Abstract
Addition of human papillomavirus (HPV) E7 CDK2/cyclin A or CDK2/cyclin E, purified from either insect cells or bacteria, dramatically upregulates histone H1 kinase activity. Activation is substrate specific, with a smaller effect noted for retinoblastoma protein (Rb). The CDK2 stimulatory activity is equivalent in high-risk (HPV type 16 [HPV16] and HPV31) and low-risk (HPV6b) E7. Mutational analyses of HPV16 E7 indicate that the major activity resides in amino acids 9 to 38, spanning CR1 and CR2, and does not require casein kinase II or Rb-binding domain functions. Synthetic peptides spanning HPV16 amino acid residues 9 to 38 also activate CDK2. Peptides containing this sequence that carry biotin on the carboxy terminus, as well as a photoactivated cross-linking group (benzophenone), also activate the complex and covalently associate with the CDK2/cyclin A complex in a specific manner requiring UV. Cross-linking studies that use protein monomers detect association of the E7 peptides with cyclin A but not CDK2. Together, our results indicate a novel mechanism whereby E7 promotes HPV replication by directly altering CDK2 activity and substrate specificity.
The human papillomaviruses (HPVs) are a large family of small (∼8 kb) DNA viruses that infect stratified squamous epithelia and cause warts (27). Certain high-risk genotypes infecting mucosal epithelia, including HPV type 16 (HPV16), HPV18, and HPV31, are implicated as critical etiological agents in cervical and other cancers (56). The E6 and E7 genes from these genotypes are potent, cooperating oncogenes. E6 binds to p53, resulting in its ubiquitin-dependent degradation (47, 51), while E7 binds and promotes degradation of the tumor suppressor pRB (21, 24, 30). However, E6 and E7 have other activities, and while their roles in the viral life cycle are not fully understood, it is believed that they play a central role in promoting the S phase in the differentiating keratinocyte so that host replication proteins are available to advance viral DNA amplification.
The HPV life cycle is regulated in a differentiation-dependent manner within stratified squamous epithelia. HPV DNA replication depends heavily upon the host cell replication machinery. HPV DNA is maintained as an episome in the basal cell layer, which contains the dividing cell population. Cells depart the basal layer, move upward through the epithelium, differentiate, and die (3, 17). Viral DNA undergoes vegetative replication (amplification) within the differentiated keratinocyte in preparation for packaging and sloughing within the dead, cornified cells of the epithelium (8). Differentiated keratinocytes are normally withdrawn from the cell cycle and are unable to support DNA synthesis. However, E7 provides an activity that promotes DNA synthesis within these cells that permits viral DNA amplification (7, 18, 29). Consequently, E7 functions within one or more pathways that regulate S-phase entry and progression.
Cell cycle progression is regulated by the cyclin-dependent kinase (CDK) family. In mammalian cells movement into and progression through the S phase are both regulated by CDK2 complexed with cyclins E and A (14). CDK activity is tightly controlled throughout the cell cycle by phosphorylation, by association with cyclins, and by association with inhibitory proteins (38, 44). CDK2 differs from CDK4/CDK6 in that it also acts independently of the retinoblastoma protein (Rb)-E2F pathway to facilitate S-phase entry and progression (33). CDK2 substrates include Rb and related pocket proteins (14, 22), p27 (48), E2F (23, 39), CDC7 (34), and NPAT (55). Histone H1 has also been identified as a CDK2 substrate whose phosphorylation in late G1 and early S phases relaxes chromatin structure in preparation for DNA synthesis (5, 26, 45).
HPV E7 binds several cell cycle-regulatory elements. E7 binding and degradation of Rb are the best-characterized functions of E7 (21, 24, 25). Previous studies have also demonstrated E7 binding to cyclin A/CDK2 (10, 50) and cyclin E/CDK2 (37) in cell extracts, but these findings are largely attributed to indirect binding mediated by p107 and related pocket proteins (see references 37 and 41 for discussion). E7 also binds the KIP family members p21 and p27 (20, 31, 54), which are critical for cell cycle advancement and S-phase entry. Binding and inhibition of proteins in these pathways may promote entry into the S phase by abolition of Rb effects on the transcription factor E2F.
Keratinocytes exit the cell cycle, differentiate, and die during the course of epithelial stratification. These events are normally irreversible, but HPV E7 is sufficient to promote an unscheduled round of DNA synthesis in differentiated keratinocytes (7, 29). E7 is also required during the viral life cycle in raft cultures for viral DNA amplification (18). How E7 performs these functions is not known. E7 is able to overcome negative growth signals in cells, including those mediated by transforming growth factor β, loss of substrate adherence, and cell cycle exit induced by serum deprivation and calcium (42, 46). These activities contribute to the immortalization and transforming potential of E7 and correlate in part with the ability of E7 to bind pRb family members. However, strong evidence exists for functions in addition to pocket protein binding that are required for immortalization and transformation (2, 12, 28, 37).
Direct binding of E7 to the CDK2 complex continues to be an attractive mechanism by which E7 promotes S-phase entry for various reasons, including repeated demonstrations of association of E7 with the CDK2 complex (10, 16, 37, 50); lack of conservation of known functional activities, such as Rb binding, among high- and low-risk forms of E7 (40); weak association of E7 Rb-binding activity with its cell cycle-promoting activity (6, 16); and lack of a requirement for Rb binding for wart formation by cottontail rabbit papillomavirus (11). Consequently, the means by which E7 promotes S-phase entry is uncertain and of high interest. In the present study, we tested if E7 could directly alter CDK2 activity in vitro. HPV E7 from both high-risk (HPV16 and HPV31) and low-risk (HPV6b) viruses, as well as E7 synthetic peptides, directly promoted cyclin A/CDK2 and cyclin E/CDK2 histone H1 kinase activity. The direct and specific association of E7 with the CDK2 complex was demonstrated by cross-linking experiments. Our results suggest a simple and direct mechanism by which HPV E7 may act in the HPV life cycle to promote the S phase.
MATERIALS AND METHODS
Recombinant E7 protein production.
