Abstract
Cell-type-specific transcription of the JC virus (JCV) promoter in glial cells initiates a series of events leading to viral replication in the brain and the development of the fatal demyelinating disease progressive multifocal leukoencephalopathy (PML) in patients with neurologic complications due to infection with human immunodeficiency virus type 1. Here we employed an in vitro infection of primary cultures of human astrocytes to compare the transcriptional profile of cellular genes after JCV infection by using an oligonucleotide-based microarray of 12,600 genes. Transcription of nearly 355 genes was enhanced and expression of 130 genes was decreased to various degrees. Many transcripts that were increased upon JCV infection were found to encode proteins with properties that suggest their involvement in cell proliferation, including cyclin A and cyclin B1; signaling pathways, such as transforming growth factor β receptor 1, platelet-derived growth factor receptor and fibroblast growth factor family receptor; and other regulatory events, such as inflammatory responses, including cyclo-oxygenase-2 (Cox-2). Microarray-based data for several cell cycle-regulatory genes were further examined by using Western blot analysis of in vitro infected astrocytes harvested early and late during the infection. Results demonstrate that protein levels of all upregulated genes were found to increase at some point during the infection time course. In parallel, immunohistochemical assessment of cell cycle proteins, including cyclins A, B1, E, and Cdk2, showed positive staining of astrocytes within PML lesions of brain tissue from patients with neuro-AIDS. Microarray analysis was found to be a useful predictor of gene expression in infected cells; however, it may not directly correlate with protein levels during infection with JCV.
The human polyomavirus, JC (JCV), is the etiologic agent for the fatal demyelinating disease of the central nervous system, progressive multifocal leukoencephalopathy (PML) (for review see references 5 and 11). Once a rare disease seen primarily in patients with impaired immune systems due to myeloproliferative and lymphoproliferative disorders, the incidence of PML has been significantly increased due to the AIDS epidemic (27). Clinical studies show that >4% of AIDS patients with neurologic problems are diagnosed with PML (1, 2, 6, 7, 10, 18, 19, 20, 26, 31, 32). JCV infects >65% of the human population by early childhood with no major clinical symptoms (16, 23, 28, 30). Reactivation of the viral genome that preferentially occurs in oligodendrocytes and astrocytes leads to cytolytic destruction of the myelin-producing oligodendrocytes and degeneration of the myelin sheath in white matter. The chief pathological features of PML are the presence of altered oligodendrocytes whose nuclei are two to three times the size of normal cells and are filled with virions and viral antigens (34-36); giant, bizarre astrocytes with pleomorphic, hyperchromatic nuclei; and occasional mitotic figures. Early studies revealed that expression of the viral genome is controlled by transcription factors that are enriched and/or active in oligodendrocytes and astrocytes, permitting production of the viral early protein, i.e., large T antigen and its various isoforms (15). Once T antigen is expressed, the virus enters into a lytic cycle that consists of active viral DNA replication and late gene transcription coding for the capsid proteins, VP1, VP2, and VP3, and the auxiliary Agnoprotein in oligodendrocytes. Astrocytic cells are semipermissive for JCV infection in that they allow for expression of T antigen and late gene expression to a limited extent but are not the major source of virion production in vivo (5, 13). However, several studies have demonstrated the ability of JCV to infect many cell types in the central nervous system (CNS) in addition to oligodendrocytes, including neurons, astrocytes, and macrophages (8, 26).
Studies that use human cell lines and animal models have provided important information on the various functions associated with the viral early protein, T antigen. For example, T antigen has the ability to associate with several cellular proteins that are implicated in the control of the cell cycle and proliferation. The association of T antigen with p53 and pRb is believed to be involved, at least in part, in the transforming ability of JCV in cell culture and its tumorigenicity in experimental animals (9, 16, 17, 24, 29). Accordingly, JCV T antigen has been reported in several human brain tumors, particularly medulloblastoma, leading to speculation about JCV involvement in CNS neoplasias. Furthermore, the JCV Agnoprotein may deregulate cell growth by altering the expression of several cell cycle-associated proteins (17). To gain more comprehensive information of the cellular events that are affected during the course of JCV infection, we employed microarray technology and compared the level of expression of cellular genes in JCV-infected and uninfected human primary astrocytes. Several of the genes whose expression was affected by JCV infection are involved in cell cycle progression and receptors for several growth factors and are classified as transcription factors and/or cofactors.
A distinction should be made between the function of JC viral proteins in transformed cells and tumor tissues versus during the course of viral infection. Of note, T antigen has been shown to interact with tumor suppressor proteins and cell cycle regulators may be dysregulated in tumor cells in vitro and in vivo. However, little information has been reported on the effect of JCV on cellular proteins during infection. While it has been previously demonstrated that p53 protein accumulates in JCV-infected cells within PML-afflicted brains and that these same cells may overexpress PCNA and Ki67, as well as cyclins A and B1 (3, 4). However, the significance of these findings has not been fully explored. In addition to T antigen, the viral Agnoprotein seems to have a regulatory role in the viral lytic cycle and may impact host function. Earlier studies have revealed that the 8-kDa Agnoprotein has a perinuclear cytoplasmic localization and may shuttle between the nucleus and cytoplasm (22). In more recent studies, it has been demonstrated that JCV Agnoprotein has the ability to associate with T antigen and controls viral gene expression and replication in human glial cells (12). As anticipated, the structural capsid protein, VP1 may also be detected in oligodendrocytes and, to a lesser extent, bizarre astrocytes (13). In order to determine the effect of JCV infection on the astrocytic cells, we performed gene expression analysis with gene array technology.
