Abstract
DsrA RNA regulates both transcription, by overcoming transcriptional silencing by the nucleoid-associated H-NS protein, and translation, by promoting efficient translation of the stress σ factor, RpoS. These two activities of DsrA can be separated by mutation: the first of three stem-loops of the 85 nucleotide RNA is necessary for RpoS translation but not for anti-H-NS action, while the second stem-loop is essential for antisilencing and less critical for RpoS translation. The third stem-loop, which behaves as a transcription terminator, can be substituted by the trp transcription terminator without loss of either DsrA function. The sequence of the first stem-loop of DsrA is complementary with the upstream leader portion of rpoS messenger RNA, suggesting that pairing of DsrA with the rpoS message might be important for translational regulation. Mutations in the Rpos leader and compensating mutations in DsrA confirm that this predicted pairing is necessary for DsrA stimulation of RpoS translation. We propose that DsrA pairing stimulates RpoS translation by acting as an anti-antisense RNA, freeing the translation initiation region from the cis-acting antisense RNA and allowing increased translation.
The small regulatory RNA, DsrA, has two rather different activities in Escherichia coli, one affecting transcription and the other affecting translation. The transcription activity, antisilencing, is seen when DsrA is overproduced; DsrA overcomes H-NS-mediated transcriptional silencing of genes, including rcsA, a positive regulator of capsule synthesis (1). In addition, DsrA stimulates the translation of the stationary phase σ factor, RpoS (2). Although both pathways of DsrA action are dependent on Hfq (also called HF-1), an RNA-binding protein originally identified as necessary for the replication of phage Qβ (D. Sledjeski, manuscript in preparation), the activities otherwise appear to be independent. The effect of DsrA on antisilencing does not require RpoS (unpublished observations) and the translational stimulation of RpoS occurs in the absence of H-NS (2). We have investigated the basis for these two activities of DsrA by mutational dissection of dsrA.
The dsrA gene product is an 85-nt RNA that is predicted to form a structure with three stem-loops (S-Ls; Fig. 1) (1). In this work, we show that DsrA regulation of transcription and translation require different regions in this small molecule. Transcriptional regulation or anti-H-NS activity is independent of the first S-L (S-L1), while translational regulation is absolutely dependent on S-L1. Localization of translational regulation within S-L1 led to an anti-antisense model for translational regulation, which is supported by the behavior of compensatory mutations within the RpoS leader and DsrA.
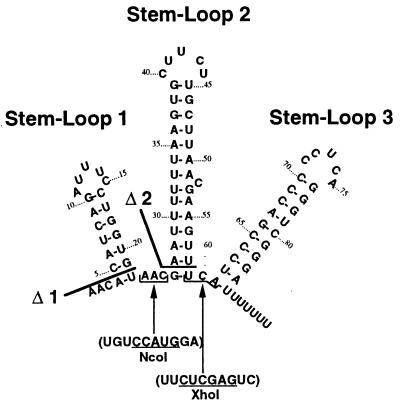
Figure 1.
Predicted structure of DsrA RNA. Mutations used in this work are indicated. The Δ1 mutation deletes the S-L above the line and similarly for the Δ2 mutation. The NcoI mutant substitutes the underlined sequence for the bracketed sequence within DsrA and similarly for the XhoI mutant. The NcoI* mutant (made by cutting, end filling, and religating the NcoI mutant DNA) had a sequence at the novel joint with a partial duplication of the site: UGUCCAUGCAUGGAA. The ΔNcoI-XhoI deletion derivative has the sequence UGGUGUCCAUGUCGAGUCC at the novel joint (underlined sequence from NcoI site, bold from XhoI site).
MATERIALS AND METHODS
Bacterial Strains.
Unless otherwise noted, strains were derivatives of either NM22180 (dsrA+) or NM22181 (ΔdsrA). These isogenic strains, derived by P1 transduction from SG20250 (3), carry deletions of the lac operon and of the ara region, introduced from LMG194 (4). The dsrA deletion was introduced by P1 transduction from strain DDS719, selecting for the linked TetR marker and screened by PCR as described previously (2). The various rpoS-lac fusions were constructed as described elsewhere and were transduced into the NM22180 and NM22181 E. coli hosts in the trp operon, selecting for the kanamycin resistance marker upstream from the fusion (5, 6).
Mutagenesis of the rpoS-lac fusions was done as described in refs. 5 and 6, starting with plasmid pTE591. The wild-type rpoS sequence between nucleotides 195 and 218 is CAAGGGATCACGGGTAGGAGCCAC. The mutagenic primers used for constructing G208C and G208U are CAAGGGATCACGGCTAGGAGCCAC and CAAGGGATCACGGTTAGGAGCCAC, respectively. In the region of 108–133 of the rpoS gene, the sequence is TCGTTACAAGGGGAAATCCGTAAACC. The mutagenic primers used to construct U124G and U124C are TCGTTACAAGGGGAAAGCCGTAAACC and TCGTTACAAGGGGAAACCCGTAAACC, respectively.