E7 from HPV6b, HPV16, and HPV31 DNA was amplified by PCR with primers with introduced restriction sites for subcloning into the pGEX4T-3 expression vector (Amersham). The following primers were used: 6BE7(BamHI) sense, 5′-CGGGATCCATGCATGGAAGACATGTT-3′; 6BE7(XhoI) antisense, 5′-CCGCTCGAGTTAGGTCTTCGGTGCGC-3′; 16E7(BamHI) sense, 5′-CGGGATCCATGCATGGAGATACACCTAC-3′; 16E7(XhoI) antisense, 5′-CCGCTCGAGTTATGGTTTCTGAGAACAGATG-3′; 31E7(BamHI) sense, 5′-CGGGATCCATGCGTGGAGAAACACCTAC-3′; and 31E7(BamHI) antisense, 5′-CGGGATCCTTACAGTCTAGTAGAACAG-3′.
Clones were identified by screening miniprep DNA, and all clones were confirmed by sequence analysis. Glutathione-S-transferase (GST)-E7 fusion proteins were overexpressed in Escherichia coli strain JM109. Bacteria were pelleted after a 4-h induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside at 30°C. The cell pellet was resuspended in lysis buffer (11.3 mM NaH2PO4, 38.7 mM Na2HPO4, 0.07% β-mercaptoethanol, 10 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.025 trypsin inhibitory units of aprotinin/ml, and 10 nM leupeptin), and supernatants were collected following centrifugation. Proteins were purified by using glutathione-agarose beads. After extensive washing of the beads with phosphate-buffered saline (PBS), fusion proteins were eluted with 50 mM Tris-HCl [pH 8.0] containing 5 mM reduced glutathione. In all cases, protein levels were quantified (4).
Carboxy-terminal truncation mutants of GST-16E7 (E7 1-87, 1-69, 1-48, 1-38, 1-27, and 1-15) were made by introducing a stop codon into pGEX-16E7 via site-directed mutagenesis by using a QuikChange kit (Stratagene). Other GST-16E7-encoding constructs (E7 9-98 and 16-98) were generated by direct subcloning via PCR into pGEX4T-3 in a manner similar to that described for full-length E7 (above). The construct encoding GST-16E7 (39-98) was made by using a QuikChange kit with PCR primers designed to exclude the relevant E7 amino-terminal region by using pGEX-16E7 as template DNA. All recombinant DNA constructs were verified by sequence analysis and examination of the apparent molecular weight of the expressed proteins.
Protein kinases.
Recombinant CDK complexes were purified following baculovirus-mediated expression in insect cells. GST-cyclin E and GST-cyclin A were coinfected with hemagglutinin (HA)-tagged CDK2 by using baculovirus vectors in Spodoptera frugiperda (Sf9) or High Five (Invitrogen) insect cells at a multiplicity of infection of 20 for cyclin viruses and 10 for CDK2. For preparation of monomers, GST-cyclin A or HA-CDK2 was transfected alone. Cells were grown at 28°C with agitation for 66 h, collected by centrifugation, and lysed by resuspension of 107 cells per ml in hypotonic lysis buffer (10 mM HEPES, pH 7.4, 10 mM NaCl, mM EDTA, 0.2 μg of pepstatin/ml, 0.2 μg of aprotinin/ml, and 0.2 mM 4-[2-amino-ethyl]-benzenesulfonyl fluoride). After 1 h, NaCl was added to a final concentration of 150 mM, the cells were sonicated on ice for 2 min with a Branson sonifier (100% duty cycle with an output of 4), and the lysate was clarified by centrifugation. GST-cyclin complexes were then isolated by affinity chromatography with glutathione-Sepharose beads (Pharmacia Biotech). Following extensive washing the complexes were eluted with 15 mM reduced glutathione in 50 mM Tris-HCl, pH 8.0. Eluted proteins were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting to confirm purity of the cyclin A/CDK2 and cyclin E/CDK2 complexes. CDK5/p25 was provided by K. Leach (Pharmacia Corp.). Bacterially expressed, reconstituted cyclin A/CDK2 kinase, mitogen-activated protein (MAP) kinase I, and myelin basic protein (MBP) were purchased from Upstate Biotechnologies (Lake Placid, N.Y.).
Pooled fractions of GST-cyclin A monomers were prepared by glutathione affinity as above, diluted 1:7 in buffer 75 (20 mM bicine, pH 8.0, 75 mM NaCl, 5% glycerol, and 1 mM dithiothreitol [DTT]), and were loaded on a Q-Sepharose column. After extensive washing with buffer 125 (20 mM bicine, pH 8.0, 125 mM NaCl, 5% glycerol, and 1 mM DTT), GST-cyclin A was eluted with buffer 200 (20 mM bicine, pH 8.0, 200 mM NaCl, 5% glycerol, and 1 mM DTT). CDK2-HA monomer was purified with the Catch and Release Immunoprecipitation System (Upstate Biotechnologies) by using the manufacturer's recommendations.
Kinase assays.
Activity of purified cyclin A/CDK2 or cyclin E/CDK2 complexes (40 ng; ∼15 nM) was measured following addition of 1 μl of 2 mCi of [γ-32P]ATP (Amersham Pharmacia Biotech)/ml and 3.75 μg of histone H1 (Boehringer Mannheim) per 30-μl assay, and incubation at 30°C for 15 min in kinase buffer (20 mM MgCl, 10 mM EGTA, and 40 mM HEPES, pH 7.0). The assay time fell within the linear portion of time course experiments (data not shown). In some assays, full-length, purified Rb (QED Bioscience, Inc.) was used as substrate at 3.75 μg per assay. Purified MBP-E1 (1 μg) and MBP-E2 (2 μg) (provided by J. Shelley and P. Wells, Pharmacia Corp.) were used as substrates in some experiments. GST-E7 (1 μg) was directly added to purified recombinant CDK2 complexes and was incubated on ice in kinase buffer for 30 min prior to assay. The kinase reactions were stopped by addition of 2× sample buffer (32), and phosphorylated proteins were analyzed by SDS-PAGE followed by autoradiography and PhosphorImager analysis. In some cases, E7 activity is represented as activation (n-fold), which was calculated by direct comparison, under identical conditions, to kinase that received no E7.
