Abstract
DsrA is an 87-nt untranslated RNA that regulates both the global transcriptional silencer and nucleoid protein H-NS and the stationary phase and stress response sigma factor RpoS (σs). We demonstrate that DsrA acts via specific RNA:RNA base pairing interactions at the hns locus to antagonize H-NS translation. We also give evidence that supports a role for RNA:RNA interactions at the rpoS locus to enhance RpoS translation. Negative regulation of hns by DsrA is achieved by the RNA:RNA interaction blocking translation of hns RNA. In contrast, results suggest that positive regulation of rpoS by DsrA occurs by formation of an RNA structure that activates a cis-acting translational operator. Sequences within DsrA complementary to three additional genes, argR, ilvIH, and rbsD, suggest that DsrA is a riboregulator of gene expression that acts coordinately via RNA:RNA interactions at multiple loci.
Keywords: hns/rpoS/antisense/small RNA/Hfq
RNA plays a variety of structural, informational, catalytic, and regulatory roles in the cell. RNA functioning as a genetic regulator, a process termed riboregulation (1), by acting either in cis or in trans, has been demonstrated widely in bacteria (2–4). Recently, riboregulation has been demonstrated in eukaryotes, including nematodes (5), plants (reviewed in ref. 6), and mammals (reviewed in ref. 7).
Various small RNAs carry out diverse functions in Escherichia coli (3, 8). As a first example, 10Sa RNA, renamed tmRNA, binds and antagonizes repressor proteins (9). This RNA also acts to remove truncated mRNAs from stalled ribosomes while tagging the abortive translation products for protein turnover (10). Second, OxyS RNA is involved in inducing a cellular response to oxidative stress and protects against DNA damage (11). Third, micF RNA regulates OmpF porin production (reviewed in ref. 12). Finally, DsrA, an 87-nt RNA (Fig. 1) first discovered through its regulatory effect on colanic acid capsule synthesis (13), is both a negative and positive regulator of gene expression in E. coli. DsrA represses either the function or synthesis of the global transcriptional silencer and nucleoid protein H-NS, allowing expression of H-NS-silenced capsular synthesis genes when DsrA is overexpressed (13, 14). DsrA also acts at a second locus, enhancing the translation of the general stress response and stationary phase sigma factor, RpoS (σs) at low temperatures (14). The mechanism of DsrA action at hns and rpoS has remained elusive.
Figure 1.
DsrA contains regions of complementarity to different genes in E. coli. Computer searches revealed complementarity to DsrA at or near the 5′ ends of the coding sequences of five E. coli genes. The DsrA nucleotide sequences complementary to indicated genes are highlighted (white letters on a black background) on the predicted structure of DsrA (13). Open arrowheads indicate potential G:U base pairs between DsrA and target sequences. A prime (′) after the gene name designates complementarity to the gene. (A) Complementarity to hns and rpoS demonstrated in this work. Two possible complementary schemes are shown for hns. (B) Putative complementarity to argR, ilvIH, and rbsD.
H-NS and RpoS are apparent antagonists that either repress or activate transcription, respectively, in response to various environmental signals (reviewed in ref. 15). H-NS silences many unlinked genes, several of which are environmental stress response proteins, through binding to curved DNA seemingly independent of sequence (reviewed in ref. 16). In contrast, RpoS accumulation favors transcription of a set of genes required for growth under stress (reviewed in refs. 15 and 17). Many stress-response genes are both repressed by H-NS and activated by RpoS, and similar kinds of conditions both relieve H-NS silencing and stimulate transcription mediated by RpoS (15, 18). Nutrient deprivation, osmotic shock, heat shock, or low temperature enhance RpoS translation or block its turnover. Of interest, H-NS also represses RpoS, not by silencing transcription but by inhibiting the translation and enhancing the turnover of RpoS (reviewed in ref. 15). H-NS is itself regulated in interesting and complex ways. H-NS negatively regulates its own transcription (19). H-NS transcription also is repressed by the low-abundance StpA protein, an H-NS paralog (intraspecies homolog). H-NS and StpA cooperate to govern certain H-NS-repressed genes (20). Conversely, H-NS transcription is enhanced coordinately with DNA synthesis and is induced by the cold-shock positive regulator CspA and the FIS protein (reviewed in ref. 16).
In this report, we demonstrate that DsrA acts through RNA:RNA base pairing interactions at the hns locus to inhibit translation. DsrA activity at rpoS also is proposed to occur by RNA:RNA interactions to stimulate translation by a separate mechanism. There is potential for RNA:RNA interactions at three additional loci: argR, ilvIH, and rbsD. Together, these results suggest that DsrA is a riboregulator that acts via specific RNA:RNA interactions to either negatively or positively regulate translational control of a broad range of biological functions.
MATERIALS AND METHODS
Bioinformatics.
Complementarity of DsrA to various genes in E. coli was determined by using the bestfit and fasta programs within the GCG suite of programs (Genetics Computer Group, Madison, WI). The significance of matching sequences was determined in GCG by Smith–Waterman alignment (21) by using Monte Carlo randomization of sequences and resampling. Matches greater than three SDs above random were considered significant.
Strains, Plasmids, and Culture Conditions.
