytidine (dFdC, gemcitabine), not attributed because of extreme toxicity. dFdC was found to be the most potent inhibitor of both replicon RNA and cells. Previous studies have shown that the intracellular metabolites of dFdC exert several antimetabolic activities, including the inhibition of ribonucleoside reductase and CTPS (14, 27).
Dynamics of the antiviral effect of inhibitors of the de novo synthesis of ribopyrimidines.
It is generally expected that exposure of cells to inhibitors of the ATC, OMPDC, and CTPS enzymes will result in reduced levels of UTP and CTP, which may subsequently lead to an arrest in logarithmic cell growth. The dynamics of HCV replicon cells exposed to these inhibitors (at their EC90s) were monitored over a 7-day period.
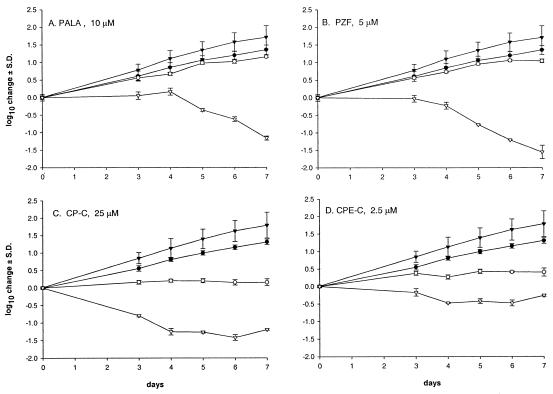
When tested at their 96-h EC90s, PALA and PZF, which are inhibitors of the early de novo pyrimidine biosynthetic steps, reduced cell proliferation only minimally, but they significantly reduced HCV RNA levels (Fig. 3A and B). On the other hand, inhibitors of the CTPS enzyme (the last biosynthetic step in the synthesis of CTP) such as CP-C (Fig. 3C), CPE-C (Fig. 3D), and 3-DU (Table 1) caused cytostatic effects on the HCV replicon cell line but also reduced HCV-replicon RNA levels. Similar levels of cytostasis were also observed with ribavirin (Fig. 2B), although CTPS enzyme inhibitors seemed more specific in reducing HCV RNA levels than inosine monophosphate dehydrogenase (IMPDH) inhibitors in this cell culture system. DFdC results were meaningless, because a noncytotoxic concentration (meaning active cell death; range of testing was 50 to 1,000 nM) could not be found (data not shown).
FIG. 3.
Dynamics of the cell growth and HCV RNA levels after exposure to selected antimetabolites. The experimental method used is described in the legend to Fig. 2. (A) PALA at 10 μM. (B) PZF at 5 μM. (C) CP-C at 25 μM. (D) CPE-C at 2.5 μM. •, cell proliferation in the absence of the compound; ○, cell proliferation in the presence of the compound; ▾, HCV RNA levels in untreated cells; ▿, HCV RNA levels in the presence of the compound. The points are averages ± standard deviations (S.D.) (error bars) for three independent experiments.
Reduction of replicon RNA copy number per cell.
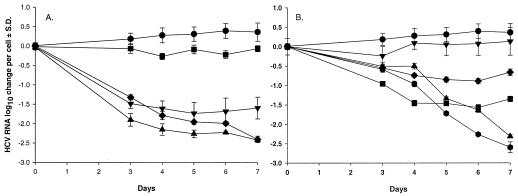
In an additional attempt to express the specificity of the antiviral action of the control compounds versus the antimetabolites, the log10 change in replicon RNA copy number per cell and per day were calculated from the above experiments. IFN-α-2a reduced the copy numbers per cell by approximately 1.6 log10 at day 3, after which a new steady-state level was achieved on days 4 to 7 (Fig. 4A). Ribavirin reduced the replicon RNA copy number per cell only minimally. In the 7-day experiments for the two control compounds, 2′-C-CH3-C and 2′-C-CH3-A, the RNA copy number per cell showed a continuous drop through day 7 and a new steady state was not reached. In fact, these control nucleoside analogues achieved a maximum reduction of 2.76 log10 in replicon HCV RNA levels per cell. In previous analyses, the replicon RNA levels were quantified at approximately 5,000 copies per cell; hence, treatment with these inhibitors reduced the copy number to approximately 10 copies per cell (data not shown).