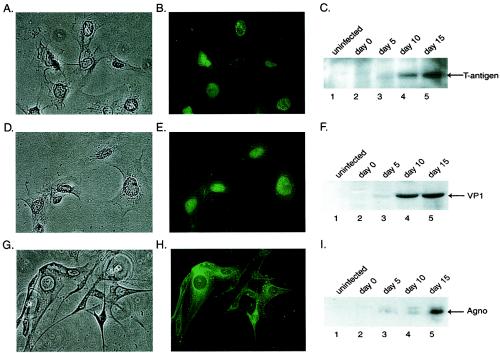
Toward this end, primary human fetal astrocytes were infected with the Mad1/SVEdelta strain of JCV, which has been described previously (29). This hybrid JCV contains the sequences for all JCV coding regions and a modified noncoding region in which the distal portion of the second 98-bp repeat sequence has been replaced with an analogous portion of a 72-bp repeat sequence of the simian virus 40 noncoding region. Infections were performed at a multiplicity of infection of 1, and production of the viral protein at 0, 5, 10, and 15 days after infection was monitored by Western blotting and immunocytochemistry. As shown in Fig. 1, T antigen was first detected at 5 days after infection and its level increased during the course of infection (Fig. 1C, compare lane 3 to lanes 4 and 5). The late gene products, including VP1 and Agnoprotein, were also detected at days 5, 10, and 15 after infection (Fig. 1F and I, respectively). Results from immunocytochemistry showed nuclear accumulation of T antigen and VP1 in the infected cells 15 days after infection (Fig. 1B and E). In accordance with earlier observations, Agnoprotein showed a strong presence in the cytoplasm around the nuclei of the infected cells (Fig. 1H).
FIG. 1.
Western blot and immunocytological analysis of viral proteins in JCV-infected primary human fetal astrocytic cells. Primary human fetal astrocytes (kindly provided by Avindra Nath) plated into tissue culture flasks were infected with the Mad1/SVEdelta strain of JCV at a multiplicity of infection of 1 in the absence of serum for 3 h at 37°C. The Mad1/SVEdelta JCV is a modification of the Mad-1 strain of JCV, which has been described previously (29). After infection, cells were washed and were refed with Dulbecco modified Eagle medium-F12 supplemented with 15% fetal calf serum. Cells were harvested on days 0, 5, 10, and 15 postinfection. Representative phase-contrast images of the cells on day 15 are shown (A, D, and G). Primary human fetal astrocytes at 15 days postinfection seeded in polylysine-coated chamber slides were fixed with ice-cold acetone, and expression of the viral proteins was analyzed by immunocytochemistry by using pAb416 antibody (1:1,000 dilution; Oncogene Research Products) for the detection of T antigen (B), rabbit polyclonal antibody for detection of VP1 (1:1,000 dilution, Lee Biomolecular) (E), and rabbit polyclonal antibody for detection of Agnoprotein (1:3,000 dilution) (14) (H). Proteins were detected with fluorescein-conjugated secondary antibodies. Western blot analysis was performed in parallel on extracts prepared from uninfected astrocytes or infected cells harvested 0, 5, 10, and 15 days postinfection by using the antibodies described above to detect T antigen (1:1,000) (C), VP1 (1:1,000 dilution) (B), and Agnoprotein (1:3,000 dilution) (C). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by transfer to a polyvinyl difluoride membrane, and bands were visualized by autoradiography by using alkaline-phosphatase conjugated secondary antibodies and CDP-Star substrate according to the manufacturer's instructions (NEN-Dupont).
Total cellular RNAs were isolated from uninfected cells as well as from cells on day 15 postinfection to determine the profile of gene expression by using the Affymetrix U95A Human GeneChip. All protocols were conducted as described in the Affymetrix GeneChip Expression Analysis Technical Manual. Total RNA was prepared by the RNeasy Total RNA Isolation Kit according to the manufacturer's instructions (Qiagen). Briefly, 20 μg of total RNA was converted to first-strand cDNA by using Superscript II reverse transcriptase primed by a poly(T) oligomer that incorporated the T7 promoter (Superscript Choice Kit; Invitrogen). Second-strand cDNA synthesis was followed by in vitro transcription for linear amplification of each transcript and incorporation of biotinylated CTP and UTP. Biotinylated cRNA was synthesized by using the High Yield RNA Transcript labeling kit (Enzo Diagnostics) by incorporating biotinylated CTP and UTP and was then purified with RNeasy affinity columns (Qiagen).