M182ΔstpA∷tetr, M182Δhns∷kanr, and M182ΔstpA∷tetrΔhns∷kanr were constructed previously from M182 (Δ[lacIPOZY]X74 galK galU strA) and are described elsewhere (20). M182 hns* (M182hns3-stu) was constructed from M182 (see below). Strain GJ74 proU∷lacZ Tn10∷ tetr (22) was provided by Henri Buc (Institut Pasteur). RO90 (MC4100 λRZ5: φ[rpoS379∷lacZ]) (23) was provided by Regine Hengge-Aronis (Univ. of Konstanz). RK33Z ΔnhaA∷(lacZ, kanr) was provided by Etana Padan (Hebrew Univ.) (24). Strains M182 proU∷lacZ Tn10∷ tetr and M182 ΔnhaA∷(lacZ, kanr) were constructed by P1 transduction (25) from GJ74 and RK33Z, respectively. The proU∷lacZ reporter strain was selected by tetracycline resistance and was screened for cotransduction of the reporter gene on inducing indicator plates as described (22). Plasmid pA is pACYC184 (26). Plasmid pDsrA, also called pDDS164, was provided by S. Gottesman (National Institutes of Health) (13). Plasmids pWKS30 (27) and pMAK705 (28) were provided by S. Kushner (Univ. of Georgia). All strains were grown at 30°C in either TBYE or minimal medium (29) except ΔnhaA strains, which were grown in a buffered medium (24). Antibiotics were used at the following concentrations: chloramphenicol (25 μg/ml), tetracycline (10 μg/ml), kanamycin (10 μg/ml), or ampicillin (200 μg/ml).
Cloning and Mutagenesis of hns.
The hns* allele was constructed by amplifying the hns gene, along with 500 bp of an upstream regulatory region between hns and tdk, by whole-cell PCR of strain M182 by using Vent Polymerase (NEB, Beverly, MA). To generate hns* (hns3-stu), external PCR primers W560 (5′-GGCGGCTCTAGAGTATATGCGTTCTCCCTTACGAAGCC-3′, XbaI site in italics) and W71 (5′-ACAAAAGCTTATTGCTTGATCAGGAAATCGTCGAGGG-3′, HindIII site in italics) were used together with internal, mutagenic primers W595 (5′-GAGATTACTACAATGAGCGAGGCCTTGAAGATTCTGAACAACATCCG-3′) and W634 (5′-CGGATGTTGTTCAGAATCTTCAAGGCCTCGCTCATTGTAGTAATCTC-3′), via recombinant PCR (30), to introduce point mutations together with a StuI site (in italics; see Fig. 2A). Recombinant PCR products were cloned via XbaI and HindIII into pWKS30 and were verified by restriction analysis and DNA sequencing.
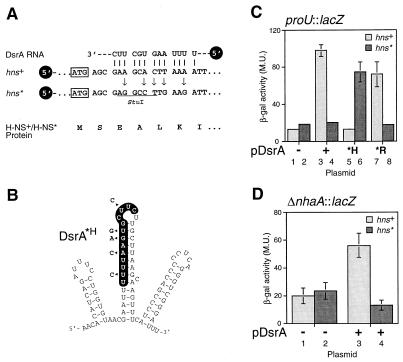
Figure 2.
DsrA acts via RNA:RNA interactions to repress H-NS. (A) Silent mutagenesis of hns to disrupt complementarity to DsrA. Five nucleotides of wild-type hns were modified by site-directed mutagenesis (arrows) to disrupt the DsrA:hns RNA base pairing (vertical lines). The mutations, which are silent in terms of the H-NS protein sequence (below), create a StuI restriction site (underlined) in the altered hns* allele. The initiator methionine codon is indicated (boxed). (B) DsrA mutations that prevent base pairing with hns but allow base pairing with hns*. Mutated nucleotides are shown next to the DsrA structure (arrowheads). (C and D) Assay of H-NS function in strains with and without hns RNA:DsrA complementarity. DsrA-induced repression of H-NS transcription silencing activity was assayed in strain M182 by using a proU∷lacZ (C) or ΔnhaA∷lacZ (D) reporter gene. β-galactosidase assays were performed as described (25), except that assays on ΔnhaA∷lacZ used a buffered growth medium (37). All assays were performed in duplicate; values given are the average of three independent experiments. Light columns, hns+ strains; dark columns, hns* strains. Plasmids used: −, pA vector; +, pDsrA; *H, pDsrA*H as in B; *R, pDsrA*R as in Fig. 3A.
Construction of Chromosomal hns Mutations.
The hns* gene was introduced into the chromosome of M182 at hns essentially as described (28). In brief, the 947-bp XbaI-HindIII fragment, containing the hns* gene along with the upstream regulatory region, was subcloned from pWKShns* into the allele exchange vector pMAK705 (28), generating pMAKhns*. M182 was transformed with this temperature-sensitive plasmid, and the chromosomal hns allele was replaced with hns*, resulting in the strain M182hns*. Integration of the plasmid into the chromosome at hns was selected by growth at the nonpermissive temperature (44°C) on chloramphenicol plates. Candidate integrants were verified by PCR by using primers W778 (within the vector) and W777 (within tdk) (primer sequences below). Simultaneous integrant resolution and plasmid curing were achieved by growth at 30°C in the absence of chloramphenicol. These events were verified by checking clones for chloramphenicol sensitivity at 30°C then by PCR of chromosomal DNA with primer pairs W778 (vector) with W71 (within hns) and W777 with W70 (within hns). Replacement of chromosomal hns+ with hns* was verified by StuI digestion of whole-cell PCR products (W560 and W71) and by Southern hybridization (data not shown). Oligonucleotides used for verification of gene replacement are W70 (5′-AAAGAATTCAGGACTACAATGAGCGAAGCAC-3′), W777 (5′-GGATAACAATTTCACACAGG-3′), W778 (5′-CGGAATAGTAGAAATATAGC-3′), and W71 (above).