The NcoI′ substitution was made using a PCR product (Stratagene Pfu pol) of the ribosome-binding site region from pTE591, and cutting the product with EagI. This fragment was cloned into pTE591, first cut with NcoI and filled in, and then cut with EagI.
Plasmid Constructions and DNA Manipulations.
For the expression of dsrA and its mutant derivatives, the base plasmid used was pNM12. This plasmid is a derivative of pBAD24 (4) that was mutagenized by the Kunkel method (7, 8) to introduce an MscI site at the −7 to −2 region upstream of the +1 transcription start site. The PCR was used to generate a promoter-less dsrA DNA fragment with an MscI site upstream and an EcoRI site downstream. This PCR product was then cloned into the same sites on pNM12, thus placing dsrA under the control of the arabinose promoter (Para), to create plasmid pNM13. pNM14 and pNM15 were also created by the Kunkel method in another vector and moved by PCR as for pNM13. Other mutant derivatives of dsrA were cloned in a similar fashion. Plasmids pNM22, 29, and 30 were constructed by introducing the mutagenic sequence into the forward primer for PCR; plasmid pNM32 had the mutagenic sequence included in the reverse primer. Introducing the XhoI site between stems 2 and 3 was also done by PCR but using pNM13 (wild-type dsrA) and pNM33 (NcoI) to generate pNM35 (XhoI) and pNM39 (NcoI-XhoI), respectively. Plasmid pNM40 (NcoI*) was derived from pNM33 by cutting with NcoI, end filling, and religating the blunt ends. pNM42 (ΔNcoI-XhoI) was derived from pNM39 by cutting with both NcoI and XhoI, end filling, and religating. The QuickChange (Stratagene) mutagenesis kit was used to generate pNM33 according to the manufacturer’s specifications. The structure of dsrA was confirmed by sequencing for all plasmids.
Other methods for DNA manipulations used were as described in Maniatis et al. (9) and Ausubel et al. (10). Sequencing was done on an Applied Biosystems 310 Genetic Analyzer (PE/Applied Biosystems) using a dye terminator or a dRhodamine terminator cycle sequencing ready reaction kit (both from PE/Applied Biosystems) according to the manufacturer’s specifications.
The Salmonella dsrA region was sequenced from a plasmid provided by R. MacNab (Yale, New Haven, CT); the accession number for this sequence is AF090431.
β-Galactosidase Assays.
Assays were performed as described elsewhere (11). Briefly, overnight cultures grown in LB with ampicillin (100 μg/ml) were diluted 100-fold and grown at 32°C with or without arabinose. Aliquots were taken at regular time intervals; assays were kinetic measurements using a SpectraMax 250 microtiter plate reader. Units represent specific activities obtained by dividing Vmax slopes by OD600. The specific activities obtained have empirically been determined to be about 25 times lower than standard Miller units.
Northern Blot Analysis.
Purification of RNA was performed using a Qiagen DNA/RNA kit tip-20 used as described by the manufacturer for the isolation of total bacterial RNA. The RNA was separated on 6% urea/acrylamide sequencing gels. Total RNA corresponding to 0.3 μg was loaded in each lane in a formamide:50 mM EDTA (5:1) sample buffer to a final volume of 20 μl. Gels were electrophoresed in 1× TBE buffer for 75–80 min at 100 V and stained in 1 μg/ml ethidium bromide. The gels were then soaked in 0.5× TBE for 5 min before being assembled in a Bio-Rad electro-transfer unit according to the manufacturer’s specifications. The RNA was transferred to a Nytran+ positively charged membrane (Schleicher & Schuell) with a pore size of 0.2 μm. Transfer was carried out overnight at 4°C and 250 mA or 16 V. The RNA was cross-linked with a UV cross-linker (Stratagene) according to the manufacturer’s specifications. The resulting blot was washed for 2–3 min in 2× SSC buffer (GIBCO/BRL) and prehybridized in Hybrisol I (Oncor) for 3–4 h at 42°C in a hybridization chamber (Techne, Cambridge, UK). A nonisotopic probe (100 ng/ml) biotinylated at the 5′ end (Bioserve Biotechnologies, Laurel, MD) was dissolved in 10 ml Hybrisol/100 cm2 of membrane. The mix was boiled for 10 min, allowed to cool to 42°C, and used to replace the prehybridization solution. Hybridization was carried out overnight at 42°C. The blots were then washed and developed using Ambion’s BrightStar Biodetect nonisotopic kit (Ambion, Austin, TX) according to the manufacturer’s specifications. The blots were exposed either on Kodak X-Omat or BioMax MR film. In vitro transcription in a T7 MaxiScript kit (Ambion) of a PCR product containing dsrA was used to generate a DsrA-sized marker for the gels.
RNA Half-Life Determination.