The subsequent steps that include fragmentation and hybridization of cRNA to probe arrays (Affymetrix U95A) containing approximately 12,600 human gene sequences were performed by Research Genetics (Huntsville, Ala.) according to Affymetrix protocols. cRNA was fragmented and was tested for quality control by using Affymetrix Test 2 arrays. The cRNA products were fragmented to 200 nucleotides or less, heated at 99°C for 5 min, and hybridized for 16 h at 45°C to human U95A microarrays. The microarrays were then washed at low (6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {ph 7.7}]) and high (100 mM morpholineethanesulfonic acid and 0.1 M NaCl) stringencies and were stained with streptavidin-phycoerythrin. Fluorescence was amplified by adding biotinylated antistreptavidin and an additional aliquot of streptavidin-phycoerythrin stain. A confocal scanner was used to collect a fluorescence signal at a 3-m resolution after excitation at 570 nm. The average signal from two sequential scans was calculated for each microarray feature.
Affymetrix Microarray Suite 5.0 was used to quantitate expression levels for targeted genes, and default values provided by Affymetrix were applied to all analysis parameters. Border pixels were removed, and the average intensity of pixels within the 75th percentile was computed for each probe. The average of the lowest 2% of probe intensities occurring in each of 16 microarray sectors was set as background and was subtracted from all features in that sector. Probe pairs were scored positive or negative for detection of the targeted sequence by comparing signals from the perfect match and mismatch probe features. The number of probe pairs meeting the default discrimination threshold (τ = 0.015) was used to assign a call of absent, present, or marginal for each assayed gene, and P was calculated to reflect confidence in the detection call. A weighted mean of probe fluorescence (corrected for nonspecific signal by subtracting the mismatch probe value) was calculated by using the One-Step Tukey's Biweight Estimate. This signal value, a relative measure of the expression level, was computed for each assayed gene. Global scaling was applied to allow comparison of gene signals across multiple microarrays: after exclusion of the highest and lowest 2%, the average total chip signal was calculated and was used to determine what scaling factor was required to adjust the chip average to an arbitrary target of 150. All signal values from one microarray were then multiplied by the appropriate scaling factor. Detecting a change of > or <2.0-fold on data normalized with Affymetrix Microarray Suite v5.0 was considered significant, in accordance with Affymetrix product specifications. Chip replicates are considered unnecessary, as repeated hybridizations with the same target have been shown to be highly reproducible (21, 33). Normalized data were then analyzed by using GeneSpring v4.2 software. Since each set of experimental conditions was created in duplicate with use of different preparations of primary cells and viral stocks, all genes shown to be classified as up- or downregulated under each set of virus-infected conditions were compared with genes under mock-infected conditions. Calculated mean changes (n-fold) of the duplicates are presented. Among the tested DNA sequences, we found that transcription of 355 genes was enhanced to various extents, while expression of nearly 130 genes was diminished due to viral infection. Among the genes whose expression was affected by viral infection, several belonged to cell cycle regulators, transcriptional activators, cytokines and immunomodulators, and genes involved in signal transduction. Table 1 represents a subset of genes annotated as cell cycle-regulatory proteins in databases supporting GeneSpring gene lists. The complete list of 355 upregulated and 130 downregulated genes can be found at http://www.temple.edu/cnvcb/supplemental.htm. Many of the genes represented in the list have unknown functions, represent cDNA clones or expressed sequence tags, or represent genes whose expression has not previously been linked to infection with JCV.
TABLE 1.
Gene and probe designations
| Gene type and probe set | GenBank accession no. | Change (n-fold) | Gene symbol | Full gene name | Description |
|---|---|---|---|---|---|
| Upregulated genes | |||||
| 1621_at | D85423 | 11.8170 | CDC5L | Cell division cycle 5-like | Transcription factor |
| 39279_atb | M60315 | 6.4661 | BMP6 | Bone morphogenetic protein 6 | Induces cartilage and bone formation, TGFβ family |
| 1968_g_at | X76079 | 5.6861 | PDGFR-A | Platelet-derived growth factor receptor alpha | PDGF receptor, tyrosine kinase |
| 1560_g_at | U24153 | 5.5000 | PAK2 | p21 (CDKN1A)-activated kinase-2 | Serine/threonine-protein kinase PAK 2 |
| 597_at | M59964 | 5.2550 | KITLIG | KIT ligand | Stimulates mast cell proliferation |
| 1536_atb | U77949 | 4.7658 | CDC6 | Cell division cycle 6 | Cell cycle regulatory protein |
| 32126_atb | M60828 | 4.4903 | FGF7 | Fibroblast growth factor 7 | Keratinocyte growth factor |