Mutations in DsrA.
Plasmid pDsrA was used as a template for inverse PCR (31) with Pwo DNA polymerase (Boehringer Mannheim). For DsrA*H (dsrA-H23), primers W820 (5′-GGAATTCAGGCCTTGAAGAAATTCGTTACACCAGG-3′) and W821 (5′-GGAATTCAGGCCTCTTGCTTAAGCAAGTTTC-3′) introduced a StuI site (in italics). For DsrA*R (dsrA-R19), W818 (5′-GGAATTCTGTACACCAGGGGGTCTGATGTGTTCATC-3′) and W819 (5′-GGAATTCTGTACAGAAGATTTTAAGTGCTTCTTGC-3′) introduced a BsrGI site (in italics). Additional point mutations were introduced concomitantly outside these sites (see Figs. 2B and 3A). For pDsrA*R, PCR products (linear plasmids) were digested with BsrGI, were ligated, and were transformed into DH5α. In the case of pDsrA*H, the oligonucleotides were phosphorylated with T4 kinase before PCR, and the PCR product plasmid was blunt-end ligated and was used to transform E. coli. Candidates were screened by restriction analysis with EcoRI or StuI, and positive clones were digested with StuI and were gel-purified to remove the short linker DNA. These plasmids then were ligated, were used to transform E. coli, and were rescreened by restriction analysis for loss of the linker, which contains EcoRI sites. Positive clones were verified by DNA sequencing.
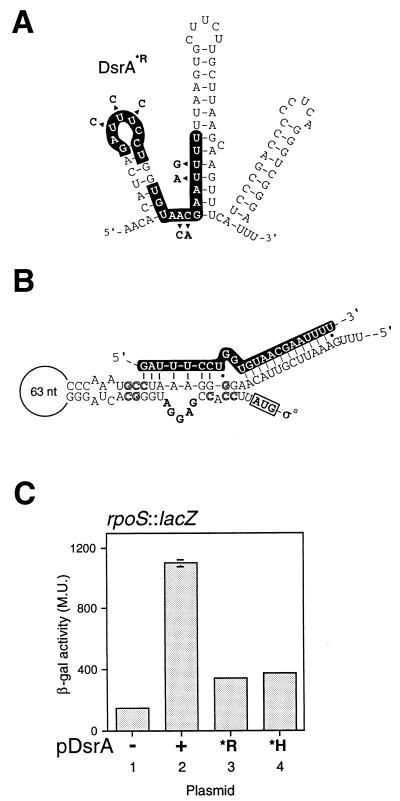
Figure 3.
DsrA activates rpoS translation: an RNA:RNA interaction model. (A) DsrA*R mutant RNA. Mutations are indicated by arrowheads. (B) Proposed structure of the rpoS mRNA translational operator. Bold letters in the bulge loop indicate the Shine–Dalgarno sequence; boxed AUG indicates the initiator methionine codon; outlined letters indicate mutations that confirm secondary structure and enable Hfq-independent translation (adapted from ref. 38 by using additional sequence of E. coli rpoS from ref. 48). The sequence of DsrA is given in white letters on a black background, indicating the complementarity to rpoS mRNA deduced in this work (Fig. 1A). Vertical lines are base pairs, dots are G:U base pairs. (C) Effect of altered DsrA on translation of rpoS mRNA. The reporter gene is a translational fusion between RpoS and LacZ in strain RO90. The RNA produced by the reporter fusion contains the region of DsrA complementarity. All values shown are the average of three independent experiments.
RNA and Statistical Variance Analyses.
RNA levels were determined by primer extension as described (29). Primer W166 (20) was used for hns, and W578 (trxA; 5′-TTTCACCG-GATACGCTTTCG-3′) was used as a loading control. Labeled cDNAs were separated on 8% polyacrylamide sequencing gels and were quantified by using a PhosphorImager and imagequant software (Molecular Dynamics). The hns RNA signal was normalized to the M182/pA sample for comparison between experiments. Three experiments were performed in duplicate, and the results were subjected to variance analysis with the program sas (procedure ANOVA). All P values <0.05 were considered significant. The differences between hns+ and hns* RNA levels were considered significant (P = 0.0044), as were the differences between hns+ and Δhns, each containing pA (P = 0.0055) and the DsrA-induced differences in a Δhns strain background (P = 0.0208). Other variances in hns RNA levels were not considered significant (Y. Ding and C. Lawrence, personal communication).
SDS and Isoelectric Focusing Gels and Western Blotting.