To determine the half-life of dsrA and dsrB message, DDS 92 cells were grown at 32°C to mid-log phase in LB medium, and rifampicin (50 μg/ml final) was added to stop transcription. Samples were taken at appropriate intervals, and RNA was isolated and detected as described previously (1). Five micrograms of RNA was loaded per lane.
RESULTS
Regulated Expression of dsrA.
To allow regulated expression of DsrA RNA, we constructed pNM13, a plasmid expressing DsrA under the control of the pBAD promoter. DsrA function was assayed in strains carrying either of two reporter fusions. RpoS translation was measured using a rpoS-lacZ translational fusion; anti-H-NS silencing was measured using a cps-lacZ transcriptional fusion. Expression of the cps-lacZ fusion is dependent on RcsA; transcription of rcsA is in turn negatively regulated by H-NS (1). We previously observed that the rpoS-lacZ fusion responded to single-copy levels of DsrA, while the cps-lacZ fusion required higher levels to be significantly stimulated.
In a dsrA− host carrying a rpoS-lacZ fusion, no activity is seen from pBAD-dsrA+ unless dsrA is induced with arabinose (Fig. 2A). Arabinose (0.0007 to 0.001%) gives activity comparable to that seen from the single-copy chromosomal gene; activity increases with increasing arabinose and saturates at an arabinose concentration of 0.02%.
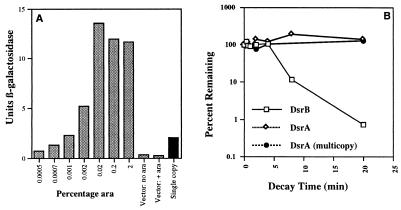
Figure 2.
(A) Expression of DsrA in response to arabinose. dsrA− cells carrying a rpoS-lacZ fusion in single copy on a λ lysogen (NM22508) were transformed with pNM13 (pBAD-dsrA+) or a vector (pNM12) control grown in LB with ampicillin broth in the presence of the indicated final percentage of arabinose at 32°C. Samples were removed at an OD600 ≈ 1 and assayed for expression of β-galactosidase from the fusion. An isogenic dsrA+ strain (strain NM22507) transformed with the vector (pNM12) was grown without arabinose and assayed under similar conditions (black bar). (B) Stability of DsrA. DsrA and DsrB messages were detected by primer extension and quantitated with a PhosphorImager. Multicopy DsrA was isolated from cells carrying pDDS164.
The stability of DsrA was estimated by growing cells at 32°C, treating the cells with rifampicin to stop additional transcription, and extracting RNA at various times after the rifampicin treatment. Both DsrA and the downstream, separately transcribed DsrB messenger RNA (1), were quantitated in a ribonuclease protection assay (Fig. 2B). Although dsrB RNA had a half-life of about 2.5 min in these experiments, DsrA showed no decay during the course of the experiment.
Mapping the Active Domains within DsrA.
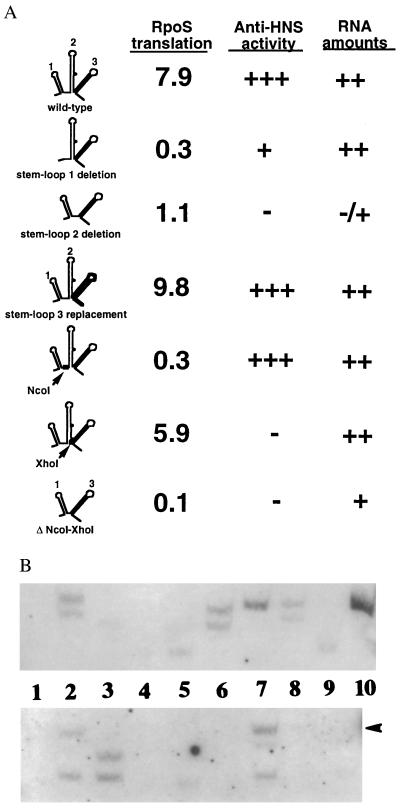
To identify sequences of DsrA that are required for its antisilencing or rpoS translational effects, we generated mutations in DsrA in the pBAD plasmids (Fig. 1). The function of the mutant plasmids was evaluated in hosts carrying either the rpoS-lacZ or cps-lacZ fusions (Fig. 3A). The amount of RNA from each of the mutants was determined by Northern blot (Fig. 3B). The results are summarized in Fig. 3A. In all cases, accumulation of the RNA was shown to be dependent upon addition of arabinose.
Figure 3.
