Abstract
Mouse mammary tumor virus (MMTV) is a betaretrovirus that infects rodent cells and uses mouse transferrin receptor 1 for cell entry. To characterize the interaction of MMTV with its receptor, we aligned the MMTV envelope surface (SU) protein with that of Friend murine leukemia virus (F-MLV) and identified a putative receptor-binding domain (RBD) that included a receptor binding sequence (RBS) of five amino acids and a heparin-binding domain (HBD). Mutation of the HBD reduced virus infectivity, and soluble heparan sulfate blocked infection of cells by wild-type pseudovirus. Interestingly, some but not all MMTV-like elements found in primary and cultured human breast cancer cell lines, termed h-MTVs, had sequence alterations in the putative RBS. Single substitution of one of the amino acids found in an h-MTV RBS variant in the RBD of MMTV, Phe40 to Ser, did not alter species tropism but abolished both virus binding to cells and infectivity. Neutralizing anti-SU monoclonal antibodies also recognized a glutathione S-transferase fusion protein that contained the five-amino-acid RBS region from MMTV. The critical Phe40 residue is located on a surface of the MMTV RBD model that is distant from and may be structurally more rigid than the region of F-MLV RBD that contains its critical binding site residues. This suggests that, in contrast to other murine retroviruses, binding to its receptor may result in few or no changes in MMTV envelope protein conformation.
Although the betaretrovirus mouse mammary tumor virus (MMTV) was identified as a transmissible agent that causes breast cancer in mice more than 70 years ago, an understanding of how the virus infects cells has lagged behind that for other retroviruses (32). This was primarily due to a lack of cell lines that produced high-titer infectious virus and of a measurable assay to determine infection. Several years ago, murine leukemia virus (MLV) pseudotypes bearing the MMTV Env protein were developed, and these have greatly facilitated the study of MMTV interaction with cells (12, 18). For example, we recently used MMTV pseudotypes to identify mouse transferrin receptor 1 (TfR1) as the cell entry receptor for this virus (40).
The use of TfR1 for virus entry is the first reported case of an enveloped virus attachment protein having a primary function that is directly related to endocytosis. In normal cell physiology, binding of transferrin to the TfR1 results in endocytosis of the complex via clathrin-coated pits that traffic to the acidic endosomal compartment, where iron is released; this is the predominant pathway used by this receptor after endocytosis (28, 36). It was previously shown that treatment of MMTV-infected mammary gland cells with acid induced cell-cell fusion (39), and we have found that MMTV pseudovirus infection is dependent on acidified endosomes for infection (40). Although neutralization of the endosomal compartment does not affect infection by most retroviruses, it does inhibit infection by avian leukosis virus (31) as well as other viruses such as influenza virus, Semliki Forest virus, and vesicular stomatitis virus (20). At least for influenza virus, it is this pH change and not receptor binding that induces the conformational change leading to fusion of the virus and plasma membranes (43). Given that MMTV uses a receptor that traffics to the acidic endosome upon ligand binding and that acid pH triggers MMTV-mediated cell-cell fusion, it may be acid pH that triggers the conformational change in the MMTV Env protein as well, thereby allowing virus-cell membrane fusion.
Although the TfR1s of different species are highly homologous, MMTV shows clear species tropism. MMTV pseudotypes infect only rodent, not hamster or human, tissue culture cells (12, 40). This tropism resides at the level of TfR1, since human and hamster cells expressing the mouse TrfR cDNA are highly susceptible to MMTV pseudotype infection (40). However, there have been reports that continued passage of MMTV on human breast cancer cell lines resulted in adapted viruses that could infect human cells (22, 26, 45). In addition, several groups have recently identified MMTV-like Env sequences in human breast cancer specimens (14, 46). The Env sequences of different MMTV strains are highly homologous to each other and to the MMTV-like env genes cloned from human breast cancer samples (see Fig. 3). Thus, any sequence differences between these various Envs, especially in the surface (SU) protein, represent candidate regions involved in receptor binding.
FIG. 3.
Amino acid sequence comparison of the SUs of MMTV(C3H), MMTV(RIII), and h-MTVs found in human tissues and cells. Boldfaced amino acid designations indicate changes unique to h-MTVs relative to all known exogenous and endogenous MMTVs. The boxed sequences show the amino acids used to make the RBS-GST fusion protein.
We thus used two approaches to determine what regions of the MMTV SU protein were involved in receptor contact and found that both pointed to a common candidate region. In the first, we aligned the primary amino acid sequence of MMTV SU with that of Friend murine leukemia virus (F-MLV), for which the crystal structure of the receptor binding domain (RBD) has been solved (15). We found strong homology between the two proteins in the sequences that make up the secondary structural elements of F-MLV, such as the beta strands and alpha helices. This alignment allowed us to identify domains important for receptor interaction, including a putative heparin-binding domain (HBD).
In the second approach, we compared the SU sequences of the MMTV-like elements cloned from several human breast cancers with those of different MMTV strains and with an MMTV passaged on the MCF-7 human breast carcinoma line. One of the MMTV-like elements had a Phe40 to Ser change in a five-amino-acid segment (Phe40-His41-Gly42-Phe43-Arg44) that could serve as a region for receptor contact for the viral receptor. Placement of this nonconservative change, as well as Phe40 to Ala40, in the SU of MMTV abolished both virus binding to and infection of cells expressing the murine TfR1 (mTfR1). In contrast, a nonconservative Gly42-to-Glu42 change present in the MCF-7-adapted virus did not affect infection when placed in the MMTV SU backbone. Importantly, neither change extended the tropism of MMTV pseudoviruses to human cells. Two different neutralizing monoclonal antibodies recognized a glutathione S-transferase (GST) fusion protein containing these five amino acids, indicating that this region is the receptor-binding site (RBS).
The critical influence of Phe40 on binding and infection as well as the presence of an HBD indicate that the region identified here constitutes the RBD of MMTV. Moreover, the inability to change the species tropism from mouse to human suggests that it is unlikely that MMTV can easily adapt to using human TfR1 for virus entry.
MATERIALS AND METHODS
Alignment and modeling.
Segments within the putative MMTV RBD that were likely to have β-strand or α-helical structure were hand aligned with the known β- and α-helices of the F-MLV RBD sequence (9, 10). The preliminary alignment was further adjusted by using a previous alignment of the F-MLV and BLV RBD (24) and then subjected to Swiss-Model for modeling of the MMTV structure. The WhatCheck and Tracelog reports from the SwissModel submission identified residues likely to be misaligned based on modeling algorithms that test a number of criteria. These include algorithms that identify residues showing unallowable side chain angles imposed by replacement of the template F-MLV RBD residues with the aligned MMTV sequence, as well as algorithms that predict the probability that residues aligned with secondary-structure elements of the template. For example, the modeling algorithms identify residues that were aligned with β-strands but are rarely or not found in β-strands of known structures. Residues that were likely to be misaligned were moved one position amino- or carboxy-terminal in the F-MLV alignment and resubmitted for modeling, and additional adjustment of the alignment was made based on the results of the second set of models. Further adjustment of the positions of residues several positions in either direction gave similar model coordinates.
Cell lines and antibodies.
293T and 293 human kidney epithelial cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). NMuMG (normal mouse mammary gland) epithelial cells were grown in DMEM containing 10% FBS and 10 μg of insulin per ml. TRH3 cells were generated by cotransfecting 293 cells with plasmids containing the mouse TfR coding region under the control of the cytomegalovirus promoter (40) and pSV2neo, containing the neomycin resistance gene under the control of the simian virus 40 promoter, followed by selection in G418 (100 μg/ml) and fluorescence-activated cell sorting (FACS) for TfR expression in a FACStar Plus (Becton Dickinson, Inc.) to derive clonal isolates. The MCF7/vp5 [derived by adapting MMTV(RIII) on MCF-7 human breast cancer cells; A. Vaidya, personal communication] and MR/C1 [MMTV(RIII)-infected mink lung] cell lines were grown in DMEM containing 10% FBS, 10 μg of insulin per ml, and 1 mM sodium pyruvate. Polyclonal goat anti-MMTV or SU antiserum was described previously (12). Polyclonal rabbit anti-GST antibody was from Sigma, Inc. (St. Louis, Mo.). Mouse monoclonal anti-gp52 antibodies Black 6, Black 8-6, 2F10, and Black 6-5D were previously described (38).