H-NS and StpA proteins were purified as described (32). Cell extracts were prepared, were diluted in SDS sample dilution buffer, and were separated as described (33, 34), except nonequilibrium tube gels were run with 2% Nonidet P-40 in a pH 3–10 gradient (Bio-Lyte, Bio-Rad), and tube gels were overlaid with 1% agarose (in SDS protein gel running buffer plus bromophenol blue) before running the second dimension. SDS gels are as described (35), except that the running buffer was 25 mM Tris⋅HCl (pH 8.3), 41 mM glycine, and 1.3% SDS. For Western blot analysis, proteins were transferred to poly(vinylidene difluoride) membranes (Bio-Rad) with a semi-dry apparatus (Bio-Rad) by using an unpublished protocol (Philippe Bertin, personal communication). In brief, the unequilibrated gel was placed on the membrane and was sandwiched between pieces of filter paper soaked in transfer buffer (25 mM Tris base, 20% methanol). Three filters above the gel also contained 40 mM ɛ-amino-n-caproic acid (Sigma); below the membrane was one filter soaked in transfer buffer, then two filters soaked in 300 mM Tris base plus 20% methanol. Proteins were transferred for 40 min at 0.8 mA/cm2 of filter. Polyclonal antibodies raised against either StpA or H-NS were prepared previously (36) (A. Zhang and M.B., unpublished data).
RESULTS
DsrA Acts Through RNA:RNA Interactions To Repress H-NS Activity.
Computer database searches revealed that the DsrA sequence contains short stretches of sequence complementary to a number of known genes in E. coli. The regions of complementarity tend to cluster either in the DsrA central stem-loop (hns′ and hns′ alt, argR′, and ilvIH′), or, alternatively, from the first stem-loop to the central stem (rpoS′ and rbsD′) (Fig. 1). Several of these regions partially overlap (e.g., hns′/rpoS′/rbsD′; hns′/argR′/ilvIH′). Of particular interest are the regions of complementarity to hns and rpoS (Fig. 1A), as DsrA has been shown to be involved in the regulation of both of these genes (13, 14). In view of the observed sequence complementarity, and because DsrA antagonizes H-NS, the basis of the regulation of H-NS could be an RNA:RNA interaction between DsrA and hns RNA. We therefore modified hns by silent mutagenesis, disrupting the putative RNA:RNA interaction while leaving the H-NS protein sequence unchanged (Fig. 2A).
The hns gene, along with 500 bp of upstream regulatory sequence (between hns and the divergently transcribed tdk), was cloned by site-directed mutagenic recombinant PCR (30) to introduce specific mutations in hns. The desired mutations introduce a unique StuI restriction site while generating the hns* (hns3-stu) allele (Fig. 2A). Recombinant PCR products were cloned, were screened by StuI digestion, and were verified by DNA sequencing. This allele was introduced into the E. coli chromosome at the hns locus by gene replacement (28). DsrA was overproduced in trans from a plasmid, pDsrA (13), with vector pA (pACYC184) serving as a control. The effect of disrupting the putative hns RNA:DsrA RNA interaction was assessed by measuring expression of two genes, proU (reviewed in ref. 16) and nhaA (37), known to be repressed by H-NS. Expression levels of proU and nhaA were assayed with lacZ fusions to these genes (Fig. 2 C and D). Strikingly, DsrA relieved repression of the proU and nhaA promoters in the wild-type strain (6-fold and 3-fold, respectively) but not in the hns* strain (Fig. 2 C columns 1–4 and D).
These results suggest that disruption of the putative RNA:RNA interaction prevents DsrA from regulating H-NS. To verify that this interaction is the essential element of regulation, the DsrA-overproducing plasmid pDsrA was mutated to produce altered DsrA molecules. The first such variant molecule, DsrA*H (produced by the dsrA-H23 allele), has lost complementarity to hns RNA but has gained putative base pairing with hns* RNA (Fig. 2B). Significantly, β-galactosidase assays with the proU∷lacZ fusion indicate that, in contrast to wild-type DsrA, DsrA*H is unable to block the wild-type hns but is capable of blocking the hns* allele (Fig. 2C, compare columns 3 and 5, 4 and 6). Thus, restoration of the base pairing between hns* and DsrA*H reestablished DsrA regulation of H-NS, confirming that this regulation occurs by RNA:RNA interaction. It is noteworthy that this alteration of DsrA does not destroy its functionality (Fig. 2C, columns 5 and 6).
DsrA also Appears To Regulate RpoS by RNA:RNA Interactions.