| 31805_at | M64347 | 4.3753 | FGFR3 | Fibroblast growth factor receptor 3 | FGF family receptor |
| 1945_at | M25753 | 3.7000 | CCNB1 | Cyclin B1 | G2/mitosis-specific cyclin B1 |
| 1069_at | U04636 | 3.5973 | PTGS2 | Prostaglandin endoperoxide synthase 2 | Cyclo-oxygenase 2 (Cox-2) |
| 40812_at | AF027153 | 3.5521 | SLC5A3 | Solute carrier family 5 member 3 | Sodium/myo-inositol cotransporter |
| 37804_at | AF038960 | 3.4047 | SDK1 | Suppressor of K+ transport defect 1 | Intracellular protein trafficking |
| 38677_at | U04735 | 3.3709 | STCH | Stress 70 protein chaperone | Microsomal stress 70 protein ATPase core |
| 1609_g_at | HT4545a | 3.3336 | HGFR | Hepatocyte growth factor receptor | MET proto-oncogene, tyrosine kinase |
| 34890_at | L09235 | 3.3230 | ATP6V1A1 | ATPase vaculolar protein pump V1 subunit A isoform 1 | H+ transporting lysosomal proton pump |
| 35547_at | AF058056 | 3.2909 | SLC16A7 | Solute carrier family 16 member 7 | Monocarboxylic acid transporter |
| 40463_at | U70322 | 3.2653 | KPNB2 | Karyopherin beta 2 (importin) | Nuclear import of ribonucleoproteins |
| 36101_s_at | M63978 | 3.1945 | VEGF | Vascular endothelial growth factor | Angiogenic, vasculogenic, endothelial growth factor |
| 1380_atb | M60828 | 3.1776 | FGF7 | Fibroblast growth factor 7 | Keratinocyte growth factor |
| 1537_at | X00588 | 2.9978 | EGFR | Epidermal growth factor receptor | EGF family receptor, v-erbB oncogene |
| 32903_atb | L11695 | 2.9686 | TGFβR1 | Transforming growth factor β type 1 receptor | TGFβR serine/threonine kinase |
| 158_at | U40992 | 2.8215 | DNAJB4 | Dnaj subfamilyB member 4 | Heat shock protein |
| 1466_s_atb | S81661 | 2.8151 | FGF7 | Fibroblast growth factor 7 | Keratinocyte growth factor |
| 38268_at | U08989 | 2.5606 | SLC1A1 | Solute carrier family 1 member 1 | Neuronal/epithelial glutamate transporter |
| 33324_s_atb | D88357 | 2.5505 | CDC2 | Cell division cycle 2 | G1-to-S, G2-to-M cell cycle regulator |
| 36508_at | AF030186 | 2.4995 | GPC4 | Glypican 4 | Heparin sulfate cell surface proteoglycan |
| 1943_at | X51688 | 2.4895 | CCNA2 | Cyclin A | G1-to-S, G2-to-M cell cycle regulator |
| 36839_atb | U77949 | 2.4867 | CDC6 | Cell division cycle 6 | Cell cycle-regulatory protein |
| 1575_at | M14758 | 2.4814 | ABCB1 | ATP-binding cassette subfamily B member 1 | P-glycoprotein, multidrug resistance protein 1 |
| 1601_s_atb | L27559 | 2.4248 | IGFBP5 | Insulin-like growth factor binding protein 5 | IGF stabilizer, IGF inhibitor/stimulator |
| 1954_at | AF035121 | 2.3478 | KDR | Kinase insert domain receptor | VEGF receptor, tyrosine kinase |
| 1828_s_at | M27968 | 2.3293 | FGF2 | Basic fibroblast growth factor | Heparin binding growth factor |
| 35812_at | AJ133769 | 2.2596 | TRN-SR | Transportin S receptor | Nucleocytoplasmic export |
| 34955_at | AF071202 | 2.2566 | ABCC4 | ATP binding cassette subfamily C member 4 | Multidrug resistance |
| 551_at | U01877 | 2.2486 | EP300 | E1A binding protein p300 | Transcriptional adapter involved in cell cycle regulation |
| 160027_s_at | Y00285 | 2.2174 | IGF2R | Insulin-like growth factor receptor 2 | Lysosomal transport protein |
| 1957_s_atb | AF054598 | 2.2097 | TGFβR1 | Transforming growth factor β type 1 receptor | TGF-βR, serine/threonine kinase |
| 1824_s_at | J05614 | 2.1776 | PCNA | Proliferating cell nuclear antigen | DNA polymerase delta auxiliary protein |
| 1565_s_at | M96995 | 2.1637 | GRB2 | Growth factor receptor-bound protein 2 | EGF and PDGF receptor signaling intermediary |
| 1677_atb | M65062 | 2.1609 | IGFBP5 | Insulin-like growth factor binding protein 5 | IGF stabilizer, IGF inhibitor/stimulator |
| 234_s_at | M57399 | 2.0998 | PTN | Pleiotrophin | Neurite growth-promoting factor |
| 41216_r_at | D13891 | 2.0909 | ID2 | Inhibitor of DNA binding 2 | Dominant-negative helix-loop-helix protein |
| 34476_r_at | D30783 | 2.0737 | EREG | Epiregulin | EGF-related cell proliferation protein |
| 1803_atb | X05360 | 2.0650 | CDC2 | Cell division cycle 2 | G1-to-S, G2-to-M cell cycle regulator |
| 41245_at | D49493 | 2.0590 | GDF10 | Growth differentiation factor 10 | Bone morphogenetic protein 3b |
| 2021_a_at | M73812 | 2.0389 | CCNE1 | Cyclin E1 | Cell cycle regulator at G1/S phase transition |
| 1733_at | M60315 | 2.0159 | BMP6 | Bone morphogenetic protein 6 | Induces cartilage and bone formation, TGFβ family |
| Downregulated genes | |||||
| 38647_at | AJ131182 | −2.5765 | COPE | Coatomer protein complex subunit epsilon | Endoplasmic reticulum-to-Golgi protein transporter |
| 40635_at | AF089750 | −2.4837 | FLOT1 | Flotillin 1 | Caveola-associated protein |
| 33301_g_at | AL031282 | −2.3164 | CDC2L2 | Cell division cycle 2-like protein 2 | Not determined |
| 37361_at | AF010187 | −2.2595 | FIBP | Fibroblast growth factor 1 intracellular binding protein | Involved in mitogenic function of FGF1 |
| 160027_s_at | Y00285 | −2.2174 | IGF2R | Insulin-like growth factor 2 receptor | Lysosomal transport protein |
Gene number from the Institute for Genomic Research.