Cloning and sequencing RIII virus env genes.
Genomic DNA from the MCF7/vp5 and MR/C1 cell lines was amplified by PCR with primers specific for the su portion of the MMTV env (P1, 5′-CTTGTGTTTTTCCACAGGATG-3′; P2, 5′-TGCGAATTCCTATCGCTTGGCTCGAATTAAATC) and sequenced directly. To clone the env genes, PCR primers were designed that included the same fragment of MMTV genomic proviral DNA present in pENVC3H [containing the full-length MMTV(C3H) env from the central HindIII site to the end of the 3′ long terminal repeat in mammalian expression vector pcDNA3.1] (12). The amplified fragments were cloned into pcDNA3.1 (Invitrogen, Inc.) to generate pENVRIII (from MR/C1 cells) and pENVRIIIM (from MCF7/vp5 cells).
Site-directed mutagenesis.
Plasmid pENVC3H was the template for mutagenesis with the QuikChange XL site-directed mutagenesis kit (Stratagene, Inc.). The codon for residue Phe40 of the envelope was mutated to Ser40, Tyr40, or Ala40, and the Gly42 codon was mutated to Glu42 in four separate mutagenic reactions. To generate the ΔHBD mutation in pENVC3H, lysine codons 123, 124, and 125 of the MMTV envelope SU region were changed to alanine.
Pseudovirus preparation and virus infection.
MMTV Env-pseudotyped MLV recombinant viruses were made by transient cotransfection of 293T cells with pHit111 (murine leukemia virus [MLV] genome with the β-galactosidase marker), pHit60 (MLV gag/pol genes) (44), and the pENV-based plasmids as previously described (18). For virus infection assays, 2 × 105 cells were grown on six-well plates for 1 day, and diluted pseudovirus supernatants containing Polybrene (8 μg/ml) were incubated with cells at 37°C for 2 h. For antibody blocking studies, pseudovirus was incubated for 10 min with polyclonal goat anti-MMTV antiserum (1:2,000) or cell supernatants from hybridoma cells (1:5) prior to addition to cells. For the heparan sulfate competition studies, medium containing viruses pseudotyped with either wild-type MMTV(C3H) envelope or the ΔHBD mutant were incubated with the indicated amounts of heparan sulfate (ICN Biochemicals 97040) at 37°C for 1 h. Polybrene (8 μg/ml) was added, and this mixture was then used to infect NMuMG cells. After incubation for 1 h, the cells were washed and refed with fresh medium. For all pseudovirus infections, the cells were stained for β-galactosidase activity 48 h after infection, and blue colonies were counted. Data are presented as LacZ-forming units (LFU) per milliliter of supernatant.
Virus binding assay.
Pseudoviruses were made as described above. The transfected cell supernatants (100 ml) were ultracentrifuged at 25,000 rpm for 2 h in an SW28 rotor, and virus pellets were resuspended in 1.5 ml of phosphate-buffered saline (PBS, pH 7.4) containing 2% FBS and 1 mM EDTA (PBS-FE), resulting in 67-fold concentration. Single-cell suspensions of NMuMG cells were prepared by incubation with PBS-FE for 15 min, followed by vigorous pipetting. Then 0.5 ml of concentrated virus stock containing equal amounts of virus particles, as determined by Western blotting analysis, was incubated with 2.5 × 105 NMuMG cells in the presence of 8 μg of Polybrene per ml at 4°C for 1 h.
For the heparan sulfate binding inhibition, the virus preparations were incubated for 30 min at 37°C with 100 μg of heparan sulfate per ml. The cells were washed three times and resuspended in 100 μl of ice-cold PBS containing 1% FBS and 0.2% sodium azide. Then 100 μl of a 1:100 dilution of goat anti-MMTV polyclonal antiserum was added and incubated at 4°C for 30 min. The cells were washed three times and incubated with 100 μl of fluorescein isothiocyanate (FITC)-conjugated rabbit anti-goat immunoglobulin antibody. The cells were washed three times, resuspended in 2% paraformaldehyde, and subjected to FACS analysis. Data were acquired on a FACScan cytometer (Becton Dickinson, Mountain View, Calif.) and analyzed with CellQuest software (Becton Dickinson Immunocytometry Systems).
TfR1 downregulation assays.
For TfR1 downregulation, TRH3 cells were incubated with MMTV(C3H) pseudovirus, MMTV(C3H) pseudovirus pretreated with goat anti-MMTV antiserum, Ser40 pseudovirus, or rat anti-mTfrF1 antibody (1 μg of MOPC-315; RDI, Flanders, N.J.) per ml by spinoculation (2-h centrifugation at 1,200 × g at room temperature) as previously described (40). The cells were washed extensively with cold PBS-1% fetal calf serum-0.2% sodium azide and then stained with rat anti-mouse CD71 (TfR1) conjugated with phycoerythrin (PharMingen). 293 cells were also incubated with pENVC3H-bearing pseudovirus, and the mTfR1-transfected cell line TRH3 was also directly stained with the phycoerythrin-conjugated anti-CD71 antibodies. Data were acquired on a FACScan cytometer (Becton Dickinson) and analyzed with CellQuest software (Becton Dickinson Immunocytometry). Dead cells were excluded by their forward scatter/side scatter properties.
Expression and purification of RBS-GST fusion protein.
A fragment encoding the RBS of the MMTV(C3H) Env (amino acids 35 to 48; boxed sequence in Fig. 3) was cloned into the BamHI site of the prokaryotic expression vector pGEX-2T (Pharmacia, Inc.). Protein was isolated from Escherichia coli JM109 transformed with the recombinant pGEX-2T or parental pGEX-2T plasmid by affinity purification with glutathione—agarose beads (Pharmacia, Inc.) according to the manufacturer's instructions.
Western blotting.
Equal volumes of pseudovirus supernatants were resolved on denaturing 10% polyacrylamide gels. The proteins were transferred to a nitrocellulose membrane and probed with polyclonal goat anti-MMTV or anti-SU (1:3,000 dilution), mouse anti-gp52 hybridoma supernatants (1:5 dilution), or rabbit anti-GST (1:3,000 dilution) and the appropriate secondary antibodies, followed by detection with an ECL kit (Amersham Biosciences, Inc.).
RESULTS
Alignment of MMTV and F-MLV SUs.
There is growing consensus that the basic mechanisms of retroviral membrane fusion are similar, primarily based on structural considerations of the transmembrane (TM) domain (13). Since TM and SU function as a heterodimer and this interaction modulates TM's fusogenic activity, it is likely that the SU structures of different retroviruses are fundamentally similar as well. Based on this concept, we hypothesized that the MMTV SU and RBD would be similar to those of other retroviruses, including the gammaretroviruses. We therefore aligned the protein sequence of MMTV Env with that of the gammaretrovirus ecotropic F-MLV 57, for which the RBD crystal structure has been solved (15).