GST-E7 (1 μg) was preincubated with p27 (200 ng) at room temperature for 1 h prior to assay in some experiments. For these assays human p27 cDNA was subcloned into pTrcHisB (Invitrogen) and was expressed in bacteria. Histidine-tagged, full-length p27 was purified by Ni2+-affinity chromatography.
Peptide synthesis.
Synthesis of the initial biotinylated peptide was at 0.25-mmol scale. Fmoc-Gly-Wang resin (0.301 g) (0.83 mmol/g; Advanced Chemtech) was used as the starting resin for the synthesis. Following removal of the Fmoc group with 20% piperidine-dimethyl formamide (DMF) and subsequent washes with DMF, N-α-Fmoc-N-ɛ-(d-biotinyl)-l-lysine-OH (1 mmol) was manually coupled to the Gly-resin with PyBop (1 mmol) and diethylpropylethylamine (2.2 mmol) for 3 h at room temperature in a small reaction vessel. This resin was then placed on an ABI 433A Peptide Synthesizer, and the remaining peptide sequence was synthesized by utilizing 433A automated Fmoc/O-benzotriazole-N-N-N′-N′-tetramethyl-uranium-hexafluorophosphate-mediated synthesis protocols. The peptide was cleaved from the resin in 10 ml of a solution containing 95% trifluoroacetic acid (TFA), 3% anisole, 1% ethanedithiol, and 1% ethyl methyl sulfide for 3 h at room temperature. The cleavage solution was filtered through a sintered glass funnel and was evaporated to near-dryness under reduced pressure. The crude peptide was collected on a sintered glass funnel, washed with diethyl ether, dissolved in dilute acetic acid, and evaporated to dryness under reduced pressure. The residue was dissolved in water and was lyophilized.
Purification of this peptide was accomplished by reverse-phase preparative high-pressure liquid chromatography (HPLC). The crude peptide was dissolved in dilute acetic acid, filtered, and loaded onto a preparative reverse-phase column (22 by 250 mm, 10 μm; Vydac C18) at 5 ml of solvent A/ml (100% water + 0.1% TFA). The linear gradient used was 0 to 40% solvent B (acetonitrile + 0.1% TFA) over 150 min. The column effluent was monitored by absorbance at 220 and 280 nm. Fractions were monitored on an analytical reverse-phase system (4.6 by 250 mm, 5 μm; Vydac C18); solvents were as described above. A linear gradient from 0 to 90% B in 19 min was employed for monitoring purposes. The chemical authenticity of the peptide was established by mass spectrometry by employing a Micromass Platform II mass spectrometer equipped with a Hewlett-Packard Series 1050 HPLC system. The identity of each peptide was confirmed by injecting 5 μl of sample into the flow of 100 μl of methanol/water (1:1) per min. The mass spectrometer was operating in electrospray ionization mode with a needle voltage of 3 kV, temperature of 120°C, and cone voltage of 30 V. The expected weight of 5,002.358 was confirmed.
The purified peptide was divided into two equal portions for addition of the benzophenone moieties. To one portion (0.030 g) of the peptide an amine-reactive probe, benzophenone-4-isothiocyanate (Molecular Probes), was added in the following manner: the peptide was dissolved in 4 ml of 0.1 M sodium bicarbonate buffer, pH 9.0, and 100 mg of the benzophenone-4-isothiocyanate in 0.5 ml of DMF was added. The reaction went for 1.5 h and was monitored by analytical HPLC (same conditions as above). The reaction mixture was evaporated to near-dryness under reduced pressure, diluted with water, and loaded onto a preparative reverse-phase column for purification (same conditions as above). Pooled fractions were lyophilized. The authenticity of the peptide was established by mass spectrometry (same as above). The expected weight of 5,241.7 was confirmed.
To the second portion of the purified parent peptide, benzophenone-4-maleimide (Molecular Probes) was added in the following manner: the peptide was dissolved in 4 ml of degassed (nitrogen bubble) KH2PO4/NaOH buffer (pH = 7.0). The benzophenone-4-maleimide was dissolved in 250 μl of dry DMF and was immediately added to the peptide/buffer mixture. The reaction was monitored by Ellman reagent and analytical HPLC (same conditions as above) and came to completion in less than 1 h. Five hundred microliters of glacial acetic acid was added to the reaction mixture, and the mixture was filtered through a fine glass fritted funnel. The peptide was then purified by HPLC (same conditions as above). Pooled fractions were lyophilized. The expected weight of 5,241.7 was confirmed (same as above).
Cross-linking experiments.
Peptides were preincubated for 1 h on ice with 0.1 μM purified recombinant CDK2/cyclin A complex in kinase buffer (below) in a final reaction volume of 30 μl. The reaction materials were then warmed to 30°C for 10 min and were spotted on Parafilm in a UV Stratalinker 2400 (Stratagene). UV cross-linking was performed for 40 s (∼180,000 μJ/cm2), and 15 μl of 3× sample buffer (32) was then added to each reaction. Samples were analyzed by SDS-PAGE with 10% polyacrylamide gels and were transferred to nylon/nitrocellulose membranes (Micron Separations Inc.), and biotinylated protein bands were detected by staining with the Vectastain Elite avidin-biotin complex kit (Vector Laboratories). In some experiments, competitor full-length 16E7 protein or 16E7 peptide spanning residues 9 to 48 was added prior to addition of the benzo-CYS-9-48-biotin (bio) peptide.
Immunoprecipitation.
Rabbit immunoaffinity-purified CDK2 antibody (Upstate Biotechnologies) was added to 120 μl of UV cross-linked reactions in which the final concentrations of CDK2/cyclin A and benzo-CYS-9-48-bio were 0.25 and 2.5 μM, respectively. PBS containing 0.2% NP-40 was then added to bring the reaction volume to 400 μl, and the sample was incubated on a rotator at 4°C for 2 h. Samples were then mixed with 25 μl of protein A/G Plus agarose beads (Santa Cruz Biotechnologies) that were washed twice in PBS-0.2% NP-40. After 2.5 h of additional incubation at 4°C, beads were pelleted and pellets were washed five times in PBS-0.2% NP-40. Laemmli sample buffer (50 μl of 2× buffer) was added, and samples were incubated at 100°C for 5 min. Beads were removed, material was run on SDS-10% polyacrylamide gels and electrophoretically blotted to nitrocellulose membranes (Micron Separations, Inc.), and biotinylated proteins were detected with the avidin-biotin complex reagent kit (Vector Laboratories).