We next chose to investigate whether an RNA:RNA interaction is involved in DsrA regulation of rpoS. The region of DsrA complementary to rpoS RNA, termed rpoS′, lies largely in the first rather than the second stem-loop of DsrA (Fig. 1A) and partially overlaps the hns′ region (Fig. 1A, compare hns′ and rpoS′). DsrA has been shown to increase RpoS (σs) at low temperatures by enhancing its translation in E. coli (14). RpoS translation in Salmonella typhimurium is governed by a cis-acting RNA sequence, with a stable secondary structure that must be disrupted to allow translation (38) (Fig. 3B). To test whether DsrA acts on rpoS in trans by an RNA:RNA interaction, we altered DsrA within its rpoS′ region (Fig. 3A), as mutations within the rpoS RNA structure can lead to enhanced translation levels (38). Although pDsrA induced rpoS expression as predicted (14) (Fig. 3C, columns 1 and 2), the mutated DsrA*R (produced from the dsrA-R19 allele) no longer induced rpoS∷lacZ translation (Fig. 3C, compare columns 2 and 3). Of interest, DsrA*H (Fig. 2B) has lost the ability to induce rpoS expression (Fig. 3C, compare columns 2 and 4). This latter effect may be attributable to one or more of the mutations in DsrA*H changing the proposed DsrA:rpoS interaction (Figs. 2B and 3B). Of importance, DsrA*R is still capable of regulating hns RNA (Fig. 2C, compare columns 7 and 8 with columns 3 and 4) and therefore has lost functionality specifically with rpoS and not with hns, consistent with a direct DsrA:rpoS interaction. This result, together with the inability of pDsrA*R to stimulate rpoS expression, supports the idea that regulation at rpoS occurs by an RNA:RNA interaction.
Effects of DsrA-RNA Interactions on hns RNA Levels.
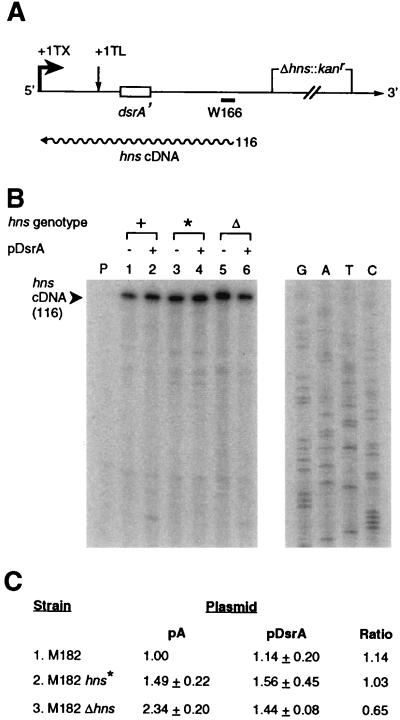
The location of the DsrA-complementary sequence proximal to the hns translation initiation sequences (Figs. 2A and 4A) suggests that DsrA might regulate H-NS production by inhibiting translation, either by occluding the translation machinery or by enhancing turnover of hns RNA. Therefore, in vivo hns RNA was examined by primer extension analysis. The primer used hybridizes upstream of the deleted region in Δhns constructs, enabling quantitation of transcripts from the deletion allele (Fig. 4A) (20).
Figure 4.
Levels of hns RNA during DsrA overexpression. (A) Diagram of the hns transcript and primer extension product. The hns primer W166 (20) hybridizes upstream of the deleted region in Δhns transcripts, enabling quantitation of RNA. +1 TX, transcriptional start; +1 TL, translational start. Size of primer extension product is given in nucleotides beside wavy line, representing cDNA. (B) Levels of hns RNA. RNA harvested from mid-log phase cells grown at 30°C was quantified by primer extension (29). Lanes and markers: P, no RNA; 1, M182hns+/pA; 2, M182hns+/pDsrA; 3, M182hns*/pA; 4, M182hns*/pDsrA; 5, M182Δhns/pA; 6, M182Δhns/pDsrA. The sequence of pTZ19U was used as a molecular weight marker (lanes G, A, T, and C). (C) Quantitation of hns cDNA primer-extension product. Values are normalized to M182hns+/pA. The average of three independent experiments, each performed in duplicate, is shown; error is reported as SD. All six data sets were subjected to variance analysis.
Changes in cDNAs reflecting changes in hns RNA levels produced by DsrA are subtle (<2-fold) but reproducible (Fig. 4B, black arrowhead, and Fig. 4C). In hns+ strains, DsrA overproduction does not, on average, appear to significantly affect hns RNA levels (Fig. 4B, compare lanes 1 and 2; Fig. 4C, line 1). In the absence of DsrA overproduction, hns* strains have a 50% higher steady-state level of transcript than wild-type strains (Fig. 4B, compare lanes 1 and 3; Fig. 4C, compare lines 1 and 2), which might suggest an enhanced hns* transcript stability. However, DsrA overproduction in hns* strains, as in hns+ strains, does not strongly perturb RNA levels (Fig. 4B, compare lanes 1 with 2, 3 with 4; Fig. 4C, line 2). More dramatically, in Δhns strains, the basal level of transcription is increased >2-fold relative to wild type (Fig. 4B, compare lanes 1 and 5; Fig. 4C, line 3), consistent with disruption of hns autoregulation (19). Furthermore, a decrease in hns RNA caused by DsrA overproduction is manifest only in Δhns strains (Fig. 4B, compare lanes 5 and 6; Fig. 4C, line 3), as considered further in Discussion.
DsrA Effects on H-NS Translation.
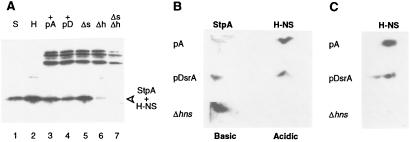
To quantify H-NS protein levels, it was first necessary to separate H-NS from the similarly sized StpA protein (15.6 kDa and 15.3 kDa, respectively), which also cross-reacts immunologically with H-NS polyclonal sera (Fig. 5A, lanes 1 and 2). Because H-NS levels predominated (Fig. 5A, compare lanes 5 and 6) and the overall level of protein (StpA+H-NS) decreased when DsrA was overproduced (Fig. 5A, compare lanes 3 and 4), a decrease in H-NS protein was suggested.