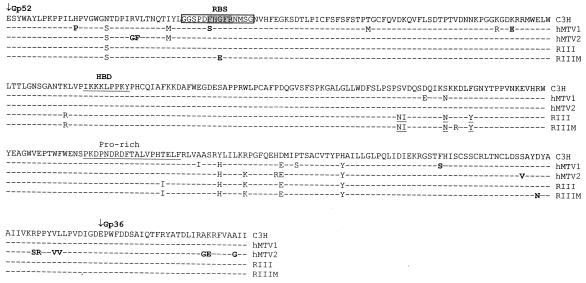
To create this alignment, a putative proline-rich region was identified at residues 230 to 245 of the MMTV SU (Fig. 1). This identification was facilitated by the location of the positively charged residue in position 246 (Arg), followed by hydrophobic amino acids (Leu247-Val248-Ala249-Ala250), a feature conserved in the beginning of the C-terminal domain of the murine gammaretroviruses and bovine leukemia virus SU (16, 24, 25). The sequence amino-terminal to the proline-rich region was designated the putative RBD of MMTV Env by analogy to the known RBD of F-MLV. The final alignment shown in Fig. 1 was used to generate the model shown in Fig. 2B.
FIG. 1.
Structural alignment of MMTV and F-MLV envelope proteins. The β-strands, α-helices, and 310 helical turns identified in the F-MLV crystal structure are underlined and numbered according to reference 15. The variable regions that change with the tropism of the different MLVs (VR regions) are marked by dotted lines above the sequences. The putative MMTV RBS is shaded, and the HBDs in MMTV and F-MLV are boxed. Amino acids in F-MLV and MMTV identified by mutation analysis to be critical to receptor interaction are in bold (1, 10, 30). N-linked glycosylation sites are in italics. An arrowhead marks the Arg codon at position 246 used to define the end of the proline-rich region in the MMTV SU.
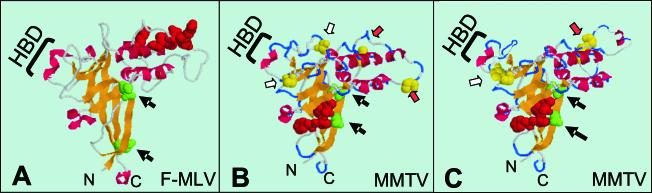
FIG. 2.
Three-dimensional model of MMTV and F-MLV SU proteins. (A) Structure of the F-MLV RBD depicted from the crystal coordinates (Protein Data Base 1AOL) (15). (B) Model of the MMTV RBD generated with SwissModel. (C) Modified model illustrating potential disulfide bonds between cysteines 62 and 73 and cysteines 133 and 156 of MMTV. The five amino acids constituting the putative RBS of MMTV (residues 40 to 44) and the amino acid residues identified by mutation analysis to be involved in the F-MLV SU-receptor interaction (see Fig. 1) are shown as space-filled red atoms. The black arrows indicate the N-linked glycosylation sites in F-MLV and putative sites in MMTV, which are depicted as space-filled green atoms. The cysteine residues with the potential for disulfide bonding are shown as space-filled yellow atoms; the red and white arrows point to cysteine residues with the potential for stabilizing the putative VRA loop and VRC loops with disulfide bonds, respectively. Abbreviations: N, amino terminus; C, carboxyl terminus; HBD, heparin-binding domain. The structures in panels A and B were depicted with RasMol 2.7.1.1 (5, 41). The diagram in panel C was drawn from the RasMol depiction in panel B.
The alignment shows the α-helices and β-strand regions of F-MLV, with the corresponding regions of MMTV that fold into similar structures, according to the SwissModel algorithms. Also shown are the positions of the variable regions of F-MLV (Fig. 1; VRA, VRB, and VRC). In the three-dimensional model, MMTV contains similar regions (compare Fig. 2A to 2B). The VRA, VRB, and VRC regions of the F-MLV RBD are thought to be stabilized by thiol bonds. Although corresponding cysteine residues are not apparent in the linear alignment shown in Fig. 1, two paired cysteine residues with the potential for disulfide bonding lie at the base of VRA and VRC in the three-dimensional model of MMTV (red and white arrows in Fig. 2B and 2C). Similarly, although the putative N-linked glycosylation sites in MMTV do not align with those of F-MLV in the linear alignment, they are found in regions similar to those in F-MLV in the model (black arrows in Fig. 2B).
An HBD has previously been found in both F-MLV (boxed sequence in Fig. 1) and the F-MLV 29 derivative PVC-211 (23). Analysis of the alignment revealed a putative HBD between residues 122 and 130 in MMTV SU (I122KKKLPPKY130) (Fig. 1). The MMTV sequences were similar to previously defined mammalian consensus sequences (XBBXBX and XBBBXXBX, where X is any amino acid and B is a basic amino acid) (21) (Fig. 1 and 3). On both the linear alignment (Fig. 1) and the three-dimensional model, the MMTV HBD mapped to a region corresponding to that of the PVC-211 HBD (compare the regions labeled HBD in Fig. 2A and 2B). The ability to align a putative structure of MMTV SU with that of F-MLV SU and the conservation of elements such as the putative HBD provided us with a model to test for virus-cell and virus-receptor interacting domains.
Heparin binding is important for virus infection.
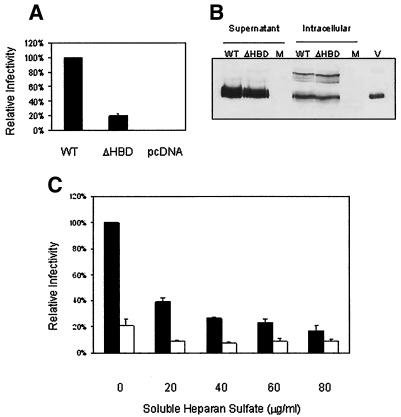
The nearly canonical HBD in the MMTV envelope (I122KKKLPPKY130) was conserved among the different exogenous and endogenous MMTVs (Fig. 3). To determine whether mutations in this putative HBD had an effect on the efficiency of infection, pseudotyped viruses that contained either the wild-type Env (pENVC3H), a mutant envelope containing lysine-to-alanine changes in the HBD (I122AAALPPKY130; ΔHBD), or no envelope (−Env) were prepared and used to infect NMuMG cells. Figure 4A shows that the relative infectivity of the ΔHBD pseudotyped virus was decreased to about 20% of that of the wild-type virus. This result indicated that mutation of these three lysine residues in the HBD resulted in a significant reduction in infectious titer. This loss of infection was not due to a reduction in the level of protein expression or incorporation of Env into particles, since Western blotting of transfected cell lysates and purified pseudovirus gave similar results for the wild-type and ΔHBD pseudoviruses (Fig. 4B).
FIG. 4.
Heparan binding enhances MMTV infection. (A) ΔHBD infection levels. Equal amounts of ΔHBD and wild-type (WT) pEnvC3H pseudovirus were used for infection. The titer for each virus was calculated and is presented as a percentage of wild-type infection levels. The data represent the averages of five independently performed experiments. (B) Virion proteins in MMTV pseudoviruses. Supernatants from equal numbers of 293T cells cotransfected with pENVC3H or ΔHBD, pHIT111, and pHIT60 were pelleted by ultracentrifugation through 30% sucrose. Equal volumes of the resuspended pellets (supernatant) or the transfected cell extracts (intracellular) were subjected to SDS-PAGE followed by Western blot analysis with anti-SU antiserum. The arrow shows the SU protein (gp52); the upper band in the extracts is the unprocessed polyprotein. Abbreviations: M, mock infected; V, purified virus. (C) Heparan sulfate treatment. Cells were treated with heparan sulfate as described in Materials and Methods. Data are presented as a percentage of wild-type infection levels without heparan sulfate and are the averages of three independent experiments. Solid bars, wild-type pseudovirus; open bars, ΔHBD.