RESULTS
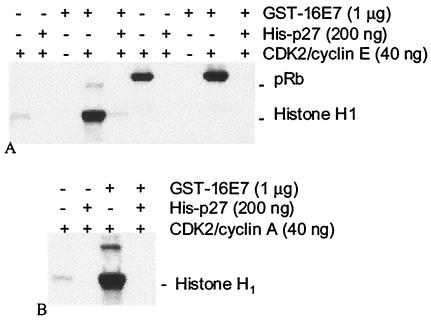
The effect of HPV E7 on CDK2 kinase activity was examined by using purified proteins. CDK2 complexed with cyclin E had low histone H1 kinase activity relative to Rb kinase activity (Fig. 1A). Addition of GST-16E7 dramatically promoted the phosphorylation of histone H1 by CDK2 complexed with cyclin E (Fig. 1A) or cyclin A (Fig. 1B), while having modest effects on Rb phosphorylation (Fig. 1A). GST alone had no effect on kinase activity (data not shown; see Fig. 3), and E7 purified away from the GST fusion tag also showed excellent CDK2-stimulatory activity (data not shown; see Fig. 5). The addition of CDK inhibitory proteins p27 (Fig. 1A and B) and p21 (data not shown) inhibited CDK2 activity in spite of excess amounts of GST-16E7 (Fig. 1) or purified 16E7 (data not shown). Activation of CDK2/cyclin A histone H1 kinase activity by GST-16E7 was dose dependent up to 640 ng of GST-16E7 (∼0.6 μM E7) (Fig. 2A and B). The CDK2/cyclin A complex isolated from bacteria was also strongly activated by purified 16E7 (Fig. 2C).
FIG. 1.
GST-16E7 activates purified cyclin E/CDK2 and cyclin A/CDK2 complexes. Autoradiographs demonstrate phosphorylation of histone H1 and Rb (pRb) by CDK2/cyclin E (A) and CDK2/cyclin A (B). (A) E7 potently upregulates CDK2/cyclin E histone H1 kinase activity while having modest effects on Rb kinase activity. Addition of p27 eliminates this effect and reduces activity to below baseline levels. (B) CDK2/cyclin A histone kinase activity is also dramatically upregulated by E7, and the effect is negated by addition of p27.
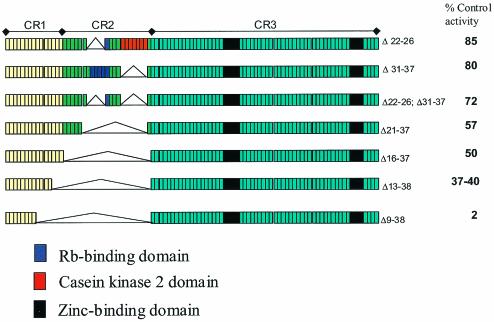
FIG. 3.
Summary of deletion mutations of E7 demonstrating that the principal CDK2 effect resides within CR1 and CR2. Initial truncation experiments showed that the CDK2-promoting activity resides within conserved regions 1 and 2 (CR1 and CR2). Here we show that deletions of amino acids 9 to 38 result in a dramatic loss of CDK2 activation. The numbers on right side of figure indicate deleted amino acids (Δ22-26 and Δ21-27, etc.) and percentage of control (full-length E7) CDK2 activation.
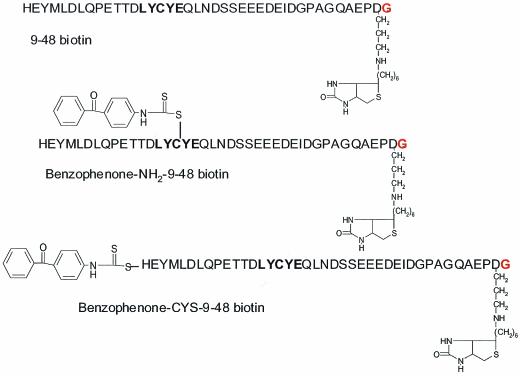
FIG. 5.
Structure of biotinylated synthetic E7 peptides. Two of the peptides contain benzophenone linked to the amino terminus (Benzophenone-NH2-9-48 biotin) or to cysteine 24 (Benzophenone-CYS-9-48 biotin).
FIG. 2.
Addition of increasing amounts of GST-16E7 to CDK2 purified from insect cells or from bacteria results in a dose-dependent increase in kinase activity. Autoradiograph (A) demonstrates E7-dependent increase in phosphorylation of histone H1 by CDK2/cyclin A. Quantification of the experiment (B) plots PhosphorImager counts versus amount of GST-16E7 added. Results indicate that E7 affect peaks at 640 ng of E7 (or approximately 0.6 μM E7). CDK2/cyclin A reconstituted from bacterially expressed proteins is also activated by purified 16E7 (C).
The effects of GST-16E7 on other kinase and substrate combinations were tested in order to further characterize E7 specificity. Table 1 summarizes results from numerous experiments. Maximum effects of E7 were noted on CDK2 complexed with either cyclin A or E, while the histone H1 kinase activity of the CDC2/cyclin B complex was unaffected by GST-16E7. Histone H1 kinase activity of the CDK5/p25 complex showed a modest stimulation by GST-16E7, while MAP kinase I was unaffected (Table 1).
TABLE 1.
Activation (n-fold) of various kinase-substrate combinations by HPV E7a
| Kinase-substrate combination | Increase (n-fold) in kinase activity with E7 addition |
|---|---|
| Cyclin A-CDK2::histone H1 | 26 |
| Cyclin E-CDK2::histone H1 | 13 |
| Cyclin E-CDK2::Rb | 1.9 |
| Cyclin B-CDC2::histone H1 | 1.0 |
| p25/CDK5::histone H1 | 4.5 |
| MAP kinase I::myelin basic protein | 1.1 |
Activation of kinase-substrate combinations is expressed as increase (n-fold) over controls that received no E7.