Figure 5.
DsrA decreases H-NS and increases StpA levels in vivo. (A) Western blot of purified proteins and extracts fractionated by SDS/PAGE. The proteins were transferred to membranes and were probed with polyclonal antisera raised against H-NS. Lanes: 1 and 2, purified StpA and H-NS proteins, respectively; 3–7, crude lysates from M182/pA (lane 3), M182/pDsrA (lane 4), M182ΔstpA (lane 5), M182Δhns (lane 6), and M182ΔstpAΔhns (lane 7). (B) H-NS and StpA proteins were separated by nonequilibrium two-dimensional isoelectric focusing and were visualized by Western blot analysis with mixed H-NS- and StpA-specific polyclonal antisera. The pH gradient is from ≈10 (left) to 5 (right). Cell extracts are from M182/pA, M182/pDsrA, and M182Δhns. (C) A separate experiment depicts H-NS separated by equilibrium two-dimensional isoelectric focusing.
Because resolution by SDS/PAGE is not straightforward, the proteins were separated by two-dimensional isoelectric focusing (33, 34) followed by electrophoretic transfer to poly(vinylidene difluoride) membranes (P. Bertin, personal communication; see Materials and Methods). Lysates from cells with and without DsrA overproduction were examined, and proteins were visualized by Western blot analysis by using mixed StpA/H-NS polyclonal sera (Fig. 5 B and C). Purified StpA and H-NS proteins served as markers (not shown). H-NS protein levels decreased in response to DsrA overproduction, with a concomitant increase in StpA protein levels (Fig. 5B, compare M182/pA with M182/pDsrA). M182Δhns was used as a control, showing the increase in StpA protein in the absence of H-NS. The average DsrA-induced decrease in H-NS levels was consistently ≈3-fold.
DISCUSSION
DsrA Function at hns and rpoS.
DsrA, a small untranslated RNA, acts at multiple loci as an inhibitor or activator of gene expression. DsrA inhibits the synthesis of the predominant nucleoid protein and global transcriptional silencer H-NS (13) and enhances the translation of the stationary phase and major stress response sigma factor RpoS (σs) (14). We have demonstrated that DsrA acts via specific RNA:RNA base pairing interactions at the hns locus. Disruption of the RNA:RNA base pairing, by altering hns RNA to hns* through silent mutagenesis, led to loss of DsrA activity whereas restoration of complementarity with DsrA*H restored function (Fig. 2). Specific mutations in the rpoS-complementary region of DsrA to generate DsrA*R similarly prevented DsrA function at rpoS (Fig. 3) without destroying the function of DsrA at the hns locus (Fig. 2C, columns 7 and 8). These findings support the idea of specific regulatory targeting by different regions of DsrA (Figs. 1 and 6).
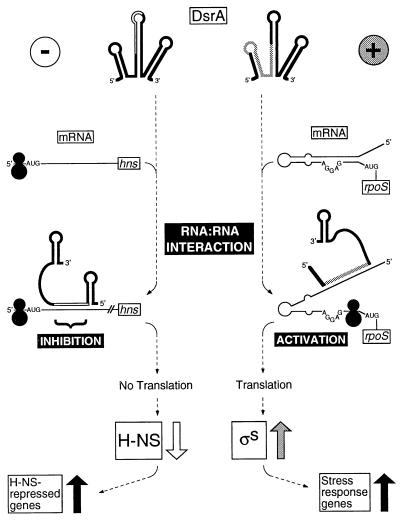
Figure 6.
Models for DsrA riboregulation. At the top, DsrA forms RNA:RNA interactions with target transcripts. On the left (−) is a model for translational repression of hns. On the right (+) is a model for translational activation of rpoS. Black circles represent ribosomes.
Negative Regulation of hns by DsrA.
H-NS levels in vivo were found to be decreased ≈3-fold by DsrA overproduction (Fig. 5 B and C). Although it has been reported that H-NS levels remain constant with DsrA overproduction (13), those findings were reported before the understanding that StpA comigrates with H-NS on SDS gels and cross-reacts with H-NS-specific polyclonal antisera. Resolution of these two proteins on two-dimensional gels clearly demonstrates that DsrA overproduction decreases H-NS protein levels in vivo (Fig. 5).
The proximity of the RNA:RNA interaction to the translational control sequences of hns RNA (Figs. 2A and 4A), together with the results outlined above, suggest a model of translational inhibition by occlusion of translation (Fig. 6 Left). DsrA might block translation by favoring an hns RNA secondary structure that occludes translation initiation or by recruiting a factor that inhibits translation. The observation that hns RNA levels are not decreased when DsrA is overproduced in wild-type hns+ cells is consistent with such an hypothesis (Fig. 4). Whereas DsrA overproduction decreases hns RNA levels produced from the Δhns allele (Fig. 4 B and C), this result must be interpreted cautiously because hns mutants are pleiotropic (reviewed in ref. 16) and Δhns strains grow poorly (20). Although we have not eliminated RNA turnover as a possibility, occlusion of H-NS translation is a more parsimonious model.