Appears on list more than once. Multiple probe sets for this gene are present on the DNA array.
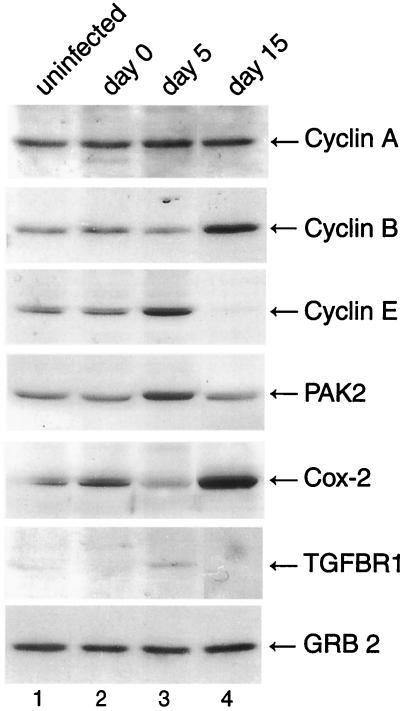
In order to validate the gene expression data, we selected a small subset of several affected genes that were involved in cell cycle or cellular proliferation such as cyclin A, cyclin B1, cyclin E, PAK2, Cox-2, and transforming growth factor βR1 (TGFβR1) to assess the level of their proteins during the course of JCV infection of primary astrocytes by Western blot analysis. As shown in Fig. 2, levels of cyclin A increased slightly at 5 days but returned to normal levels at 15 days after infection, while RNA levels were found to be increased 2.5-fold at 15 days postinfection. A significant increase in the levels of cyclin B1 and Cox-2 was observed at 15 days postinfection, which is consistent with the observed 3.7- and 3.6-fold increases in RNA levels, respectively. Cyclin E and PAK2 showed a subtle increase at the early phase of viral infection followed by a decrease at the late phase of infection. This is in contrast with the results from RNA analysis by microarray, where 2- and 5.5-fold increases in cyclin E and PAK2 RNA, respectively, at 15 days postinfection were observed. A similar pattern was also observed when TGFβR1 protein levels were examined during the course of JCV infection. As seen in Fig. 2, the levels of the protein at 5 days were increased while a decrease was observed at day 15 as the infection cycle progressed; RNA levels for TGFβR1 were increased 2.2-fold. Surprisingly, the level of a nonspecific protein such as Grb-2 remained fairly constant during the course of infection, even though an increase in RNA levels of approximately 2.2-fold was observed at 15 days postinfection. This may be due to stability of the mRNA or to posttranscriptional regulation of Grb-2. In summary, with the exception of Grb-2, all proteins examined showed an increase either at 5 or 15 days postinfection, though the increase was not sustained during the course of the infection. Several of the proteins, including cyclin A, cyclin E, PAK2, and TGFβR1, showed an increase on day 5 and a decrease on day 15 postinfection, with increases of >2-fold in RNA levels at day 15, suggesting that either these RNAs are not efficiently translated at the later stages of viral infection and/or that the proteins are rapidly degraded at the later times of the infection cycle. Cyclin B1 and Cox-2, however, showed increases at both the RNA and protein levels 15 days postinfection.
FIG. 2.
Western blot analysis of extracts from JCV-infected astrocytes for gene products of selected upregulated genes. We carried out Western blot analysis of protein extracts (50 μg) from uninfected primary astrocytes or cells infected with JCV 0, 5, and 15 days after infection with the following mouse monoclonal antibodies: anti-cyclin A (1:1,000, BF683), anti-cyclin B1 (1:1,000, GNS1), and anti-Grb-2 (1:1,000, no. 81). Alternatively we used the following polyclonal antibodies: anti-cyclin E (1:1,000, M20), anti-PAK2 (1:1,000, V-19), anti-Cox-2 (1:1,000, C-20), and anti-TGFβR1 (1:500, R-20). All antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.), with the exception of Grb-2, which was purchased from BD Transduction Laboratories. Western blot analysis was performed as described in the Fig. 1 legend.