FIG. 4.
Replicon RNA changes per cell. The plots were obtained from collected data as shown in Fig. 2 and 3, in which log10 changes for cell count and replicon RNA levels were subtracted from each other. (A) Control antiviral compounds. •, no drug control; ▾, IFN-α-2a at 100 IU/ml; ▪, ribavirin at 100 μM; ⧫, 2′-C-CH3-C at 100 μM; ▴, 2′-C-CH3-A at 20 μM. (B) Selection of the most important antimetabolites. •, no drug control; ▾, dFdC at 50 nM; ▪, CP-C at 25 μM; ⧫, CPE-C at 2.5 μM; ▴, PALA at 10 μM; , PZF at 5 μM.
The reductions in replicon RNA copy numbers per cell for the antimetabolites CP-C and CPE-C showed a profile similar to that seen for IFN-α-2a, i.e., a new steady state with a potential rebound at day 7. In contrast, PALA and PZF treatment resulted in a time-dependent reduction in replicon RNA levels, with a reduction of 2.96 log10 after 7 days' treatment with 5 μM PZF (Fig. 4B).
Prevention studies.
A series of experiments was then conducted to study the possibility of preventing the observed antiviral and cytostatic effects. Cells were incubated simultaneously with the test compound (at the 96-h EC90) and the natural ribo- or 2′-deoxynucleosides (at 50 μM). The antiviral effect of IFN-α-2a and 2′-C-CH3-A could not be prevented by any of the natural nucleosides at this concentration. As expected for the IMPDH inhibitors, 2′-deoxyguanosine and guanosine prevented the effects of ribavirin on cell growth and HCV replicon RNA replication. For dFdC, the observed toxicities and antiviral effects were prevented by 2′-deoxycytidine. In line with expectations for CTPS inhibitors (CPE-C, CP-C, and 3-DU), the addition of cytidine to the culture medium compensated for the inhibitory effects. Surprisingly, the antimetabolic effects of CPE-C could be prevented by both 50 μM uridine and cytidine in the medium. The effects of PALA and PZF could be prevented by the addition of uridine to the culture medium. These results indeed indicate that (i) the intracellular levels of uridine and cytidine (most likely at the 5′ triphosphate form) are indeed responsible for the antiviral activity and for the cytostatic effect, and/or (ii) the activity of the antimetabolites depends up steps in the uridine-cytidine pathway.
From all the above, the most important observation made is that, although most of the compounds induced cytostasis, not all the antimetabolites possessed the ability to reduce HCV RNA replicon copy number per cell. Typically, IMPDH inhibitors showed minimal reduction, while CTPS inhibitors were more potent. Thus, intracellular nucleoside pools play an important role in maintaining steady-state levels of HCV RNA copy number. When cells enter antimetabolite-induced cytostasis, reductions in UTP and CTP levels seem to have a greater impact on reducing HCV RNA turnover than the level of GTP (or purines).
Replicon RNA turnover is determined by an equilibrium between active production through RdRP and HCV replicon RNA half-life. The data suggest that certain de novo pyrimidine ribonucleoside inhibitors may have the capacity to mimic conditions present in confluent cells, e.g., the rapid degradation of replicon RNA pools under cytostasis. High levels of cytoplasmic pyrimidine nucleosides and/or the de novo synthesis of pyrimidines seems to be of crucial importance to replicon replication, because inhibiting these synthetic steps results in measurable reductions in viral RNA. If the limited availability of intracellular UTP and CTP were responsible for the loss of replicon RNA steady state in confluent, untreated cells, then our observations with many of these antimetabolite inhibitors in replicon cells could be interpreted as nonspecific with respect to antiviral activity.