Mutational analysis of DsrA. (A) Summary of effects of DsrA mutants on rpoS-lac and cps-lac fusions. To assay activity on rpoS-lac fusions, plasmids carrying the various pBAD-dsrA derivatives were introduced into the dsrA− rpoS-lac host NM22186 grown and assayed as described in Materials and Methods. Plasmids were those indicated in the legend of B. The activity on the cps-lac fusion was estimated from color on lactose MacConkey plates. Estimates of RNA amounts come from the data in B. (B) Accumulation of DsrA RNA. NM22186 transformed with pNM13 (pBAD-dsrA+) and various mutant plasmids as indicated below were grown at 32°C to an OD600 ≈ 0.7 either in the presence or absence of arabinose. RNA was extracted and processed as described in Materials and Methods. The upper panel was probed with a 5′-biotinylated probe specific for the first S-L of DsrA (S-L1 probe: TTCGTTACACCAGGAAATCTGATGTGTT); the biotinylated probe for the lower panel was specific for the third S-L (S-L3 probe: ATCCCGACCCTGAGGGGGTCGGGA). Note that the S-L1 probe would not be expected to detect DsrA RNA deleted for the first S-L, and the S-L3 probe should not detect DsrA with the altered terminator loop and has less homology with DsrA carrying the XhoI substitution or the NcoI-XhoI deletion than with the wild-type DsrA. We have not determined whether the multiple bands observed are in vivo products or due to cleavage during RNA isolation. Lane 1, pNM13 (dsrA+), no arabinose; lane 2, pNM13 with 0.02% arabinose; lane 3, pNM 14 (Δ1); lanes 4 and 5, pNM15 (Δ2) (lane 4, 1×; lane 5, 10× RNA added); lane 6, pNM32 (trpA terminator substituted for S-L3); lane 7, pNM33 (NcoI); lane 8, pNM35 (XhoI); lane 9, pNM42 (ΔNcoI-XhoI); lane 10, 5 ng of in vitro made DsrA product (upper panel only). Arrow to right of lower panel indicates position of in vitro made DsrA marker.
A deletion of the sequence that corresponds to the predicted first S-L (Δ1, Fig. 1) resulted in loss of the ability to induce rpoS-lacZ translation but retention of weak but detectable anti-H-NS activity; RNA amounts were close to wild-type levels (Fig. 3B, lane 3). Therefore, even though the cps-lacZ fusion requires high levels of DsrA to show stimulation, deletion of the full first stem and loop retained at least some anti-H-NS activity. Deletion of the sequences corresponding to the second predicted S-L (Δ2, Fig. 1) remained partially active for rpoS-lacZ activation but lost anti-H-NS activity. RNA amounts were also significantly decreased for this mutant; we could only detect the RNA when 10-fold more was loaded onto the gels (Fig. 3B, lane 5). Therefore, the partial activity for rpoS-lacZ may be entirely due to decreased amounts of the RNA rather than any requirement for the deleted sequences for rpoS translation activity. Because of the lower sensitivity of the cps-lacZ fusion, it is not possible to accurately measure anti-H-NS activity from the low levels of RNA accumulating in this mutant.
The third S-L of DsrA appears to encode a factor-independent transcription terminator, followed by the characteristic run of U nucleotides (Fig. 1). A point mutation in the predicted S-L3 does block termination, resulting in accumulation of unusually long RNA that has decreased function (1). In plasmid pNM32, the terminator S-L of dsrA was replaced with that of the trpA terminator (12, 13). The S-L3 replacement retains both wild-type activities (Fig. 3A) and wild-type RNA amounts, suggesting that sequences within S-L3 are not necessary for DsrA activity beyond their role in transcription termination.
Two restriction sites were introduced into the dsrA gene. An NcoI site inserted between S-L1 and 2 (NcoI, Figs. 1 and 3) yields DsrA with a phenotype similar to Δ1. The mutant plasmid (pNM33) had lost the ability to induce rpoS-lacZ expression but retained anti-H-NS activity. A derivative of this plasmid, pNM40 (NcoI*), has four additional base pairs, but a similar activity (data not shown). Even though shorter RNA products were not seen as with other derivatives, wild-type levels of full-length RNA accumulated for both the NcoI (Fig. 3B, lane 7) and the NcoI* mutant (data not shown). Insertion of an XhoI site between S-L2 and 3 (XhoI, Fig. 1) was phenotypically similar to Δ2, with partial activity for rpoS-lacZ translation but no anti-H-NS activity. However, more RNA accumulated for this mutant than for the Δ2 mutant (Fig. 3B, lane 8). A plasmid deleted for the region between the two sites lost both activities but again had RNA amounts less than the wild type, but significantly more than the Δ2 mutant (Fig. 3B, lane 9). Therefore, the first S-L and the region changed by the insertion of an NcoI site are necessary for translational regulation of RpoS but are not essential for anti-H-NS activity. Both activities are independent of S-L3. If S-L1 and S-L3 are both unnecessary for anti-H-NS activity, it seems highly likely that the remaining parts of the molecule are needed for antisilencing activity. Mutations tested thus far in these regions (Δ2, XhoI, and ΔNcoI-XhoI) are consistent with this, retaining some rpoS stimulation but abolishing antisilencing; these mutations also impair stability of the RNA to varying degrees.
A Model for Translational Regulation of rpoS by Sequences in S-L1 of dsrA.