Another high-risk E7 (GST-31E7), as well as a low-risk E7 (GST-6bE7), was expressed and were purified to determine if other E7 types might exhibit similar activities. Table 2 summarizes the effects of these proteins on CDK2/cyclin A complexes in experiments that use either histone H1 or Rb as substrate. In this particular experiment CDK2/cyclin A was stimulated 18-fold by GST-16E7, while 16E7 purified away from its fusion tag stimulated kinase activity by 14-fold. GST-31E7 and GST-6bE7 fusion proteins also had potent stimulatory effects on CDK2 histone H1 kinase activity. GST-6bE7 displayed an intermediate effect relative to GST-16E7 and GST-31E7, while all three GST-E7 proteins had similar, smaller effects on Rb kinase activity (Table 2).
TABLE 2.
Activation (n-fold) of cyclin A-CDK2 histone and pRb kinase activity by HPV E7a
| Form of E7 | Kinase activity of:
|
|
|---|---|---|
| Histone H1 | pRb | |
| 16E7b | 14.0 | NDc |
| GST-16E7 | 18.2 | 2.3 |
| GST-31E7 | 12.6 | 1.6 |
| GST-6bE7 | 15.8 | 2.2 |
Kinase activity is expressed as increase in kinase activity over controls not receiving E7.
HPV16 E7 purified away from GST tag.
Activity not determined.
The identity of the required E7 domain(s) was next sought by using mutational analysis. An initial series of 16E7 deletion mutants examined the effects of removal of sequence from the amino and carboxy termini. These experiments demonstrated that GST-16E7 tolerates substantial truncation of C-terminal E7 residues (i.e., residues 39 to 98 or CR3) with only a moderate loss of CDK2 histone H1-promoting activity, while removal of residues from the amino terminus (i.e., within CR1) results in substantial losses of activity (data not shown). Therefore, the effects of deletions within CR1 and CR2 were examined (Fig. 3). Deletions of the Rb-binding and casein kinase 2 domains resulted in only modest loss of activity. However, progressively larger deletions between amino acids 9 and 38 resulted in major losses in the ability to activate CDK2, with the 9-to-38-amino-acid deletion resulting in nearly complete loss of activity (Fig. 3). In summary, the core CDK2-promoting activity of E7 resides within sequences contained in CR1 and CR2 amino acids 9 to 38 (Fig. 3).
Synthetic peptides were tested next for their ability to activate CDK2. Initial experiments with a peptide encompassing HPV16 residues 9 to 48 indicated that it activated CDK2, albeit at a higher concentration than required for recombinant E7 protein. A dose-response curve with the peptide showed that peak activation of CDK2 occurred at approximately 50 μM (Fig. 4A and B). Several peptides of various lengths were then tested at 100 μM. These studies clearly showed that truncation of the peptides at either end significantly affected their ability to activate CDK2. For instance, truncation of amino-terminal residues 9 to 13 completely eliminated activity (Fig. 4C). Truncation of carboxy-terminal residues 34 to 38 also caused a nearly complete loss of activity. Consequently, the minimal peptide required for CDK2 activation in the studies is encompassed in HPV16 residues 9 to 38.
FIG. 4.
Synthetic E7 peptides spanning residues 9 to 38 activate CDK2. (A) 16E7 (9-48) activated CDK2 in a dose-dependent manner, with peak activation at approximately 50 μM. (B) PhosphorImager quantification of data from panel A. (C) Truncation of the 16E7 (1-48) peptide sequence from the amino and carboxy termini and subsequent testing at 100 μM (A and B) demonstrate that sequences 1 to 9 and 39 to 48 are not required for CDK2 activation. Thus, 16E7 (9-38) is the minimal peptide for CDK2 activation.
The observation that E7 and E7-derived peptides activate purified CDK2 implies a direct interaction with the kinase complex. We pursued cross-linking studies to test this hypothesis and to determine if E7 might bind a CDK2 inhibitory protein. HPV16 9-48 peptides were designed with a biotin moiety on the carboxy terminus and a photoactivated benzophenone group on either the amino terminus (benzo-NH2-9-48 bio) or on cysteine 24 (benzo-CYS-9-48 bio) (Fig. 5). All peptides stimulated CDK2 kinase activity (Fig. 6A). Peptide-kinase mixtures were next exposed to UV light, and Western blotting was performed. Both benzophenone-containing peptides were found to incorporate into two bands believed to be phosphorylated variants of CDK2 and into a single band corresponding to GST-cyclin A (Fig. 6B). In both cases the biotin-containing bands showed increases in molecular mass of ∼5 kDa over what was expected, suggesting that both CDK2 and cyclin A were cross-linked to a single biotin-containing peptide. A ladder of biotin-reactive bands, which might suggest nonspecific binding or multiple binding sites, was never observed. No cross-linking to histone H1 was detected for either peptide (Fig. 6B).
FIG. 6.
Cross-linking of E7 peptides to CDK2/cyclin A complex. (A) HPV16 E7 (1.8 μM) and E7-derived peptides (50 μM) stimulate CDK2/cyclin A histone kinase activity. (B) Benzo-CYS-9-48-bio and benzo-NH2-9-48-bio peptides associate with cyclin A/CDK2 complex following UV irradiation. Histone H1 is not cross-linked under identical conditions. (C) CDK2/cyclin A (0.1 μM) and benzo-CYS-9-48-bio (0.5 μM) were with or without competitor E7 or E7 peptide. Both full-length 16E7 and the 9-48 16E7 peptide compete with the benzo-CYS-9-48-bio peptide for binding to the cyclin A/CDK2 complex. These results suggest that labeling of the CDK2/cyclin A complex by benzo-CYS-9-48-bio is specific and is not due to indiscriminate labeling of the complex. (D) Immunoprecipitation (IP) followed by Western blotting for incorporation of biotin-containing peptide into immunoprecipitated target protein(s) demonstrates that biotin-reactive bands in Western blots are cyclin A and CDK2 and that reactivity of these bands with benzo-CYS-9-48-bio peptide is dependent upon UV. (E) Cross-linking of benzophenone-containing peptides to increasing amounts of GST, cyclin A, or CDK2 demonstrates a strong preference of the peptide for cyclin A.