To confirm that ΔHBD mutation reduced MMTV infectivity through the loss of interaction with this proteoglycan, we tested whether pseudovirus infection was affected by soluble heparan sulfate. Wild-type and ΔHBD pseudovirus-containing supernatants were incubated in the presence or absence of increasing amounts of soluble heparan sulfate for 1 h at 37°C and then used to infect NMuMG cells. Treatment of wild-type virus with soluble heparan sulfate resulted in a dose-dependent decrease in infectious titer (Fig. 4C). Additionally, the titer of the wild-type virus at saturating levels of heparan sulfate was strikingly similar to the titer of the untreated ΔHBD pseudovirus (Fig. 4C). In contrast, treatment of the ΔHBD pseudovirus with soluble heparan sulfate caused a minor decrease in the infectious titer, but the titer was not decreased further with increasing amounts of heparan sulfate (Fig. 4C). Taken together, these data indicate that binding to heparan sulfate on the cell surface plays a role in MMTV infection.
Identification of altered RBS sequences in MMTVs found in human cells and breast cancer samples.
In addition to aligning the MMTV SU with that of F-MLV, we undertook a second approach to identifying SU sequences important for receptor interaction, based on our previous observation that MLV pseudotypes prepared with the MMTV(C3H) Env did not infect human tissue culture cells, including those engineered to overexpress the human transferrin receptor (12, 40). This observation indicated to us that virus-receptor interaction determined at least one level of species-specific tropism. However, several groups recently reported finding MMTV Env-like sequences in primary human breast cancer tissue samples and cell lines but not normal tissue (14, 29). Moreover, it was previously shown that MMTV could be adapted to replicate in the human BT474 and MCF-7 mammary carcinoma cell lines (22, 26), indicating that there was the potential for zoonotic transmission of MMTV.
If MMTV were able to adapt to using the human TfR1 molecule for entry, this would most likely occur as the result of sequence alterations in the receptor-binding domain of SU. Identification of such altered residues should provide candidate residues constituting the RBS. We therefore compared the SU sequences from several MMTV-like elements (h-MTVs) isolated from primary human breast cancer samples. We also analyzed the SU sequences from MMTV(RIII) virus that was adapted to the human breast cancer cell line-MCF-7 (RIIIM) and compared it to the wild-type MMTV(RIII) and MMTV(C3H) genes. Because the MMTV(RIII) used to infect the MCF-7 cells came from an infected mink lung cell line, we sequenced the su gene of the virus present in the MR/C1 cell line and found that it was identical to the published sequence for this strain (37).
Most of the polymorphisms in the h-MTV and RIIIM sequences were not unique to these viruses but were also found in other strains of MMTV (Fig. 3 and not shown). Moreover, none of the polymorphisms were found in all the human cell-associated viruses. However, two single-nucleotide alterations that resulted in nonconservative amino acid changes were found in a region that could constitute an RBS (Fig. 3) (see next paragraph). One alteration (Phe40 to Ser40) was identified in the su sequence from one human breast cancer sample (h-MTV1) but not the other (h-MTV2). The second polymorphism (Gly42 to Glu42) was found in the su sequence from the MMTV(RIII)/MCF7-adapted virus (RIIIM) but not the parental virus. Only one other polymorphism was found in the RIIIM virus, close to the C terminus of SU (a semiconservative Asp-to-Asn change).
Phe40 and Gly42 are in a five-amino-acid stretch of polar and hydrophobic residues directly adjacent to a glycosylation site, features often found at sites of protein-protein interaction in soluble proteins. When the Ser40 and Glu42 polymorphisms were placed on the three-dimensional model, they mapped to the outer surface of the molecule and formed a concave surface, consistent with their proposed role in receptor interaction (space-filled red atoms in Fig. 2B). Thus, it was possible that these represented alterations in a putative RBS that extended SU tropism for the human TfR1.
Amino acid residue 40 is critical for virus infectivity on mouse cells.
Because both the Ser40 and Glu42 polymorphisms mapped to the putative RBS region, we introduced them into the wild-type MMTV(C3H) Env construct used for producing pseudoviruses, pENVC3H. We also changed Phe40 to Ala40 (nonconservative) and Tyr40 (conservative) in pENVC3H to determine whether this residue functioned in receptor interaction. The wild-type and variant Envs were used to make pseudoviruses, and the viruses were tested for the ability to infect mouse NMuMG cells.
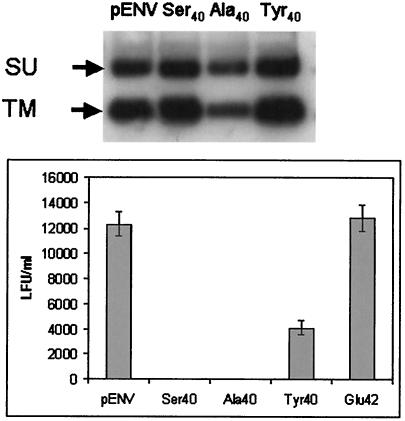
We first demonstrated that the mutant Envs were efficiently processed and incorporated into virions. Total cell extracts from wild-type- and mutant pENVC3H-transfected 293T cells (not shown) and pseudoviruses pelleted by ultracentrifugation (Fig. 5, top panel) were subjected to Western blot analysis with anti-MMTV antiserum to determine the level of viral proteins and their stable association in virions. The mutant envelopes were processed into mature SU and TM and stably integrated into virions to the same extent as wild-type Env, indicating that no gross changes in these processes had occurred as a result of the sequence alterations.
FIG. 5.
Mutation of a single amino acid in the RBD abolishes infectivity. (Top panel) Virion proteins in MMTV pseudoviruses. Supernatants from equal numbers of 293T cells cotransfected with pENVC3H or the mutant pENVs (Ser40, Ala40, and Tyr40), pHIT111, and pHIT60 (MLV gag and genome) were pelleted by ultracentrifugation. Then 50 μl of the resuspended pellets was subjected to SDS-PAGE followed by Western blotting analysis with anti-MMTV antiserum. The arrows point to SU (gp52) and TM (gp36). (Bottom panel) Pseudovirus infection of NMuMG cells. Triplicate infections were performed with equal amounts of supernatant. The data are presented as the titer and the standard deviation for each infection.
The pseudoviruses were next tested for their ability to infect NMuMG cells. The Ser40 and Ala40 mutations completely abolished infectivity, whereas the Glu42 mutation did not have a significant effect on infection; the Tyr40 mutation caused a modest decrease in infection levels, consistent with the conservative nature of this change (Fig. 5, bottom panel). These data indicated that amino acid Phe40 was critical for virus infectivity in mouse cells and indicated that this region might indeed constitute part of the RBS.
Virus binding to mouse cells is dependent on amino acid Phe40.
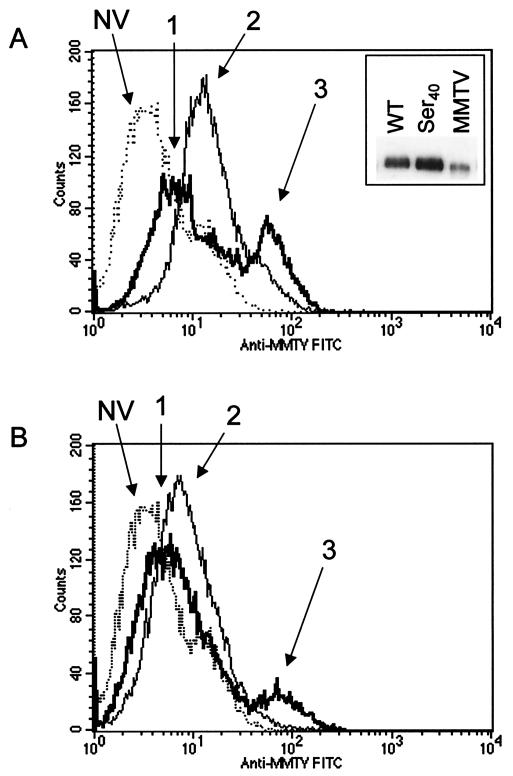
The loss of infectivity that accompanied the nonconservative Ser40 and Ala40 mutations could be due to abolition of Env binding to its receptor or to alterations that affected subsequent steps in the infection pathway, such as membrane fusion. To differentiate between these two possibilities, we performed virus-binding assays. NMuMG cells were incubated with equal amounts of wild-type or Ser40 pseudovirions, stained with anti-MMTV antibodies, and then analyzed by FACS to detect bound virus. There was a population of cells that bound high levels of wild-type pseudovirions (mean channel fluorescence [MCF] = 66.1; thick line, arrow 3 in Fig. 6A), as well as a population that bound fewer virions (MCF = 9.8; arrow 1). In contrast, although there was a low virus-binding population of cells with the Ser40 mutant pseudovirus (MCF = 18.7; thin line, arrow 2), there were no cells that bound the larger amounts of virus. Cells stained with anti-MMTV antibodies in the absence of virus had an MCF of 6.2 (dotted line, arrow NV).