Any model of DsrA action on hns RNA must consider the complexity of H-NS and StpA cross-regulation and autoregulatory loops (20) as well as the effects of other H-NS and StpA activators and repressors such as FIS, Lrp, and CspA (16). The reduction in H-NS translation was accompanied by an increase in StpA levels (Fig. 5B), which can be attributed to decreased transcriptional repression of StpA by H-NS (ref. 20; R.L. and M.B., unpublished data). Furthermore, because H-NS and StpA coordinately regulate some genes while independently regulating others, small perturbations in H-NS levels can cause global changes in gene expression.
Positive Regulation of rpoS by DsrA.
RpoS translation requires Hfq (HF-I), a putative RNA chaperone (40, 41) and global regulator of transcription (42, 43). Hfq mutants do not support rpoS translation (23). The annealing of DsrA to the rpoS translational operator region (Fig. 3B) functionally resembles a set of mutations in this region that enable translation in an hfq mutant strain (38) (Fig. 3B). Indeed, compensating mutations in rpoS that restore structure also prevent translation in the absence of Hfq (38). In light of these considerations, an “unzippering” model is proposed (Fig. 6 Right). DsrA base pairing to the inhibitory strand of the rpoS translational operator prevents rpoS intrastrand base pairing, freeing the Shine–Dalgarno sequence and enabling translation.
Although negative regulation by DsrA at hns and positive regulation by DsrA at rpoS appear to be mediated by RNA:RNA interactions at or near translational control regions, these are apparently mechanistically distinct processes. Of interest, the two regulatory modes involve different regions of the DsrA molecule. Whereas the RNA:RNA interaction that results in negative regulation at hns leaves the DsrA 5′ stem-loop intact, the positive regulation at rpoS is proposed to disrupt this structure (Figs. 1–3 and 6). It is tempting to speculate that the 5′ stem-loop is the determinant of negative versus positive control, presenting the recognition/binding site for the effectors of negative regulation (Fig. 6).
DsrA RNA as a Riboregulator at Multiple Loci.
Cells have evolved numerous systems for sensing and adapting to changes in their environment. Adverse conditions, including nutrient deprivation, temperature and pH changes, and osmotic shock elicit responses to environmental stimuli via genetic regulatory networks called stimulons (44), which can activate one set of genes while repressing others. DsrA may therefore be a stimulon regulator that uses multiple antisense sequences to fine-tune global regulatory responses to stress by conditionally preventing or enabling expression of target genes such as hns and rpoS.
Regions of complementarity within the DsrA molecule (Fig. 1B) hint that, in addition to hns and rpoS, three other genes, argR, ilvIH, and rbsD, also may be regulated coordinately in a fashion similar to hns or rpoS. The regions of DsrA complementarity within these genes all map near translation start sequences (data not shown). Again, the regions of complementarity can be mapped roughly into two distinct groups: those that lie in the central stem-loop (hns, argR, and ilvIH) and those that lie in the 5′ stem-loop and central stem (rpoS and rbsD). If the location of the region of complementarity within DsrA corresponds to whether regulation at a specific locus is negative or positive, as for hns and rpoS (Fig. 6), this would predict that argR and ilvIH, like hns, will be regulated negatively by DsrA whereas rbsD, like rpoS, will be regulated positively.
All three of the remaining DsrA-complementary genes are involved in metabolic pathways in the cell. IlvIH is an enzyme that represents a control point in the biosynthesis of isoleucine, leucine, and valine (45). ArgR is a well studied regulon repressor that affects pyrimidine, arginine, and polyamine synthesis (46). The rbs gene, which is of unknown function, is in a polycistronic operon that encodes a multi-subunit active transport system for ribose (47). Clearly, DsrA could be important for coordinate regulation of these pathways and those controlled by H-NS and RpoS, thereby acting as a global orchestrator of a broad sweep of biological functions.
Acknowledgments
We thank H. Buc, R. Hengge-Aronis, E. Padan, S. Gottesman, and S. Kushner for strains and plasmids and B. Cousineau, V. Derbyshire, M. Parker, and M. Stapleton for useful comments on the manuscript. We also thank Y. Ding and C. Lawrence for help with statistical variance analysis, V. Maples and S. Kushner for help with the gene replacement protocol, S. Lawrence and D. Smith for technical assistance, M. Ryan for advice on tube gels, and P. Bertin for contribution of an unpublished protocol and for helpful suggestions on Western blots. We appreciate the expert assistance of M. Belisle and M. Carl with the figures and manuscript, respectively. The Molecular Genetics Core facility at the Wadsworth Center provided oligonucleotide synthesis and DNA sequencing services. This work was supported by National Institutes of Health Grants GM44844 and GM39422 to M.B. and GM18542 to R.L.