The translation of RpoS has been shown to be regulated, increasing under a variety of stress conditions and, we had previously shown, increasing at low temperatures in a DsrA-dependent fashion (2, 14). RpoS translation was also found to be dependent on the Hfq host factor (15, 16). The requirement for Hfq can be bypassed by mutations which disrupt structures within the long upstream leader of rpoS (6) (Fig. 4B). The first S-L of DsrA and regions nearby are necessary and sufficient for DsrA to stimulate RpoS translation; this region also is complementary to the RpoS leader region implicated in translational regulation (Fig. 4). Since weakening the hairpin structure of the RpoS leader by mutation allows high-level expression of RpoS, we predicted that DsrA pairing with this upstream region of the RpoS leader would also eliminate pairing of the two regions of RpoS message, freeing the Shine-Dalgarno region for efficient translation (Fig. 4C). Because DsrA and the RpoS leader need to pair, the model predicts that sequences and not simply structure within the first S-L of DsrA will be important, and that sequence changes should be tolerated if changes in the target also occur so that pairing between DsrA and the RpoS leader is maintained.
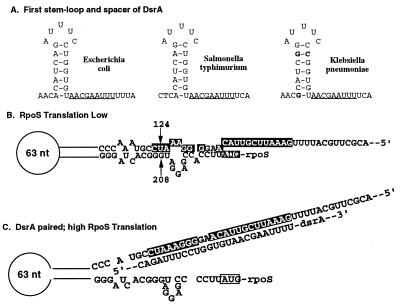
Figure 4.
Model for DsrA Action. (A) First S-L of DsrA. The sequence of the first S-L in E. coli, Klebsiella pneumoniae, and Salmonella typhimurium are shown. The changes in Klebsiella within the first S-L and the region of complementarity to the E. coli RpoS leader are shown in bold; no changes in this region are found in the Salmonella sequence. (B) Paired structure in RpoS leader. The structure for the RpoS leader deduced from mutations that overcome Hfq dependence (6) is shown. The bases predicted to pair with DsrA are highlighted in black with white lettering. Base pair 124:208 (arrows) is used in experiments described in Fig. 6. (C) Proposed opening of RpoS structure to allow high levels of RpoS translation as a result of sequestering of the upper strand by DsrA. Regions of pairing of RpoS and DsrA are shown.
We tested whether the secondary structure within the RpoS leader is the target for DsrA action by asking if mutations which bypass the requirement for Hfq by disrupting the structure also bypass the requirement for DsrA. A series of mutations in the leader region, previously tested for degree of Hfq dependence, were tested in dsrA+ and dsrA− hosts, and the ratio of activities in these two hosts compared with that seen with and without Hfq. For the wild-type RpoS leader, the dependence on Hfq or on DsrA was about six-to ninefold. In most of the mutants tested, those that showed reduced dependence on Hfq were generally also less dependent on DsrA. For instance, mutations that disrupt a G:C pairing at position 126 and 206 of the leader (SD2 and SD3) increased expression of an rpoS-lacZ fusion and lessened the Hfq dependence from sixfold to two- to threefold, while combining the mutations (SD2 + SD3) restored a low basal level and 10-fold Hfq dependence (6). The response to DsrA was similar; SD2 or SD3 alone showed about a twofold DsrA dependence, while the SD2 + SD3 construct was stimulated ninefold by DsrA, similar to that seen with the wild type (data not shown). Therefore, disrupting the structure sufficiently to free rpoS translation from Hfq dependence also leads to DsrA independence, consistent with the idea that both Hfq and DsrA act at a similar step, opening this structure.
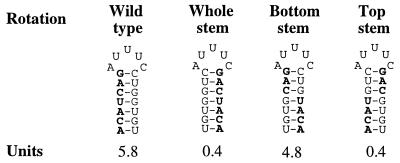
Mutants in the 7 bp stem 1 of DsrA that would be expected to retain structure but change sequence were constructed. DsrA with an inversion of the four pairs of nucleotides at the base of the stem rotated (bottom stem inversion, Fig. 5) retained wild-type activity. However, if the three pairs of nucleotides at the top of the stem (top stem inversion, Fig. 5) or if all of the nucleotides within stem 1 (full stem inversion, Fig. 5) were rotated, DsrA was unable to stimulate RpoS-lacZ translation. All mutants had wild-type levels of RNA (data not shown). Therefore, the sequence itself is important, at least for portions of stem 1.
Figure 5.
Mutations within the first stem of DsrA. The structure of the wild-type sequence for the first stem of E. coli DsrA, with the left side bases shown in bold, as well as the structure of stems with regions inverted are shown. Below the structure are values for expression of a rpoS-lac fusion in cells carrying the wild-type or mutant plasmids. Wild-type plasmid, pNM13; whole-stem inversion, pNM22; bottom stem inversion, pNM29; top stem inversion, pNM30.
Mutational Analysis of Loop 1 of DsrA.