FIG. 6.
Phe residue at position 40 is required for high-level virus binding to NMuMG cells. Equal amounts of MMTV pseudotypes prepared with either the wild-type (WT) envelope or envelope containing the Ser40 mutation (inset, panel A) were incubated on ice with NMuMG cells. The cells were stained with anti-MMTV antiserum, followed by FITC-labeled secondary antibodies, and subjected to FACS analysis. Dead cells were excluded by their forward scatter/side scatter properties. The experiment was performed three to four times with similar results; a representative experiment is shown. (A) Histograms showing the fluorescence of cells incubated with wild-type (thick line) or Ser40 mutant virus (thin line) or without virus (dotted line). The inset shows a Western blot of 10 μl of each concentrated virus preparation or milk-borne MMTV (MMTV) as a positive control, with polyclonal anti-MMTV SU antiserum used for detection. NV, no virus. (B) Histograms showing the fluorescence of cells incubated with wild-type or mutant virus in cells pretreated with 100 μg of heparan sulfate per ml.
We next tested whether either of the peaks (low and high virus binding) was the result of proteoglycan interactions with the HBD in the MMTV Env. The same virus-binding assays were performed in the presence of 100 μg of heparan sulfate per ml. In the presence of heparan, the high-level virus-binding peak was still seen with the wild-type pseudovirus (MCF = 78.9), although the number of cells in this population was diminished by about 2.5-fold (Fig. 6B). However, heparan sulfate reduced the fluorescence intensity of the low-virus-binding peaks to an intensity similar to that seen with cells that were not incubated with virus for both the wild-type (MCF = 7.6) and Ser40 (MCF = 10.3) viruses. These results indicated that the low-level binding of virus to cells was due at least in part to proteoglycan interactions but that high-level binding was due to other interactions, possibly through a receptor.
Virus downregulation of surface TfR1.
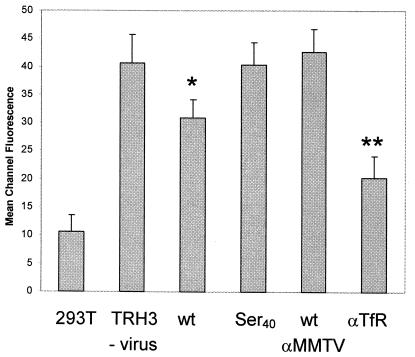
To determine whether the wild-type or Ser40 mutant virus interacted with receptors on cells, we tested whether they blocked surface expression of TfR1. We showed previously that binding of wild-type pseudovirus to cells partially blocked binding of an anti-TfR1 monoclonal antibody (40). TRH3 cells (a clonal isolate of 293T cells stably expressing mouse TfR1) were incubated with wild-type or Ser40 pseudovirions for 2 h at room temperature, shifted to 4°C, and washed with buffer containing sodium azide to prevent further internalization. The cells were then examined by FACS analysis for surface mTfR1. As controls, wild-type virus was treated with goat anti-MMTV antiserum and TRH3 cells were incubated with the rat C2 monoclonal antibody, known to downregulate surface expression of the receptor.
There was a significant decrease in TfR1 expression on TRH3 cells incubated with wild-type virus compared with cells incubated with the Ser40 mutant (compare the MCF of TRH3 cells with those for no virus, labeled TRH3 versus TRH3 cells incubated with virus, labeled wt in Fig. 7). Incubation of the wild-type virus with anti-MMTV antiserum blocked this effect, whereas the C2 antibody caused greater downregulation of the receptor on the surface than did virus, as we showed previously (40). These data indicate that the Ser40 mutant was unable to bind to the MMTV entry receptor TfR1, consistent with its constituting part of the RBS.
FIG. 7.
Wild-type but not mutant MMTV blocks surface expression of the transferrin receptor. Cells were incubated with virus by spinoculation and then stained with FITC-labeled anti-mouse TfR antibodies. Each experiment was performed four to five times; shown is the MCF value ± standard deviation. Abbreviations: 293T, untransfected 293T cells incubated with MMTV(C3H) pseudotypes; TRH3/no virus, clonal isolate of 293T stably transfected with mTfR1; wt, TRH3 cells incubated with MMTV(C3H)-pseudotyped virus; Ser40, TRH3 cells incubated with the Ser40 mutant; wt/αMMTV, TRH3 cells incubated with MMTV(C3H) pretreated with goat anti-MMTV polyclonal antiserum; αTfR, TRH3 cells incubated with anti-mouse TfR monoclonal antibody and MMTV(C3H). P values were calculated with Student's t test. *, P ≤ 0.05; **, P ≤ 0.005 compared to TRH3 cells not bound to virus.
Monoclonal antibodies that block infection recognize the RBS.
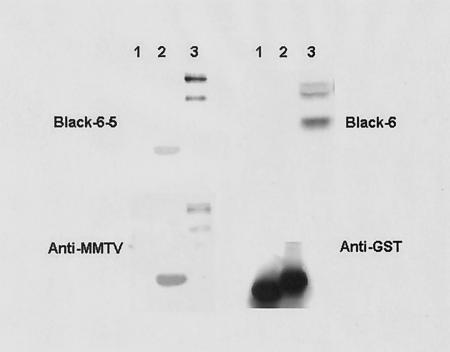
These data suggested that residue Phe40 was indispensable for receptor binding. We tested a recently developed panel of anti-Env monoclonal antibodies for their ability to block pseudovirus infection and found two, Black 6-5D and Black 8-6, that efficiently blocked infection (Table 1). In contrast, two other monoclonal antibodies, Black 6 and 2F10, were unable to block infection, although they both recognized the MMTV Env by enzyme immunoassay and Western blot analysis (38) (Fig. 8).
TABLE 1.
Anti-SU monoclonal antibodies that block infectiona
| Antibody | Avg % of control ± SD |
|---|---|
| None | 100 |
| Black 6 | 113.6 ± 26.5 |
| 2F10 | 118.0 ± 41 |
| Black 6-5D | 7.4 ± 5.7 |
| Black 8-6 | 2.4 (2.8, 2.0) |
| Goat anti-MMTV | 0 |
A 1:5 dilution of each hybridoma supernatant or 1 μl of polyclonal goat anti-MMTV antiserum was incubated with MMTV pseudotypes for 10 min at room temperature prior to addition to NMuMG cells. All of the experiments were performed three to four times except for the Black 8-6 blocking, which was performed twice (values shown in parentheses). Data are presented as the average percentage of colonies obtained with antibody compared to the number obtained with untreated virus ± standard deviation.
FIG. 8.
Blocking monoclonal antibodies bind to the RBS. Equal amounts of GST alone (lane 1), the RBS-GST fusion protein (lane 2), or extract from pEnvC3H-transfected 293T cells (lane 3) were subjected to SDS-PAGE followed by Western blot analysis with monoclonal Black 6, Black 6-5D, polyclonal anti-MMTV SU, or anti-GST antibody.