The specificity of cross-linking was examined by competition with the unlabeled HPV16 9-48 peptide and with purified full-length E7. Both completely blocked cross-linking of the benzophenone-containing peptide (Fig. 6C), suggesting that they recognize similar or closely associated binding sites on the kinase. To eliminate the possibility that the benozophenone-containing peptide might be reacting with contaminating proteins other than cyclin A and CDK2, we performed immunoprecipitation-Western blot experiments. Both cyclin A and CDK2 antibodies precipitated the same biotin-containing bands, thus verifying that the E7 peptides are cross-linked directly to the CDK2/cyclin A complex (Fig. 6D). The preference of E7 peptides for monomeric cyclin or CDK2 was tested next. Cyclin A, GST, or CDK2 was tested at various concentrations for cross-linking to the benzophenone-containing peptides. Only cyclin A (Fig. 6E) showed evidence of cross-linking with the E7 peptide.
DISCUSSION
Alleviation of the S-phase checkpoint is a major function of HPV E7 in the viral life cycle and cancer. An association of E7 with CDK2 in cells has been demonstrated by several laboratories (10, 16, 37, 50), but it has remained unclear whether E7 can associate directly with the kinase. This report shows that HPV E7 dramatically and specifically promotes the histone H1 kinase activity of CDK2 complexed with either cyclin A or cyclin E. Furthermore, we demonstrate close association of E7 with the CDK2 complex by cross-linking, with a preference for cyclin A over CDK2 when these proteins are used in their monomeric form. These observations suggest that E7 peptides bind primarily to cyclin but that the benzophenone group is near enough to CDK2 within the complex to cross-link efficiently. Our observations point to a simple, direct mechanism by which E7 may advance the cell cycle progression via CDK2. In addition, our results suggest that E7 may act to redirect CDK2 activity toward targets that promote aspects of the viral life cycle.
Previous studies have implicated E7 interactions with p21 and p27 in regulating CDK2 activity (20, 24, 31, 54). Several lines of evidence suggest that CDK2 activation by E7 in our system is not dependent upon these CDK inhibitors (CDKIs). First, we have used purified CDK2 complex. Because we could not completely rule out some contamination by insect cell CDKIs, we repeated our findings with kinase purified from bacteria. E7 also activated this CDK2/cyclin A kinase, indicating that CDKIs are not involved in CDK2 activation by E7. Our polyacrylamide gel analyses of the insect cell purified complex also show that, if these proteins are present, they are a minor contaminant and are not sufficient to explain the large E7-dependent increases in CDK2 activity. Second, addition of p21 or p27 to our kinase assays at levels well below those of E7 is sufficient to eliminate the E7 effect, again suggesting that the effect is not mediated via these proteins. Finally, cross-linking of E7 to the kinase complex suggests a direct interaction with the kinase, specifically with cyclin A, that is not mediated via another protein. If interaction of E7 with a CDKI was mediating CDK2 activation, then we might be expected to observe cross-linking of E7 to such a protein. This was not observed.
Histone H1 phosphorylation and subsequent relaxation of chromatin structure have been demonstrated in W138 fibroblasts and in cells lacking Rb, with entry into the S phase (26). These finding suggest that E7 may initiate host and viral DNA synthesis by directly stimulating CDK2 phosphorylation of histone H1. Consistent with this suggestion is the observation that E7 induces DNA synthesis in quiescent cells by way of sequences residing in CR1 and CR2 (2, 43). While histone H1 is normally a poor CDK2 substrate, relative to Rb, based on differences in Kcat/Km (53), our study suggests that E7 enhances its phosphorylation by CDK2. Alternatively, since E7 is likely to alter CDK2 phosphorylation of substrates in addition to histone H1, our observations may reflect a wider-ranging change in CDK2 substrate preference.
Many of the known biological functions of E7 are related to its role as an oncogene, while less is known of E7 function in the papillomavirus life cycle. E7 has a number of protein binding functions, including the ability to bind TATA-binding proteins (35, 36) and AP-1 (1). Of most significance to the present study, E7 binds Rb family members (9, 13, 52), p21 (20, 31), and p27 (54), which all act in the CDK phosphorylation pathway to inhibit cell cycle progression. Since low-risk E7 is attenuated in these binding functions, it is unclear which, if any, are key for the HPV life cycle. Our data suggest that activation of CDK2 by E7 may be a key viral life cycle event. Importantly, CDK2 is a principal regulator of the G1/S transition (14, 38) and E7 promotes cell transit through this checkpoint into the S phase (7, 19, 29). The demonstration that E7 directly alters cyclin A/CDK2 and cyclin E/CDK2 kinase activity but not cyclin B/CDC2 activity suggests a mechanism by which E7 might specifically disengage the G1/S checkpoint. In addition, CDK2 activation may be a conserved functional requirement of the viral life cycle, since it is conserved in both high- and low-risk forms of E7.
The E7 amino terminus contains two conserved regions (CR1 and CR2) with similarity to two noncontiguous portions of the adenovirus E1A protein and simian virus 40 T-antigen. Similar to E1A and T-Ag, CR1 (amino acids 1 to 16) and CR2 (amino acids 17 to 38) of E7 are implicated as important mediators of abnormal growth control (40, 41). Our analyses of E7 find that the predominant CDK2 effect resides within CR1 and CR2, indicating that CDK2 activation may be conserved among other viral proteins. The CR1 and CR2 regions of E7 also overlap with biological activities that include immortalization, transformation, and promotion of DNA synthesis (reviewed in reference 41), suggesting CDK2 activation as a candidate for function in these activities as well.
A central complexity encountering HPV is replication of its genome within a cell, the differentiated keratinocyte, which under normal conditions does not support DNA synthesis. Previous studies have shown the E7 alone is sufficient for this activity (7, 29) and is required for HPV vegetative replication in raft cultures (18). Our results suggest a means by which HPV E7 may promote onset of host cell S-phase through direct activation of CDK2. Activation of CDK2 histone H1 kinase activity may play an important role in preparing either host cell DNA (26) or papillomavirus DNA (15, 49) for replication. In addition, HPV E7 may act to redirect CDK2 activity toward virally encoded substrates and other yet unknown substrates that facilitate amplification of the virus.