Fig. 4 indicates that the 5 nucleotides predicted to form loop 1 are conserved in Klebsiella and Salmonella and are all complementary to the RpoS message. When all five positions were randomized, a minority retained activity. All those that were active had G or A at the first position, and a large percentage also had the wild-type nucleotide, C at the fifth position. No particular patterns were observed for other positions within the loop.
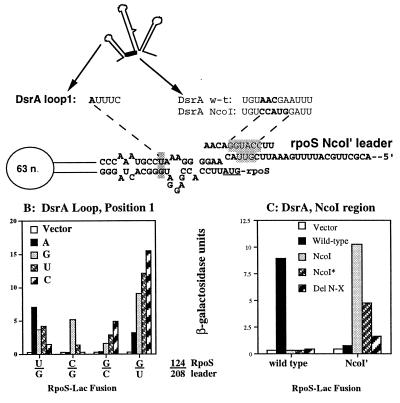
To test the importance of complementarity more directly, we chose position 1 of the DsrA S-L1 because it had given a restricted set of sequences among active clones in the randomization experiment and because it should pair with U124 of the RpoS upstream leader. U124 is predicted to pair with G208 of the downstream leader (see Fig. 4), and both are far from the translation initiation sequence. All four bases were introduced into DsrA at this position, and these were tested for activity on hosts deleted for dsrA and carrying rpoS-lacZ fusions with various changes for the 124:208 bp, all of which retained pairing.
The β-galactosidase activity for strains carrying either the vector or one of these plasmids grown with 0.02% arabinose are shown in Fig. 6B. In all four cases, the most active DsrA derivative was the one which would be expected to be complementary to the nucleotide at position 124 of the RpoS leader. When the wild-type U is at position 124 (predicting a U:G pairing with the downstream leader), A at position 1 in the loop of DsrA is most active; G also has significant activity, consistent with the finding from the randomization experiments that both A and G could be found among high activity plasmids. Surprisingly, U or C at this position also gave activity significantly above the vector background level, suggesting that pairing between DsrA and the RpoS leader at this position is not absolutely critical when the rest of the sequence complementarity between DsrA and the RpoS leader is preserved.
Figure 6.
Testing complementarity with compensatory mutants. (A) Changes made in DsrA and the RpoS leader. (B) The wild-type S-L1 position 1 (A) is predicted to pair with the U at position 124 of the RpoS leader; alternatively, this U is predicted to pair with a G at position 208 of the leader. The wild-type and three mutant versions of this U:G pair within rpoS-lacZ were tested: wild-type U:G (strain NM22187), C:G (NM22201), G:C (NM22203), and G:U (NM22207). These four strains were transformed with vector (pNM12, white bars), the wild-type pBAD-dsrA+ plasmid (pNM13, black bars), or derivatives with changes at position 1 of the loop to G (pNM43, slight gray columns), T (pNM44, narrow striped bars), or C (pNM45, broad striped bars). Transformed strains were grown in the presence of 0.02% arabinose to an OD600 ≈1 and assayed for β-galactosidase activity. (C) An NcoI site introduced into DsrA between S-L1 and S-L2 is predicted to disrupt pairing of DsrA with the unpaired leader of RpoS. NM22204 (NcoI′) carries a derivative of the rpoS-lacZ fusion which restores complementarity to the NcoI DsrA. Cells carrying the wild-type or NcoI′ rpoS-lac fusion were transformed with vector (pNM12, white bars), pBAD-dsrA+ (pNM13, black bars), pBAD-dsrA-NcoI (pNM33, light gray bars), pBAD-dsrA-NcoI*, in which the NcoI site was cut, end-filled, and religated (pNM40, narrow striped bars), or pBAD-dsrA-ΔNcoI-XhoI, in which the NcoI and XhoI sites were cut, end-filled, and religated (pNM42, broad striped bars). Cells were grown in the presence of 0.02% arabinose at 32°C and assayed for β-galactosidase.
However, there was a much more stringent requirement for correct DsrA/RpoS pairing at this position when the rpoS-lacZ fusion contained a C:G pair in place of the wild-type U:G pair. In this case, none of the DsrA plasmids except that with G at position 1 gave activity significantly above the vector control (Fig. 6B). This result both supports the role of pairing between DsrA and the RpoS leader and suggests that the relaxed specificity of the DsrA requirements for wild-type RpoS leader reflects the relatively weak pairing of the U:G pair in the leader.
Somewhat surprisingly, the stringency of DsrA/RpoS leader interaction is not entirely symmetric. When the C:G pair was changed to a G:C pair, DsrA derivatives predicted to be unable to pair at this position still gave activities better than vector, although the most activity was seen with DsrA carrying C (the correct pairer) at this position. Finally, a G:U pair at this position of the RpoS leader is again more permissive than the G:C derivative; a higher basal level of activity was seen with the vector and all of the DsrA derivatives gave higher activities than the G:C pair, with the correctly paired C derivative again giving the best activity.
Therefore, maintaining complementarity of position 124 of RpoS and position 1 of the loop of DsrA was necessary for optimum translation of RpoS, independent of the precise sequences; changes in the nature of the pairing within the RpoS leader change the importance of correct DsrA pairing at this position.