The monoclonal antibodies could block infection by direct interaction with the RBS, by steric hindrance, or by changing the conformation of SU so that it was unable to contact the receptor. To determine if the blocking monoclonal antibodies were interacting with the RBS, a GST-RBS fusion protein was made (residues 35 to 48; Fig. 3), and the monoclonal antibodies were used in Western blot analysis. Both the Black 6-5D (Fig. 8) and Black 8-6 (not shown) neutralizing monoclonal antibodies specifically recognized the RBS-GST fusion protein but not GST alone; the nonneutralizing Black 6 (Fig. 8) and 2F10 (not shown) monoclonal antibodies did not bind the fusion protein. Taken together with the finding that residue Phe40 is indispensable for both infectivity and receptor binding, these data suggest that this region of the MMTV SU contains the RBS.
MMTV pseudotypes do not infect human tissue culture cells.
We next tested whether changing Phe40 to Ser40 or Gly42 to Glu42 in the MMTV(C3H) Env protein changed the tropism of pseudoviruses from mouse to human cells. Either there was no infection of human cells by MMTV(C3H) pseudovirus (MCF-7), or the level of infection was two to three logs lower than it was on mouse cells (293T and T47D), and none of the genetically modified variants showed titers on any human cell (Table 2). We also tested whether the MCF7/vp5 cell line that was infected with the RIIIM virus had undergone genetic changes that rendered it susceptible to infection with MMTV pseudotypes. MMTV-infected cells are not subject to superinfection interference and thus can be infected with MMTV pseudotypes (12). Again, none of the viruses were able to efficiently infect this cell line (Table 2).
TABLE 2.
Infection with wild-type and variant MMTV pseudovirusesa
| Pseudovirus | Avg LFU/ml ± SD
|
||||
|---|---|---|---|---|---|
| MCF-7 | MCF-7/vp5 | T47D | 293T | NMuMG (10−4) | |
| pENVC3H | 0 | 0 | 17.3 ± 5.5 | 49.9 ± 49.0 | 1.56 ± 0.11 |
| pENVC3H + Black 6-5D | ND | ND | 4.0 ± 6.9 | 106.7 ± 92.4 | 0.19 ± 0.05 |
| Tyr40 | 0 | 0 | 0 | 0 | 0.41 ± 0.06 |
| Ser40 | 0 | 0 | 0 | 0 | 0 |
| Ala40 | 0 | 0 | 0 | 0 | 0 |
| Glu42 | 0 | 0 | 0 | 0 | 1.28 ± 0.11 |
| pENVRIII | 0 | 0 | 34.0 ± 19.9 | 120.7 ± 108.0 | 0.43 ± 0.06 |
| pENVRIII + Black 6-5D | ND | ND | 60.0 ± 36.0 | 210.7 ± 164.8 | 0.06 ± 0.03 |
| pENVRIIIM | 2, 0 | 12, 0 | 60.8 ± 24.2 | 142.3 ± 173.4 | 1.04 ± 0.13 |
| pENVRIIIM + Black 6-5D | ND | ND | 64 ± 30.2 | ND | 0.16 ± 0.05 |
| No Env | ND | ND | 13.75 ± 4.9 | 34.17 ± 22.7 | 0 |
Data are presented as the averages for supernatant from four to fifteen experiments ± standard deviation with the exception of the RIIIM infection of MCF-7 and MCF-7/vp5 cells, which was performed twice. All pseudoviruses were checked by Western blot analysis (i.e., see Fig. 3) to ensure that equal amounts of viral proteins were produced. ND, not determined.
The MMTV(C3H) and MMTV(RIII) Env sequences showed additional polymorphic differences, although all are found in other strains of exogenous or endogenous MMTVs (Fig. 3 and not shown). To determine whether these other differences contributed to Env function or tropism, the MMTV(RIII) and MMTV(RIIIM) env genes were cloned into the same mammalian expression vector as pENVC3H and used to infect NMuMG, 293T, MCF-7, MCF-7/vp5, and T47D cells. Pseudoviruses bearing either EnvRIII or EnvRIIIM efficiently infected NMuMG and TRH3 cells but not the other cell lines (Table 2). There was low-level infection of both 293T and T47D human cells (around two orders of magnitude less than was seen with cells expressing the mouse TfR1) with both EnvRIII- and EnvRIIIM- as well as ENVC3H-bearing viruses, but this infection occurred in the presence of blocking monoclonal antibody Black 6-5D (Table 2) or polyclonal anti-MMTV antibodies (not shown). Additionally, infection of human but not mouse cells was independent of specific attachment to the receptor, since it occurred with virions lacking Env (Table 2). In contrast, Env-negative virus showed no infection, and the anti-MMTV blocking antibody caused an 80 to 90% reduction in infection of mouse cells with all the Env-bearing pseudoviruses.
DISCUSSION
Retroviruses exhibit a high degree of host range and cell type specificity. This specificity is partly determined by the presence of cell surface entry receptors on target cells. A number of retroviral receptors have now been cloned, and from the molecules that have been identified as receptors, it is clear that retroviruses take advantage of host-encoded cell surface proteins that play important roles in cellular function and metabolism (33). MMTV's use of TrfR1 for cell entry suggests that it interacts with its receptor differently than do other retroviruses with their receptors. TfR1 is a type II transmembrane protein with a single membrane-spanning domain and a very short cytoplasmic tail (36) and thus is structurally very different from the ecotropic, xenotropic, and amphotropic MLV receptors, which have multiple (10 to 14) membrane-spanning domains (33). Unlike most other retroviral receptors, TfR1 requires endocytosis and trafficking to the acidic endosome to fulfill its metabolic function, that is, delivery of iron to the cell.
Although MMTV uses a different type of entry receptor, our model predicts that many of the structural elements found in F-MLV are likely to occur in MMTV. We found a similar arrangement of β-sheets and α-helices in MMTV as well as segments corresponding to the regions of F-MLV that are referred to as VRA, VRB, and VRC. Additionally, while the putative N-linked glycosylation sites in MMTV do not appear to correspond in the linear alignment (Fig. 1), they are found in similar physical locations on the model (Fig. 2). Finally, there are six thiol pairs essential to folding of VRA (C46 to C98, C72 to C83, and C73 to C87), VRB (C178 to C184), and VRC (C121 to C141 and C133 to C146) in the F-MLV RBD. In contrast, there are only two potential thiol pairs in MMTV. Again, although the two potential thiol pairs (C62 to C73 [white arrows in Fig. 2B and 2C] and C133 to C156 [red arrows]) do not directly align with those in F-MLV, the model predicts that they occur in positions that have the potential to stabilize the putative VRA and VRC regions of the MMTV RBD (Fig. 2C). Interestingly, only one of the VRA disulfide bonds (C46 to C98) is found in all gammaretroviruses (the other two are found only in ecotropic MLVs), and only ecotropic MLV, amphotropic MLV, gibbon-ape leukemia virus, and feline leukemia virus but not xenotropic MLV have the VRB-stabilizing pair.
A number of approaches have shown that VRA contains amino acids critical for interaction of different MLV Envs with their cognate entry receptors. These include site-directed mutagenesis and comparison of the primary Env sequences of highly related members of the MLV family that use different entry receptors (1-3, 10, 30, 34). We used similar approaches to identify critical amino acids in the MMTV Env protein. Although the alignment predicts that there is a segment in the MMTV RBD corresponding to the VRA loop in F-MLV, our functional analysis showed that the site involved in receptor interaction maps not to VRA but to a different part of the SU protein, predicted to be on the face of the molecule (Fig. 2). This five-amino-acid stretch consisting of polar and hydrophobic residues and flanked by an N-linked glycosylation site appears to form a concave structure on the surface of Env that could serve as a binding site for the receptor.