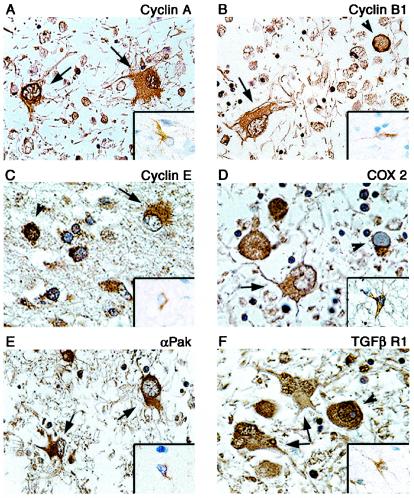
To further determine the ability of microarray in a cell culture infection model to predict up- or downregulation of genes in vivo, immunohistochemistry was performed on paraffin-embedded, formalin-fixed tissue sections of brain tissue from a PML patient. Examination of cyclins A, B1, E, and Cox-2 showed enhanced cytoplasmic staining of these proteins in the bizarre astrocytes of PML lesions compared to results from non-PML normal brain, though nuclear staining in PML astrocytes was notably absent (Fig. 3A to D). Similarly, increased expression of PAK2 and TGFβR1 was evident in astrocytes of PML compared to that in normal astrocytes of non-PML samples (Fig. 3E and F). Interestingly, punctate immunoreactivity to TGFβR1 was also detected in the nuclei of some astrocytes (Fig. 3F). Oligodendrocyte inclusion bodies also showed staining in the perinuclear region or cytoplasm for most proteins analyzed, while nuclear staining of inclusion bodies was also seen with antibodies to cyclin A, cyclin E, PAK2, and TGFβR1. These staining patterns in astrocytes corroborate the results from in vitro infection of primary astrocytes by JCV and indicate that infection of the astrocytes by JCV both in cell culture and in the brains of PML patients leads to dysregulation of cell cycle regulators.
FIG. 3.
Immunohistochemical analysis of cellular proteins in demyelinated plaques of PML and normal brain tissue. Paraffin-embedded sections of brain tissue lesions from a patient with PML or from normal brain were analyzed for the expression of cyclin A (A), cyclin B1 (B), cyclin E (C), Cox-2 (D), PAK2 (E), and TGFβR1 (F). Insets show staining of normal brain tissue depicting a representative astrocyte with various antibodies as indicated in each panel. Bizarre astrocytes exhibit cytoplasmic immunoreactivity for cyclin A, cyclin B1, cyclin E, Cox-2, and PAK2 (arrows). Bizarre astrocytes exhibit cytoplasmic and punctate nuclear immunoreactivity when tested with an antibody for TGFβR1 (arrows). Oligodendrocyte inclusion bodies show cytoplasmic or perinuclear immunoreactivity for cyclin E, PAK2, TGFβR1, and Cox-2 and nuclear immunoreactivity for cyclin A, cyclin E, and TGFβR1 (arrowheads). Immunohistochemistry was performed by using the ABC Vector Elite system and was detected with DAB chromogen as described previously (14). All panels, original magnification ×1,000.
The upregulation of a host of cell cycle regulatory proteins has been previously demonstrated in JCV-transformed human tumor cells in vitro and in vivo. However, a distinction should be made between the study of transformed cells, as is the case for most cell lines and certainly for tumor tissues and primary cultures of nontransformed cells. In this regard, limited information on cellular gene expression in JCV-infected nontransformed cells or within PML brain lesions is known. In one report, cyclins A and B1 were shown to be overexpressed in PML astrocytes and oligodendrocytes, as well as positive Ki67 staining (4). While the results presented here may suggest increased cycling of the cells, one must cautiously consider that myelin-producing oligodendrocytes are thought to be terminally differentiated and that no present evidence suggests that inclusion-bearing oligodendrocytes are undergoing cell division. However, the bizarre astrocytes of PML do contain multilobulated nuclei or may even appear multinucleated, suggesting that the cells may be passing through some steps of the cell cycle. In any case, the advantage for JCV in controlling the cell cycle is clear. It may be necessary for a double-stranded DNA virus to control the cellular replication machinery normally present during S phase. In this regard, JCV T antigen can orchestrate viral DNA replication and complete a productive infection cycle.
In summary, microarray technology was employed to investigate the expression of cellular genes during the course of infection with JCV. We chose to carry out our study in highly purified human primary astrocytes instead of mixed primary cultures of human fetal brain, which are commonly used for JCV infection in vitro, due to the inherent variations in the cell-type context of each preparation that can significantly affect the efficacy of infection in each experiment and thus the reproducibility of the outcome. Also, it is extremely difficult to obtain highly purified cultures of oligodendrocytes in the large quantities necessary to yield a sufficient amount of RNA for analysis. Thus, our results may not provide a complete picture of events related to cellular gene expression in JCV-infected brain cells, particularly oligodendrocytes. Nevertheless, our results illuminate some of the changes that occur in astrocytes upon JCV infection. The role of astrocytes in PML has not been elucidated, though it is widely accepted that they represent an abortive infection. Many questions remain to be answered, such as whether astrocytes provide any protection to infected oligodendrocytes or may even contribute toward a favorable environment for viral replication. The present study has suggested a number of proteins of known and unknown function whose gene expression has been altered upon infection with JCV. Further study toward understanding different mechanisms that may affect astrocyte function during the course of PML may lead us to understand astrocyte responses to viral infections in the CNS.
Acknowledgments
We thank past and present members of the Center for Neurovirology and Cancer Biology for their insightful discussions and sharing of ideas and reagents. We thank Avindra Nath (Johns Hopkins University) for providing purified cultures of human fetal astrocytes, Walter Atwood (Brown University) for providing Mad1/SVEdelta, and Susan Morgello (Mount Sinai School of Medicine) and the Manhattan HIV-1 Brain Bank for providing human brain tissue samples. We also thank C. Schriver for preparation of the manuscript.