Analysis of the Unpaired Region at the 5′ End of the RpoS Leader Structure.
We had noted above that insertion of an NcoI site between S-L1 and 2 specifically abolished stimulation of rpoS-lacZ translation. The sequence match between DsrA and the upstream leader of RpoS extends into this region as well (Fig. 4 B and C), and because this is in a region of the leader predicted to be unpaired (6), changes in the leader should not disrupt the predicted leader–leader pairing interactions. A rpoS-lacZ fusion that restores pairing to the DsrA NcoI mutant was constructed (NcoI′) and used as a second test of the importance of complementarity for DsrA regulation. Fig. 6C shows that wild-type DsrA has an activity 10- to 12-fold lower on the rpoS-lacZ fusion carrying the NcoI′ mutation than on the wild-type rpoS-lacZ. In contrast, a mutant DsrA with the NcoI sequence stimulates translation from the complementary mutant rpoS-lacZ NcoI′ 9- to 10-fold better than it does from the wild-type rpoS-lacZ (where activity is not much above that of a vector). The NcoI* plasmid, which shortens the region of complementary nucleotides with the NcoI′ target region but still retains a stretch of 8 bp of pairing (compared with 12 for wild-type DsrA with wild-type RpoS and 15 for DsrA-NcoI with NcoI′ RpoS), is also able to stimulate activity of NcoI′ but not wild-type rpoS-lacZ. A plasmid with the region between the NcoI site and the XhoI site deleted, which retains most of the NcoI sequence (see Fig. 1 legend), retained some activity despite reduced levels of RNA (see Fig. 3B), consistent with our earlier suggestion that there is no essential role of S-L2 for DsrA activity on rpoS translation. Therefore, as predicted by the model, complementarity between the upstream leader of RpoS and DsrA is necessary for efficient stimulation of translation by DsrA.
DISCUSSION
Our molecular dissection of the small RNA, DsrA, clearly indicates that different parts of this small molecule are involved in the translational regulation of RpoS and transcriptional antisilencing. The third S-L can be replaced by another terminator. Sequences from the first S-L and from the region early in the second stem appear to be unnecessary for overcoming H-NS silencing. This leaves the second S-L and the region up to the third stem (and maybe the few nucleotides at the 5′ end of the molecule) as likely to be critical for antisilencing.
We present evidence that DsrA can regulate the translation of RpoS by direct interaction with the 5′ end of the RpoS leader mRNA. This upstream leader RNA pairs with the leader just before the translation initiation site, serving as a cis-acting antisense inhibitor of translation. DsrA acts as an anti-antisense RNA, allowing translation by relieving the inhibitory pairing. An extended region of complementarity between the first S-L of DsrA and the RpoS message allows efficient pairing. We found that disrupting the pairing interfered with the ability of DsrA to stimulate RpoS translation and re-establishing pairing by compensating mutations in the RpoS leader restored efficient translation.
Most previously known regulatory RNAs act either as modulators of replication or translation, generally inhibiting these processes (for review, see refs. 17 and 18). Frequently they are antisense RNAs, encoded directly by the opposite strand of the regulated gene and therefore are complementary over a long region. However, within this fully complementary region, some structures and sequences are more critical than others in initiating and stabilizing the interaction with the target RNA. DsrA RNA is complementary to the RpoS leader mRNA over a stretch of about 20 nucleotides (Fig. 4), and even within this stretch, complementarity is not complete. As with previously studied antisense RNAs, our work suggests that not all nucleotides or pairing interactions are equally important for regulation, and that it is not just the sum of interactions that matters. Exchanging the four pairs of nucleotides at the base of stem 1 leads to the loss of three possible matches with the RpoS leader but results in three potential new matches. This mutant is neither less nor more active than wild type. In contrast, exchange of the three nucleotide pairs at the top of stem 1 generates a net loss of two possible matches; this mutant is completely inactive. The Klebsiella pneumoniae dsrA homolog contains a G9:C17 pairing in stem 1 while Escherichia coli carries an A:U pair at the same position. The latter do not match with their respective partners in the E. coli RpoS leader (Fig. 4). However, changing the E. coli A9:U17 pairing to that found in Klebsiella (G9:C17) did not improve DsrA function (data not shown).
We did find two cases in which loss of a single match between DsrA and the RpoS leader led to almost complete loss of activity. The first of these is a mutation of U124 to C124, creating a C:G bp within the RpoS leader. As seen in Fig. 6B, the only DsrA derivative which had activity on this leader was that in which position 1 of loop 1 was changed to a G, able to pair with C124. We note that when the RpoS leader pairing at this position is U:G rather than C:G, all bases within the pairing position of DsrA give significant activity. Therefore, in the wild-type case, there is significant flexibility for the trans-acting DsrA RNA. The significance of this for the biology of expression of RpoS under conditions other than those involving DsrA is unknown. A second highly selective pairing requirement was found for the fifth position of the DsrA loop. In randomization experiments, this was almost always C for active clones and rarely C for the fully inactive isolates. In addition, a CUUUC loop is reasonably active on the wild-type host and even more active on a host with G:C pair at 124:208 (Fig. 6B), but DsrA with a CUUUA loop was totally inactive (data not shown).