Acknowledgments
We thank David Morgan (University of California at San Francisco) for cyclin and CDK2 baculoviruses. A. Koff (Memorial Sloan-Kettering Cancer Center) provided p27 cDNA. We thank Karen Leach and Paula Munns (Pharmacia) for p25/CDK5. Robert Garlick (Pharmacia) is acknowledged for providing cleaved, purified E7. Paul Lambert (University of Wisconsin at Madison) provided HPV16 DNA. Peggy Garner-Hamrick (Pharmacia) gave help with the GST-31E7 expression construct.
REFERENCES
- 1.Antinore, M. J., M. J. Birrer, D. Patel, L. Nader, and D. J. McCance. 1996. The human papillomavirus type 16 E7 gene product interacts with and trans-activates the AP1 family of transcription. EMBO J. 15:1950-1960. [PMC free article] [PubMed] [Google Scholar]
- 2.Banks, L., C. Edmonds, and K. H. Vousden. 1990. Ability of the HPV16 E7 protein to bind RB and induce DNA synthesis is not sufficient for efficient transforming activity in NIH3T3 cells. Oncogene 5:1383-1389. [PubMed] [Google Scholar]
- 3.Bedell, M. A., J. B. Hudson, T. R. Golub, M. E. Turyk, M. Hosken, G. D. Wilbanks, and L. A. Laimins. 1991. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J. Virol. 65:2254-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Buttinelli, M., G. Panetta, D. Rhodes, and A. Travers. 1999. The role of histone H1 in chromatin condensation and transcriptional repression. Genetica 106:117-124. [DOI] [PubMed] [Google Scholar]
- 6.Caldeira, S., E. M. de Villiers, and M. Tommasino. 2000. Human papillomavirus E7 proteins stimulate proliferation independently of their ability to associate with retinoblastoma protein. Oncogene 19:821-826. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, S., D. C. Schmidt-Grimminger, T. Murant, T. R. Broker, and L. T. Chow. 1995. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 9:2335-2349. [DOI] [PubMed] [Google Scholar]
- 8.Chow, L. T., and T. R. Broker. 1994. Papillomavirus DNA replication. Intervirology 37:150-158. [DOI] [PubMed] [Google Scholar]
- 9.Ciccolini, F., G. Di Pasquale, F. Carlotti, L. Crawford, and M. Tommasino. 1994. Functional studies of E7 proteins from different HPV types. Oncogene 9:2633-2638. [PubMed] [Google Scholar]
- 10.Davies, R., R. Hicks, T. Crook, J. Morris, and K. Vousden. 1993. Human papillomavirus type 16 E7 associates with a histone H1 kinase and with p107 through sequences necessary for transformation. J. Virol. 67:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Defeo-Jones, D., G. A. Vuocolo, K. M. Haskell, M. G. Hanobik, D. M. Kiefer, E. M. McAvoy, M. Ivey-Hoyle, J. L. Brandsma, A. Oliff, and R. E. Jones. 1993. Papillomavirus E7 protein binding to the retinoblastoma protein is not required for viral induction of warts. J. Virol. 67:716-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demers, G. W., E. Espling, J. B. Harry, B. G. Etscheid, and D. A. Galloway. 1996. Abrogation of growth arrest signals by human papillomavirus type 16 E7 is mediated by sequences required for transformation. J. Virol. 70:6862-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyson, N., P. M. Howley, K. Münger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 14.Ekholm, S. V., and S. I. Reed. 2000. Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr. Opin. Cell Biol. 12:676-684. [DOI] [PubMed] [Google Scholar]
- 15.Favre, M., F. Brietburd, O. Croissant, and G. Orth. 1977. Chromatin-like structures obtained after alkaline disruption of bovine and human papillomaviruses. J. Virol. 21:1205-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finzer, P., C. Kuntzen, U. Soto, H. zur Hausen, and F. Rosl. 2001. Inhibitors of histone deacetylase arrest cell cycle and induce apoptosis in cervical carcinoma cells circumventing human papillomavirus oncogene expression. Oncogene 20:4768-4776. [DOI] [PubMed] [Google Scholar]
- 17.Fisher, C. 1994. The cellular basis for development and differentiation in mammalian keratinizing epithelia, p. 131-150. In I. Leigh, B. Lane, and F. Watt (ed.), The keratinocyte handbook. Cambridge University Press, Cambridge, United Kingdom.