This work was made possible by grants awarded by the NIH to L.D.V. and K.K.
REFERENCES
- 1.Anders, K. H., W. E. Guerra, U. Tomiyasu, M. A. Verity, and H. V. Vinters. 1986. The neuropathology of AIDS: UCLA experience and review. Am. J. Pathol. 124:537-558. [PMC free article] [PubMed] [Google Scholar]
- 2.Antinori, A., A. Ammassari, M. L. Giancola, A. Cingolani, S. Grisetti, R. Murri, L. Alba, B. Ciancio, F. Soldani, D. Larussa, G. Ippolito, and A. De Luca. 2001. Epidemiology and prognosis of AIDS-associated progressive multifocal leukoencephalopathy in the HAART era. J. Neurovirol. 7:323-328. [DOI] [PubMed] [Google Scholar]
- 3.Ariza, A., J. L. Mate, A. Fernandez-Vasalo, C. Gomez-Plaza, J. Perez-Piteira, M. Pujol, and J. J. Navas-Palacios. 1994. p53 and proliferating cell nuclear antigen expression in JC virus-infected cells of progressive multifocal leukoencephalopathy. Hum. Pathol. 25:1341-1345. [DOI] [PubMed] [Google Scholar]
- 4.Ariza, A., J. L. Mate, M. Isamat, A. Calatrava, A. Fernandez-Vasalo, and J. J. Navas-Palacios. 1998. Overexpression of Ki-67 and cyclins A and B1 in JC virus-infected cells of progressive multifocal leukoencephalopathy. J. Neuropathol. Exp. Neurol. 57:226-230. [DOI] [PubMed] [Google Scholar]
- 5.Berger, J. R., and M. Concha. 1995. Progressive multifocal leukoencephalopathy: the evolution of a disease once considered rare. J. Neurovirol. 1:5-18. [DOI] [PubMed] [Google Scholar]
- 6.Berger, J. R., B. Kaszovitz, J. D. Post, and G. Dickinson. 1987. Progressive multifocal leukoencephalopathy associated with human immunodeficiency virus infection: a review of the literature with a report of sixteen cases. Ann. Intern. Med. 107:78-87. [DOI] [PubMed] [Google Scholar]
- 7.Berger, J. R., A. Chauhan, D. Galey, and A. Nath. 2001. Epidemiological evidence and molecular basis of interactions between HIV and JC virus. J. Neurovirol. 7:329-338. [DOI] [PubMed] [Google Scholar]
- 8.Boldorini, R., S. Cristina, L. Vago, A. Tosoni, S. Guzzetti, and G. Costanzi. 1993. Ultrastructural studies in the lytic phase of progressive multifocal leukonecephalopathy in AIDS patients. Ultrastruct. Pathol. 17:599-609. [DOI] [PubMed] [Google Scholar]
- 9.Chesters, P. M., J. Heritage, and D. J. McCance. 1983. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. J. Infect. Dis. 147:676-684. [DOI] [PubMed] [Google Scholar]
- 10.Cinque, P., C. Pierotti, M. G. Vigano, A. Bestetti, C. Fausti, D. Bertelli, and A. Lazzarin. 2001. The good and evil of HAART in HIV-related progressive multifocal leukoencephalopathy. J. Neurovirol. 7:358-363. [DOI] [PubMed] [Google Scholar]
- 11.Clifford, D. B., and E. O. Major. 2001. The biology of JC virus and progressive multifocal leukoencephalopathy. J. Neurovirol. 4:279. [DOI] [PubMed] [Google Scholar]
- 12.Darbinyan, A., N. Darbinian, M. Safak, S. Radhakrishnan, A. Giordano, and K. Khalili. 2002. Evidence for dysregulation of cell cycle by human polyomavirus, JCV, late auxiliary protein. Oncogene 21:5574-5581. [DOI] [PubMed] [Google Scholar]
- 13.Del Valle, L., S. Croul, S. Morgello, S. Amini, J. Rappaport, and K. Khalili. 2000. Detection of HIV-1 Tat and JCV capsid protein, VP1, in AIDS brain with progressive multifocal leukoencephalopathy. J. Neurovirol. 6:221-228. [DOI] [PubMed] [Google Scholar]
- 14.Del Valle, L., J. Gordon, S. Enam, S. Delbue, S. Croul, S. Abraham, S. Radhakrishnan, M. Assimakoupoulou, C. D. Katsetos, and K. Khalili. 2001. Expression of human neurotropic polyomavirus JCV late gene product AGNO protein in human medulloblastoma. J. Natl. Cancer Inst. 94:267-273. [DOI] [PubMed] [Google Scholar]
- 15.Frisque, R. J. 2001. Structure and function of JC virus T proteins. J. Neurovirol. 7:293-297. [DOI] [PubMed] [Google Scholar]
- 16.Gardner, S. D. 1977. The new human papovaviruses: their nature and significance, p. 93-115. In A. P. Waterson (ed.), Recent advances in clinical virology. Churchill Livingstone, Edinburgh, United Kingdom.