In light of our observations, most of the antimetabolites evaluated nonspecifically destroyed an established equilibrium of active cytoplasmic replication versus the half-life of the replicon RNA molecule. However, antimetabolites such as PALA and PZF did show some specific antiviral activity. PZF was considered a potential anticancer agent in the 1970s, but clinical trials were abandoned because of lack of efficacy at the maximum tolerated dose and unwanted side effects such as mucositis (4, 5, 8, 10, 12, 13, 24). Whether PZF shows any therapeutic anti-HCV potential at a relevant antiviral dose will need further investigation.
Acknowledgments
We thank Leanne Cartee for useful commentary. We thank NIH for providing PZF.
This study was supported in part by NIH grant 1 R43 AI 52686-01. R. F. Schinazi is supported in part by the Department of Veterans Affairs.
R.F.S. is a consultant to Pharmasset, Ltd., and his particulars have been reviewed by Emory University's Conflict of Interest Committee.
REFERENCES
- 1.Balzarini, J. 2000. Effect of antimetabolite drugs of nucleoside metabolism on the anti-human immunodeficiency virus activity of nucleoside reverse transcriptase inhibitors. Pharmacol. Ther. 87:175-187. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2001. Novel cell culture systems for the hepatitis C virus. Antivir. Res. 52:1-17. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 4.Budman, D., V. Currie, and R. Wittes. 1977. Phase II trial of pyrazofurin in malignant melanoma. Cancer Treat. Rep. 61:1733-1734. [PubMed] [Google Scholar]
- 5.Carroll, D. S., N. E. Kemeny, and R. J. Gralla. 1979. Phase II evaluation of pyrazofurin in patients with advanced colorectal carcinoma. Cancer Treat. Rep. 63:139-140. [PubMed] [Google Scholar]
- 6.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 7.Cheney, I. W., V. C. Lai, W. Zhong, T. Brodhag, S. Dempsey, C. Lim, Z. Hong, J. Y. Lau, and R. C. Tam. 2002. Comparative analysis of anti-hepatitis C virus activity and gene expression mediated by alpha, beta, and gamma interferons. J. Virol. 76:11148-11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, E., V. Currie, and R. E. Wittes. 1979. Phase II trial of pyrazofurin in advanced head and neck cancer. Cancer Treat. Rep. 63:2047-2048. [PubMed] [Google Scholar]
- 9.Chung, R. T., and L. M. Kaplan. 1999. Heterogeneous nuclear ribonucleoprotein I (hnRNP-I/PTB) selectively binds the conserved 3′ terminus of hepatitis C viral RNA. Biochem. Biophys. Res. Commun. 254:351-362. [DOI] [PubMed] [Google Scholar]
- 10.Creagan, E. T., J. Rubin, C. G. Moertel, A. J. Schutt, M. J. O'Connell, R. G. Hahn, R. J. Reitemeir, and S. Frytak. 1977. Phase II study of pyrazofurin in advanced colorectal carcinoma. Cancer Treat. Rep. 61:491-493. [PubMed] [Google Scholar]
- 11.De Clercq, E., J. Murase, and V. E. Marquez. 1991. Broad-spectrum antiviral and cytocidal activity of cyclopentenylcytosine, a carbocyclic nucleoside targeted at CTP synthetase. Biochem. Pharmacol. 41:1821-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gralla, R. J., V. E. Currie, R. E. Wittes, R. B. Golbey, and C. W. Young. 1978. Phase II evaluation of pyrazofurin in patients with carcinoma of the lung. Cancer Treat. Rep. 62:451-452. [PubMed] [Google Scholar]
- 13.Gralla, R. J., P. P. Sordillo, and G. B. Magill. 1978. Phase II evaluation of pyrazofurin in patients with metastatic sarcoma. Cancer Treat. Rep. 62:1573-1574. [PubMed] [Google Scholar]