MMTV may use a different region of its RBD for receptor interaction because it utilizes TfR1 for cell entry. Binding of the MLV RBD to the receptor is thought to trigger the conformational change that results in exposure of the fusion peptide and initiation of virus-cell membrane fusion at the cell surface. In contrast, because MMTV uses a receptor that traffics to the acidic endosome and is dependent on acid pH for infection, virus-cell membrane fusion may occur in this compartment. Thus, by using a different region of the RBD for receptor contact than is used for MLV, binding of MMTV to TfR1 may not induce the conformational changes in Env that are required for membrane fusion events, thereby allowing trafficking to the compartment and infection to proceed there. In support of this concept, the sequences in the MMTV RBS correspond to a strand of the β-barrel in F-MLV that is thought to be the rigid core structure of RBD.
The data presented here support the notion that the Phe40His41Gly42Phe43Arg44 peptide constitutes the RBS and that the Phe40 residue is critical to the interaction with the receptor. First, nonconservative changes in the Phe40 residue completely abolished infectivity but not Env levels on virions. Second, pseudovirions bearing the Ser40 mutation showed only low-level binding to cells, and this binding was inhibited by heparan sulfate. In contrast, there was a subset of cells that exhibited about 10-fold-higher binding of wild-type pseudovirus that was not completely inhibited by heparan. Although not all cells showed this high level of binding, this is probably due to variable levels of TfR1 on the surface of NMuMG cells; TfR1 is known to be expressed at a low level or not at all on nondividing cells and at a high level on activity dividing cells (36). Moreover, unlike wild-type pseudovirions, Ser40 mutant virions did not cause loss of antibody-recognizable TfR1 on the cell surface. Finally, we identified two blocking monoclonal antibodies that recognized a peptide containing the RBS. Although there are alternative explanations for all of these findings, including second-site conformational changes by the introduction of nonconservative mutations, the data presented here are consistent with the hypothesis that amino acids 40 to 44 are part of the RBS.
The results presented here also indicate that heparan binding plays a role in the interaction of MMTV with cells. Heparan sulfate is a proteoglycan found on the cell surface and is involved in cell-cell and cell-protein interactions (21). These proteoglycans are typically highly negatively charged and hence have a propensity to interact with sequences rich in positively charged residues. Many viruses, including human respiratory syncytial virus (19), human parainfluenza virus type 3 (8), herpes simplex virus (42), human papillomavirus (17), human herpesvirus 8 (6), and adenovirus (11), have been shown to bind cells via proteoglycans. In the case of human immunodeficiency virus (HIV), heparan sulfate on cell surface syndecan has been shown to be important for Env-mediated binding to cells (7).
Our data demonstrate that the MMTV HBD is also necessary for efficient infection, and because infection by wild-type virus was inhibited by soluble heparan sulfate, this suggests that proteoglycans play a role in the MMTV-cell interaction. Since low-level infection did occur with both the ΔHBD pseudovirus and wild-type virus in the presence of soluble heparan, cell surface heparan sulfate is not absolutely required but may enhance infection, perhaps by attracting or concentrating virus particles to the cell surface to increase the chance of receptor attachment. Indeed, our data show that the Ser40 mutant virus, which does not infect cells, still shows low-level binding that could be blocked by heparan sulfate. In contrast, the high-level binding seen with wild-type virus was still detectable in the presence of this proteoglycan. Heparan sulfate at 100 μg/ml did reduce the number of cells that showed high-level binding, albeit not as efficiently as it inhibited infection. This is probably due to the different amounts of virus used in these experiments (approximately 335 times more virus was used in the binding experiment than in the infection assay). Interestingly, more heparan-blocked, low-level binding occurred with the Ser40 mutant than with the wild-type virus. This could be due to conformational changes in the mutant SU that make the HBS more accessible.
Changes in the dependence of a virus on heparan sulfate can alter its pathogenesis. For example, Ruscetti and colleagues demonstrated that the increase in neuropathogenesis of PVC-211 is the result of increased heparin binding (23). In contrast, mutations in the E protein of the flavivirus Murray Valley encephalitis virus that increase affinity for cell surface glycosamines result in the loss of neuroinvasiveness (27). Similarly, mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus that result in heparan sulfate-dependent virus entry in cultured cells lead to viral attenuation and rapid clearance in vivo (4). Finally, HIV binding to heparan sulfate on syndecan has been proposed to play a role in vivo in the stability of virus bound to endothelia and in the transfer of virus from nonpermissive to permissive cells (7). Whether proteoglycan interaction plays a role in in vivo infection awaits the introduction of the ΔHBD mutation into infectious MMTV.
The mouse and human TfR1 amino acid sequences show 76% identity and 86% homology. Using our MMTV Env-pseudotyped MLV, we were unable to demonstrate efficient infection of human cells (12, 18) even when cells overexpressed the human TfR1 (40), indicating that the human version of this molecule does not function as a receptor. Although pseudoviruses prepared in 293T cells showed low-level infection of some human cells, this infection was not inhibited by blocking anti-SU antibodies, nor was it dependent on Env on virions.
Although it was reported that continuous passage of MMTV on human breast cancer cell lines resulted in adapted viruses that could infect human cells (22, 26, 45) and that MMTV-like sequences are found in human breast cancer but not normal tissue (14, 29, 35, 46), we show here that sporadic changes found in the RBS of some “human-adapted” MMTVs did not after the species tropism. Although we found low levels of MMTV in the MCF-7/vp5 cell line by PCR, this virus does not seem to have spread in the population because viral sequences were not detected by Southern blot analysis of DNA isolated from these cells (P. Popken-Harris, personal communication). Thus, it does not appear that MMTV can adapt to using the human TfR1 for entry, and if it does infect human cells, it probably does so through a non-receptor-mediated pathway.
Acknowledgments
The first two authors contributed equally to this work.
We thank Tatyana Golovkina for providing the anti-Env monoclonal antibodies, Beatriz Pogo for sharing sequence data, and Akhil Vaidya for the MMTV(RIII)-infected MCF-7 and MR/C1 cell lines.
This work was supported by PHS grant R01 CA73746 to S.R.R. and PHS grant R01 AI43310 to L.M.A. J.C.R. was supported by NRSA grant F32 CA90037.