- 17.Grinnell, B. W., B. L. Padgett, and D. L. Walker. 1983. Comparison of infectious JC virus DNAs cloned from human brain. J. Virol. 45:299-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krupp, L. B., R. B. Lipton, M. L. Swerdlow, N. E. Leeds, and J. Llena. 1985. Progressive multifocal leukoencephalopathy: clinical and radiographic features. Ann. Neurol. 17:344-349. [DOI] [PubMed] [Google Scholar]
- 19.Kure, K., J. L. Llena, W. D. Lyman, R. Soeiro, K. M. Weidenheim, D. Hirano, and W. Dickson. 1991. Human immunodeficiency virus-1 infection of the nervous system: an autopsy study of 268 adult, pediatric, and fetal brains. Hum. Pathol. 22:700-710. [DOI] [PubMed] [Google Scholar]
- 20.Lang, W., J. Miklossy, J. P. Deruaz, G. P. Pizzolato, A. Probst, T. Schaffner, E. Gessage, and P. Kleihues. 1989. Neuropathology of the acquired immune deficiency syndrome (AIDS): a report of 135 consecutive autopsy cases from Switzerland. Acta Neuropathol. 77:379-390. [DOI] [PubMed] [Google Scholar]
- 21.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittman, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 22.Okada, Y., S. Endo, H. Takahashi, H. Sawa, T. Umemura, and K. Nagashima. 2001. Distribution and function of JCV agnoprotein. J. Neurovirol. 7:302-306. [DOI] [PubMed] [Google Scholar]
- 23.Padgett, B. L., and D. L. Walker. 1973. Prevalence of antibodies in human sera against JC virus, an isolate from a case of progressive multifocal leukoencephalopathy. J. Infect. Dis. 127:467-470. [DOI] [PubMed] [Google Scholar]
- 24.Padgett, B. L., C. M. Rogers, and D. L. Walker. 1977. JC virus, a human polyomavirus associated with progressive multifocal leukoencephalopathy: additional biological characteristics and antigenic relationships. Infect. Immun. 15:656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Safak, M., B. Sadowska, R. Barrucco, and K. Khalili. 2002. Functional interaction between JC virus late regulatory Agnoprotein and cellular Y-box binding transcription factor, YB-1. J. Virol. 76:3828-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shinohara, T., K. Nagashima, and E. O. Major. 1997. Propagation of the human polyomavirus, JCV, in human neuroblastoma cell lines. Virology 228:269-277. [DOI] [PubMed] [Google Scholar]
- 27.Snider, W. D., S. M. Simpson, S. Nielson, J. W. M. Gold, C. E. Metroka, and J. B. Posner. 1983. Neurological complications of acquired immune deficiency syndrome. Analysis of 50 patients. Ann. Neurol. 14:403-418. [DOI] [PubMed] [Google Scholar]
- 28.Taguchi, R., J. Kajioka, and T. Miyamura. 1982. Prevalence rate and age of acquisition of antibodies against JC virus and BK virus in human sera. Microbiol. Immunol. 26:1057-1064. [DOI] [PubMed] [Google Scholar]
- 29.Vacante, D. A., R. Traub, and E. O. Major. 1989. Extension of JC virus host range to monkey cells by insertion of a simian virus 40 enhancer into the JC virus regulatory region. Virology 170:353-361. [DOI] [PubMed] [Google Scholar]
- 30.Walker, D. L., and B. L. Padgett. 1983. The epidemiology of human papovaviruses, p. 99-106. In J. L. Sever and D. L. Madden, (ed.), Polyomaviruses and human neurological disease. Alan R. Liss, New York, N.Y.
- 31.Wiley, C. A., and J. A. Nelson. 1990. Human immunodeficiency virus: infection of the nervous system. Curr. Top. Microbiol. Immunol. 160:157-172. [DOI] [PubMed] [Google Scholar]
- 32.Wiley, C. A., M. Grafe, C. Kennedy, and J. A. Nelson. 1988. Human immunodeficiency virus (HIV) and JC virus in acquired immune deficiency syndrome (AIDS) patients with progressive multifocal leukoencephalopathy. Acta Neuropathol. 76:338-346. [DOI] [PubMed] [Google Scholar]
- 33.Wodicka, L., H. Dong, M. Mittmann, M. H. Ho, and D. J. Lockhart. 1997. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15:1359-1367. [DOI] [PubMed] [Google Scholar]
- 34.Zu Rhein, G. M. 1972. Virions in progressive multifocal leukoencephalopathy, p. 2893-2912. In J. Minkler (ed.), Pathology of the nervous system, vol. 3. McGraw-Hill, New York, N.Y.
- 35.Zu Rhein, G. M. 1969. Association of papova-virions with a human demyelinating disease (progressive multifocal leukoencephalopathy). Prog. Med. Virol. 11:185-247. [PubMed] [Google Scholar]
- 36.Zu Rhein, G. M., and S. M. Chou. 1968. Papova virus in progressive multifocal leukoencephalopathy. Res. Publ. Assoc. Nerv. Ment. Dis. 44:307-362. [PubMed] [Google Scholar]