- 14.Heinemann, V., L. Schulz, R. D. Issels, and W. Plunkett. 1995. Gemcitabine: a modulator of intracellular nucleoside and deoxynucleoside metabolism. Semin. Oncol. 22:11-18. [PubMed] [Google Scholar]
- 15.Inoue, Y., M. Miyazaki, R. Ohashi, T. Tsuji, K. Fukaya, H. Kouchi, T. Uemura, K. Mihara, and M. Namba. 1998. Ubiquitous presence of cellular proteins that specifically bind to the 3′ terminal region of hepatitis C virus. Biochem. Biophys. Res. Commun. 245:198-203. [DOI] [PubMed] [Google Scholar]
- 16.Ito, T., and M. M. Lai. 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 71:8698-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanford, R. E., B. Guerra, H. Lee, D. R. Averett, B. Pfeiffer, D. Chavez, L. Notvall, and C. Bigger. 2003. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(I)-poly(C), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. J. Virol. 77:1092-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau, J. Y., R. C. Tam, T. J. Liang, and Z. Hong. 2002. Mechanism of action of ribavirin in the combination treatment of chronic HCV infection. Hepatology 35:1002-1009. [DOI] [PubMed] [Google Scholar]
- 19.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 22.Markland, W., T. J. McQuaid, J. Jain, and A. D. Kwong. 2000. Broad-spectrum antiviral activity of the IMP dehydrogenase inhibitor VX-497: a comparison with ribavirin and demonstration of antiviral additivity with alpha interferon. Antimicrob. Agents Chemother. 44:859-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrey, J. D., D. F. Smee, R. W. Sidwell, and C. Tseng. 2002. Identification of active antiviral compounds against a New York isolate of West Nile virus. Antivir. Res. 55:107-116. [DOI] [PubMed] [Google Scholar]
- 24.Nichols, W. C., L. K. Kvols, J. N. Ingle, J. H. Edmonson, D. L. Ahmann, J. Rubin, and M. J. O'Connell. 1978. Phase II study of triazinate and pyrazofurin in patients with advanced breast cancer previously exposed to cytotoxic chemotherapy. Cancer Treat. Rep. 62:837-839. [PubMed] [Google Scholar]
- 25.Petrik, J., H. Parker, and G. J. Alexander. 1999. Human hepatic glyceraldehyde-3-phosphate dehydrogenase binds to the poly(U) tract of the 3′ non-coding region of hepatitis C virus genomic RNA. J. Gen. Virol. 80:3109-3113. [DOI] [PubMed] [Google Scholar]
- 26.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plunkett, W., P. Huang, Y. Z. Xu, V. Heinemann, R. Grunewald, and V. Gandhi. 1995. Gemcitabine: metabolism, mechanisms of action, and self-potentiation. Semin. Oncol. 22:3-10. [PubMed] [Google Scholar]
- 28.Stuyver, L. J., S. Lostia, S. E. Patterson, J. Clark, K. A. Watanabe, M. J. Otto, and K. W. Pankiewicz. 2003. Inhibitors of the IMPDH enzyme as potential anti-bovine viral diarrhea virus agents. Antiviral Chem. Chemother. 13:49-56. [DOI] [PubMed] [Google Scholar]
- 29.Stuyver, L. J., T. Whitaker, T. R. McBrayer, B. I. Hernandez-Santiago, S. Lostia, P. M. Tharnish, M. Ramesh, C. K. Chu, R. Jordan, J. Shi, S. Rachakonda, K. A. Watanabe, M. J. Otto, and R. F. Schinazi. 2003. A ribonucleoside analogue that blocks the replication of bovine viral diarrhea and hepatitis C viruses in culture. Antimicrob. Agents Chemother. 47:244-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuchihara, K., T. Tanaka, M. Hijikata, S. Kuge, H. Toyoda, A. Nomoto, N. Yamamoto, and K. Shimotohno. 1997. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J. Virol. 71:6720-6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker, M. P., and Z. Hong. 2002. HCV RNA-dependent RNA polymerase as a target for antiviral development. Curr. Opin. Pharmacol. 2:534-540. [DOI] [PubMed] [Google Scholar]