- 18.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive phase of the viral life cycle. J. Virol. 74:6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster, S. A., and D. A. Galloway. 1996. Human papillomavirus type 16 E7 alleviates a proliferation block in early passage human mammary epithelial cells. Oncogene 12:1773-1779. [PubMed] [Google Scholar]
- 20.Funk, J. O., S. Waga, J. B. Harry, E. Espling, B. Stillman, and D. A. Galloway. 1997. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 11:2090-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez, S. L., M. Stremlau, X. He, J. R. Basile, and K. Munger. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 75:7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harbour, J. W., R. X. Luo, A. Dei-Santi, A. A. Postigo, and D. C. Dean. 1999. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 98:859-869. [DOI] [PubMed] [Google Scholar]
- 23.He, Y., and W. D. Cress. 2002. E2F-3B is a physiological target of cyclin A. J. Biol. Chem. 277:23493-23499. [DOI] [PubMed] [Google Scholar]
- 24.Helt, A. M., J. O. Funk, and D. A. Galloway. 2002. Inactivation of both the retinoblastoma tumor suppressor and p21 by the human papillomavirus type 16 E7 oncoprotein is necessary to inhibit cell cycle arrest in human epithelial cells. J. Virol. 76:10559-10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helt, A. M., and D. A. Galloway. 2001. Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J. Virol. 75:6737-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrera, R. E., F. Chen, and R. A. Weinberg. 1996. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb-deficient fibroblasts. Proc. Natl. Acad. Sci. USA 93:11510-11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howley, P. M. 1996. Papillomavirinae: the viruses and their replication, p. 2045-2076. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 28.Jewers, R. J., P. Hildebrandt, J. W. Ludlow, B. Kell, and D. J. McCance. 1992. Regions of human papillomavirus type 16 E7 oncoprotein required for immortalization of human keratinocytes. J. Virol. 66:1329-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jian, Y. C., D. C. Schmidt-Grimminger, W. M. Chien, X. Wu, T. R. Broker, and L. T. Chow. 1998. Post-transcriptional induction of p21cip1 protein by human papillomavirus E7 inhibits unscheduled DNA synthesis reactivated in differentiated keratinocytes. Oncogene 17:2027-2038. [DOI] [PubMed] [Google Scholar]
- 30.Jones, D. L., R. M. Alani, and K. Münger. 1997. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 11:2101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, D. L., D. A. Thompson, and K. Münger. 1997. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology 239:97-107. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 33.Leone, G., J. DeGregori, L. Jakoi, J. G. Cook, and J. R. Nevins. 1999. Collaborative role of E2F transcriptional activity and G1 cyclin-dependent kinase activity in the induction of S phase. Proc. Natl. Acad. Sci. USA 96:6626-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masai, H., E. Matsui, Z. You, Y. Ishimi, K. Tamai, and K.-i. Arai. 2000. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a critical threonine residue of Cdc7 by Cdks. J. Biol. Chem. 275:29042-29052. [DOI] [PubMed] [Google Scholar]
- 35.Massimi, P., D. Pim, and L. Banks. 1997. Human papillomavirus type 16 E7 binds to the conserved carboxy-terminal region of the TATA box binding protein and this contributes to E7 transforming activity. J. Gen. Virol. 78:2607-2613. [DOI] [PubMed] [Google Scholar]
- 36.Massimi, P., D. Pim, A. Storey, and L. Banks. 1996. HPV-16 E7 and adenovirus E1a complex formation with TATA box binding protein is enhanced by casein kinase II phosphorylation. Oncogene 12:2325-2330. [PubMed] [Google Scholar]
- 37.McIntyre, M. C., M. N. Ruesch, and L. A. Laimins. 1996. Human papillomavirus E7 oncoproteins bind a single form of cyclin E in a complex with cdk2 and p107. Virology 215:73-82. [DOI] [PubMed] [Google Scholar]
- 38.Morgan, D. O. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261-291. [DOI] [PubMed] [Google Scholar]
- 39.Morris, L., K. E. Allen, and N. B. La Thangue. 2000. Regulation of E2F transcription by cyclin E-Cdk2 kinase mediated through p300/CBP co-activators. Nat. Cell Biol. 2:232-239. [DOI] [PubMed] [Google Scholar]
- 40.Munger, K., J. R. Basile, S. Duensing, A. Eichten, S. L. Gonzalez, M. Grace, and V. L. Zacny. 2001. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 20:7888-7898. [DOI] [PubMed] [Google Scholar]
- 41.Münger, K., and A. L. Halpern. 1997. HPV 16 E7: primary structure and biological properties, p. 17-36. In G. Myers, F. Sverdrup, C. Baker, A. McBride, K. Münger, H.-U. Bernard, and J. Meissner (ed.), Human papillomaviruses 1997 compendium, part III. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 42.Pei, X. F., L. Sherman, Y. H. Sun, and R. Schlegel. 1998. HPV-16 E7 protein bypasses keratinocyte growth inhibition by serum and calcium. Carcinogenesis 19:1481-1486. [DOI] [PubMed] [Google Scholar]
- 43.Rawls, J. A., R. Pusztai, and M. Green. 1990. Chemical synthesis of human papillomavirus type 16 E7 oncoprotein: autonomous protein domains for induction of cellular DNA synthesis and for trans activation. J. Virol. 64:6121-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts, J. M. 1999. Evolving ideas about cyclins. Cell 98:129-132. [DOI] [PubMed] [Google Scholar]
- 45.Roth, S. Y., and C. D. Allis. 1992. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem. Sci. 17:93-98. [DOI] [PubMed] [Google Scholar]
- 46.Ruesch, M. N., and L. A. Laimins. 1998. Human papillomavirus oncoproteins alter differentiation-dependent cell cycle exit on suspension in semisolid medium. Virology 250:19-29. [DOI] [PubMed] [Google Scholar]
- 47.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 48.Sheaff, R. J., M. Groudine, M. Gordon, J. M. Roberts, and B. E. Clurman. 1997. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 11:1464-1478. [DOI] [PubMed] [Google Scholar]
- 49.Swindle, C. S., and J. A. Engler. 1998. Association of the human papillomavirus type 11 E1 protein with histone H1. J. Virol. 72:1994-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tommasino, M., J. P. Adamczewski, F. Carlotti, C. F. Barth, R. Manetti, M. Contorni, F. Cavalieri, T. Hunt, and L. Crawford. 1993. HPV16 E7 protein associates with the protein kinase p33CDK2 and cyclin A. Oncogene 8:195-202. [PubMed] [Google Scholar]
- 51.Werness, B. A., A. J. Levine, and P. M. Howley. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76-79. [DOI] [PubMed] [Google Scholar]
- 52.Wu, E. W., K. E. Clemens, D. V. Heck, and K. Münger. 1993. The human papillomavirus E7 oncoprotein and the cellular transcription factor E2F bind to separate sites on the retinoblastoma tumor suppressor protein. J. Virol. 67:2402-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, X., T. Nakano, S. Wick, M. Dubay, and L. Brizuela. 1999. Mechanism of Cdk2/Cyclin E inhibition by p27 and p27 phosphorylation. Biochemistry 38:8713-8722. [DOI] [PubMed] [Google Scholar]
- 54.Zerfass-Thome, K., W. Zwerschke, B. Mannhardt, R. Tindle, J. W. Botz, and P. Jansen-Durr. 1996. Inactivation of the cdk inhibitor p27KIP1 by the human papillomavirus type 16 E7 oncoprotein. Oncogene 13:2323-2330. [PubMed] [Google Scholar]
- 55.Zhao, J., B. K. Kennedy, B. D. Lawrence, D. A. Barbie, A. G. Matera, and E. Harlow. 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14:2283-2297. [PMC free article] [PubMed] [Google Scholar]
- 56.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342-350. [DOI] [PubMed] [Google Scholar]