We find that DsrA is quite stable, not too surprising given its secondary structure and the stability of many other small RNAs. However, its stability raises questions about regulation of its activity in vivo. Does it continue to act, on multiple RpoS leaders, once made, or must it be continuously made to continue to act? We can imagine that it may act stoichiometrically, binding and then either remaining bound to a given message or possibly causing cleavage of the annealed DsrA/RpoS RNA complex, freeing the downstream truncated message for efficient translation (until the Rpos message is itself degraded), but destroying DsrA in the process. We have noted some evidence of more efficient stimulation of anti-HNS silencing by DsrA defective in promoting RpoS translation, consistent with any sort of stoichiometric model that consumes DsrA (N. Majdalani, unpublished observations).
Studies on the translation of RpoS have previously suggested the requirement for the RNA-binding protein Hfq for efficient translation (15, 16). Mutations that disrupt the structure shown in Fig. 4A become independent of the Hfq requirement and, simultaneously, become independent of DsrA. However, we have previously demonstrated that the DsrA dependence of RpoS translation is most profound at low temperatures, conditions where the synthesis of DsrA is elevated (2). As the temperature rises, the dependence of RpoS translation on Hfq remains, while the dependence on DsrA is not as dramatic (unpublished observations). We can consider two types of explanations for these observations. It seems possible that, whatever the mechanism by which Hfq increases RpoS translation, at low temperatures but not at high temperatures DsrA is necessary to help stabilize the open structure. In this model, environmental signals would act directly on Hfq and/or the RpoS leader to turn up translation at elevated temperatures. A second model would suggest that there are other DsrA-like molecules in the cell, made in response to some of the stress signals other than low temperature known to increase RpoS translation. In this second model, Hfq may not itself mediate any of the responses to environmental stress, but may work together with regulated RNAs, and changes in the amounts or activities of these RNAs would mediate the stress response. At least one other RNA does affect RpoS translation. OxyS, an RNA induced after oxidative stress, down-regulates RpoS. As with DsrA, regulation is at the level of translation, but in this case the RNA appears to act as a competitive inhibitor for Hfq (19). Interestingly, OxyS, like DsrA, has more than one target within the cell (20). It seems likely that OxyS and DsrA represent the beginning of what promises to be a growing number of trans-acting regulatory RNAs with multiple functions, providing a new and flexible mode of global regulation.
Acknowledgments
We thank Gisela Storz, Sankar Adhya, Bob Weisberg, Michael Yarmolinsky, YanNing Zhou, and Francis Repoila for their comments on the manuscript. Funding was provided in part by National Institutes of Health Grants GM40403 (to T.E.) and GM56448 (to D.D.S).
ABBREVIATION
- S-L
stem-loop
Note Added in Proof
In this issue of the Proceedings, Lease et al. (21) have proposed a similar model for DsrA stimulation of RpoS translation and have shown that a multiple mutant of S-L1 is defective for this activity.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF090431).
References
- 1. Sledjeski D, Gottesman S. Proc Natl Acad Sci USA. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sledjeski D D, Gottesman S. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 3.Gottesman S, Trisler P, Torres-Cabassa A S. J Bacteriol. 1985;162:1111–1119. doi: 10.1128/jb.162.3.1111-1119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guzman L-M, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott T. J Bacteriol. 1992;174:245–253. doi: 10.1128/jb.174.1.245-253.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown L, Elliott T. J Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 9.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 10.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. In: Current Protocols in Molecular Biology. Chanda V B, editor. New York: Wiley; 1996. [Google Scholar]
- 11.Zhou Y-N, Gottesman S. J Bacteriol. 1998;180:1154–1158. doi: 10.1128/jb.180.5.1154-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keiler K C, Waller P R H, Sauer R T. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 13.Farnham P J, Platt T. Cell. 1980;20:739–748. doi: 10.1016/0092-8674(80)90320-7. [DOI] [PubMed] [Google Scholar]
- 14.Lange R, Hengge-Aronis R. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 15.Brown L, Elliott T. J Bacteriol. 1996;178:3763–3770. doi: 10.1128/jb.178.13.3763-3770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muffler A, Fischer D, Hengge-Aronis R. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 17.Gerdes K, Gultyaev A P, Franch T, Pedersen K, Mikkelsen N D. Annu Rev Genet. 1997;31:1–31. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Wagner E G H, Simons R W. Annu Rev Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, A., Altuvia, S., Tiwari, A., Argaman, L., Hengge-Aronis, R. & Storz, G. (1998) EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 20.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 21.Lease R A, Cusick M E, Belfort M. Proc Natl Acad Sci USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]