Note Added in Proof
Results presented independently by Majdalani et al. in this issue of the Proceedings (49) confirm our model of translational regulation of rpoS via RNA:RNA interactions with DsrA.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Rastinejad F, Conboy M J, Rando T A, Blau H M. Cell. 1993;75:1107–1117. doi: 10.1016/0092-8674(93)90320-p. [DOI] [PubMed] [Google Scholar]
- 2.Simons R W, Kleckner N. Annu Rev Genet. 1988;22:567–600. doi: 10.1146/annurev.ge.22.120188.003031. [DOI] [PubMed] [Google Scholar]
- 3.Delihas N. Mol Microbiol. 1995;15:411–414. doi: 10.1111/j.1365-2958.1995.tb02254.x. [DOI] [PubMed] [Google Scholar]
- 4.Henkin T M. Annu Rev Genet. 1996;30:35–57. doi: 10.1146/annurev.genet.30.1.35. [DOI] [PubMed] [Google Scholar]
- 5.Moss E G, Lee R C, Ambros V. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 6.Jorgensen R A, Atkinson R G, Forster R L S, Lucas W J. Science. 1998;279:1486–1487. doi: 10.1126/science.279.5356.1486. [DOI] [PubMed] [Google Scholar]
- 7.Panning B, Jaenisch R. Cell. 1998;93:305–308. doi: 10.1016/s0092-8674(00)81155-1. [DOI] [PubMed] [Google Scholar]
- 8.Inouye M, Delihas N. Cell. 1988;53:5–7. doi: 10.1016/0092-8674(88)90480-1. [DOI] [PubMed] [Google Scholar]
- 9.Retallack D M, Friedman D I. Cell. 1995;83:227–235. doi: 10.1016/0092-8674(95)90164-7. [DOI] [PubMed] [Google Scholar]
- 10.Keiler K C, Waller P R H, Sauer R T. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 11.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki T, Ueguchi C, Mizuno T. J Bacteriol. 1996;178:3650–3653. doi: 10.1128/jb.178.12.3650-3653.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sledjeski D, Gottesman S. Proc Natl Acad Sci USA. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sledjeski D D, Gupta A, Gottesman S. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 15.Hengge-Aronis R. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 16.Williams R M, Rimsky S. FEMS Microbiol Lett. 1997;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 17.Loewen P C, Hengge-Aronis R. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 18.Atlung T, Ingmer H. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 19.Ueguchi C, Kakeda M, Mizuno T. Mol Gen Genet. 1993;236:171–178. doi: 10.1007/BF00277109. [DOI] [PubMed] [Google Scholar]
- 20.Zhang A, Rimsky S, Reaban M E, Buc H, Belfort M. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]
- 21.Lipman D J, Wilbur W J, Smith T F, Waterman M S. Nucl Acids Res. 1984;12:215–226. doi: 10.1093/nar/12.1part1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gowrishankar J. J Bacteriol. 1985;164:434–445. doi: 10.1128/jb.164.1.434-445.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muffler A, Fischer D, Hengge-Aronis R. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 24.Karpel R, Alon T, Glaser G, Schuldiner S, Padan E. J Biol Chem. 1991;266:21753–21759. [PubMed] [Google Scholar]
- 25.Miller J H. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 72–74. [Google Scholar]
- 26.Chang A C Y, Cohen S N. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang R F, Kushner S R. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 28.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belfort M, Ehrenman K, Chandry P S. Methods Enzymol. 1990;181:521–539. doi: 10.1016/0076-6879(90)81149-o. [DOI] [PubMed] [Google Scholar]
- 30.Higuchi R. In: PCR Protocols. A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 177–183. [Google Scholar]
- 31.Ochman H, Medhora M M, Garza D, Hartl D L. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 219–227. [Google Scholar]
- 32.Cusick M E, Belfort M. Mol Microbiol. 1998;28:847–857. doi: 10.1046/j.1365-2958.1998.00848.x. [DOI] [PubMed] [Google Scholar]
- 33.O’Farrell P H. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 34.Garrels J I. J Biol Chem. 1979;254:7961–7977. [PubMed] [Google Scholar]
- 35.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Zhang A, Derbyshire V, Galloway Salvo J L, Belfort M. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]
- 37.Dover N, Higgins C F, Carmel O, Rimon A, Pinner E, Padan E. J Bacteriol. 1996;178:6508–6517. doi: 10.1128/jb.178.22.6508-6517.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown L, Elliott T. J Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Draper D E. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtis R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 902–908. [Google Scholar]
- 40.Tsui H T, Feng G, Winkler M E. J Bacteriol. 1997;179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuppli D, Miranda G, Tsui H T, Winkler M E, Sogo J M, Weber H. Proc Natl Acad Sci USA. 1997;94:10239–10242. doi: 10.1073/pnas.94.19.10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsui H T, Leung H C, Winkler M E. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 43.Muffler A, Traulsen D D, Fischer D, Lange R, Hengge-Aronis R. J Bacteriol. 1998;179:297–300. doi: 10.1128/jb.179.1.297-300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neidhardt F C, Savageau M A. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Linn E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1310–1324. [Google Scholar]
- 45.Umbarger H E. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 442–457. [Google Scholar]
- 46.Glansdorff N. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Linn E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 408–433. [Google Scholar]
- 47.Boos W, Lucht J M. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1175–1209. [Google Scholar]
- 48.Takayanagi Y, Tanaka K, Takahashi H. Mol Gen Genet. 1994;243:525–531. doi: 10.1007/BF00284200. [DOI] [PubMed] [Google Scholar]
- 49.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. Proc Natl Acad Sci USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]