REFERENCES
- 1.Bae, Y., S. M. Kingsman, and A. J. Kingsman. 1997. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J. Virol. 71:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battini, J. L., O. Danos, and J. M. Heard. 1995. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J. Virol. 69:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini, J. L., J. M. Heard, and O. Danos. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J. Virol. 66:1468-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard, K. A., W. B. Klimstra, and R. E. Johnston. 2000. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 276:93-103. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, H. J. 2000. Recent changes to RasMol, recombining the variants. Trends Biochem. Sci. 25:453-455. [DOI] [PubMed] [Google Scholar]
- 6.Birkmann, A., K. Mahr, A. Ensser, S. Yaguboglu, F. Titgemeyer, B. Fleckenstein, and F. Neipel. 2001. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8. J. Virol. 75:11583-11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobardt, M. D., A. C. S. Saphire, H.-C. Hung, X. Yu, B. Van der Schueren, Z. Zhang, G. David, and P. A. Gallay. 2003. Syndecan captures, protects and transmits HIV to T lymphocytes. Immunity 18:27-39. [DOI] [PubMed] [Google Scholar]
- 8.Bose, S., and A. K. Banerjee. 2002. Role of heparan sulfate in human parainfluenza virus type 3 infection. Virology 298:73-83. [DOI] [PubMed] [Google Scholar]
- 9.Davey, R. A., C. A. Hamson, J. J. Healey, and J. M. Cunningham. 1997. In vitro binding of purified murine ecotropic retrovirus envelope surface protein to its receptor, MCAT-1. J. Virol. 71:8096-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davey, R. A., Y. Zuo, and J. M. Cunningham. 1999. Identification of a receptor-binding pocket on the envelope protein of friend murine leukemia virus. J. Virol. 73:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dechecchi, M. C., P. Melotti, A. Bonizzato, M. Santacatterina, M. Chilosi, and G. Cabrini. 2001. Heparan sulfate glycosaminoglycans are receptors sufficient to mediate the initial binding of adenovirus types 2 and 5. J. Virol. 8772-8780. [DOI] [PMC free article] [PubMed]
- 12.Dzuris, J. L., W. Zhu, T. V. Golovkina, and S. R. Ross. 1999. Lack of receptor interference by endogenous expression of the mouse mammary tumor virus envelope protein. Virology 263:418-426. [DOI] [PubMed] [Google Scholar]
- 13.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 14.Etkind, P., J. Du, A. Khan, J. Pillitteri, and P. H. Wiernik. 2000. Mouse mammary tumor virus-like ENV gene sequences in human breast tumors and in a lymphoma of a breast cancer patient. Clin. Cancer Res. 6:1273-1278. [PubMed] [Google Scholar]
- 15.Fass, D., R. A. Davey, C. A. Hamson, P. S. Kim, J. M. Cunningham, and J. M. Berger. 1997. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science 277:1662-1666. [DOI] [PubMed] [Google Scholar]
- 16.Gatot, J.-S., I. Callebaut, C. Van Lint, D. Demonte, P. Kerkhofs, D. Portetelle, A. Burny, L. Willems, and R. Kettmann. 2002. Bovine leukemia virus SU protein interacts with zinc, and mutations within two interacting regions differently affect viral fusion and infectivity in vivo. J. Virol. 76:7956-7967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giroglou T., L. Florin, F. Schafer, R. E. Streeck, and M. Sapp. Human papillomavirus infection requires cell surface heparan sulfate. J. Virol. 75:1565-1570. [DOI] [PMC free article] [PubMed]
- 18.Golovkina, T. V., J. L. Dzuris, B. van den Hoogen, A. B. Jaffe, P. C. Wright, S. M. Cofer, and S. R. Ross. 1998. A novel membrane protein is a mouse mammary tumor virus receptor. J. Virol. 72:3066-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallak, L. K., D. Spillmann, P. L. Collins, and M. E. Peeples. 2000. Glycosaminoglycan sulfaction requirements for respiratory syncytial virus infection. J. Virol. 74:10508-10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 21.Hileman, R. E., J. R. Fromm, J. M. Weiler, and R. J. Linhardt. 1998. Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. BioEssays 20:156-167 [DOI] [PubMed] [Google Scholar]
- 22.Howard, D. K., and J. Schlom. 1980. Isolation of a series of novel variants of murine mammary tumor viruses with broadened host range. Int. J. Cancer 25:647-654. [DOI] [PubMed] [Google Scholar]
- 23.Jinno-Oue, A., M. Oue, and S. K. Ruscetti. 2001. A unique heparin-binding domain in the envelope protein of the neuropathogenic PVC-211 murine leukemia virus may contribute to its brain capillary endothelial cell tropism. J. Virol. 75:12439-12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston, E. R., L. M. Albritton, and K. Radke. 2002. Envelope proteins containing single amino acid substitutions support a structural model of the receptor-binding domain of bovine leukemia virus surface protein. J. Virol. 76:10861-10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, F. J., I. Seiliez, C. Denesvrei, D. Lavillette, F.-L. Cosset, and M. Sitbon. 2000. Definition of an amino-terminal domain of the human T-cell leukemia virus type 1 envelope surface unit that extends the fusogenic range of an ecotropic murine leukemia virus. J. Biol. Chem. 275:23417-23420. [DOI] [PubMed] [Google Scholar]
- 26.Lasfargues, E. Y., W. G. Coutinho, and A. S. Dion. 1979. A human breast tumor cell line (BT474) that supports mammary tumor virus replication. In Vitro 15:723-728. [DOI] [PubMed] [Google Scholar]
- 27.Lee, E., and M. Lobigs. 2002. Mechanism of virulence attenuation of glycosaminoglycan-binding variants of Japanese encephalitis virus and Murray Valley encephalitis virus. J. Virol. 76:4901-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, J. H., J. A. Engler, J. F. Collawn, and B. A. Moore. 2001. Receptor mediated uptake of peptides that bind the human transferrin receptor. Eur. J. Biochem. 268:2004-2012. [DOI] [PubMed] [Google Scholar]
- 29.Liu, B., Y. Wang, S. M. Melana, I. Pelisson, V. Najfeld, J. F. Holland, and B. G. Pogo. 2001. Identification of a proviral structure in human breast cancer. Cancer Res. 61:1754-1759. [PubMed] [Google Scholar]
- 30.MacKrell, A. J., N. W. Soong, C. M. Curtis, and W. F. Anderson. 1996. Identification of a subdomain in the moloney murine leukemia virus envelope protein involved in receptor binding. J. Virol. 70:1768-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679-689. [DOI] [PubMed] [Google Scholar]
- 32.Nandi, S., and C. M. McGrath. 1973. Mammary neoplasia in mice. Adv. Cancer Res 17:353-414. [Google Scholar]
- 33.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65:371-389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, B. H., B. Matuschke, E. Lavi, and G. N. Gaulton. 1994. A point mutation in the env gene of a murine leukemia virus induces syncytium formation and neurologic disease. J. Virol. 68:7516-7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pogo, B. G. T., S. M. Melana, J. F. Holland, J. F. Mandeli, S. Polotti, P. Casalini, and S. Menard. 1999. Sequences homologous to the mouse mammary tumor virus env gene in human breast cancer correlate with overexpression of laminin receptor. Clin. Cancer Res. 5:2108-2111. [PubMed] [Google Scholar]
- 36.Ponka, P., and C. N. Lok. 1999. The transferrin receptor: role in health and disease. Int. J. Biochem. Cell Biol. 31:1111-1137. [DOI] [PubMed] [Google Scholar]
- 37.Popken-Harris, P., L. Pliml, and L. Harris. 2001. Sequence and genetic analyses of the 3′ terminus and integration sites of the RIII/Sa mouse mammary tumor (MMTV) exogenous provirus. Virus Genes 23:35-43. [DOI] [PubMed] [Google Scholar]
- 38.Purdy, A., L. Case, M. Duvall, M. Overstrom-Coleman, N. Monnier, A. Chervonsky, and T. Golovkina. 2003. Unique resistance of I/LnJ mice to a retrovirus is due to sustained IFN-gamma dependent production of virus-neutralizing antibodies. J. Exp. Med. 197:233-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Redmond, S., G. Peters, and C. Dickson. 1984. Mouse mammary tumor virus can mediate cell fusion at reduced pH. Virology 133:393-402. [DOI] [PubMed] [Google Scholar]
- 40.Ross, S. R., J. J. Schofield, C. J. Farr, and M. Bucan. 2002. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc. Natl. Acad Sci. USA 99:12386-12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomeolecular graphics for all. Trends Biochem. Sci. 20:374-376. [DOI] [PubMed] [Google Scholar]
- 42.Shukla, D., and P. G. Spear. 2002. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Investig. 108:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 44.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaidya, A. B., E. Y. Lasfargues, G. Heubel, J. C. Lasfargues, and D. H. Moore. 1976. Murine mammary tumor virus: characterization of infection of nonmurine cells. J. Virol. 18:911-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, Y., J. F. Holland, I. R. Bleiweiss, S. Melana, X. Liu, I. Pelisson, A. Cantarella, K. Stellrecht, S. Mani, and B. G. T. Pogo. 1995. Detection of mammary tumor virus ENV gene-like sequences in human breast cancer. Cancer Res. 35:5173-5179. [PubMed] [Google Scholar]