Abstract
Instability of repetitive sequences, both in intronic sequences and within coding regions, has been demonstrated to be a hallmark of genomic instability in human cancer. Understanding how these mutational events arise may provide an opportunity for prevention or early intervention in cancer development. To study the source of this instability, we have identified a region of the β-lactamase gene that is tolerant to the insertion of fragments of exogenous DNA as large as 1,614 bp with minimal loss of enzyme activity, as determined by antibiotic resistance. Fragments inserted out-of-frame render Escherichia coli sensitive to antibiotic, and compensatory frameshift mutations that restore the reading frame of β-lactamase can be selected on the basis of antibiotic resistance. We have utilized this site to insert a synthetic microsatellite sequence within the β-lactamase gene and selected for mutations yielding frameshifts. This assay provides for detection of one frameshift mutation in a background of 106 wild-type sequences. Mismatch repair deficiency increased the observed frameshift frequency ≈300-fold. Exposure of plasmid containing microsatellite sequences to hydrogen peroxide resulted in frameshift mutations that were localized exclusively to the microsatellite sequences, whereas DNA damage by UV or N-methyl-N′-nitro-N-nitrosoguanidine did not result in enhanced mutagenesis. We postulate that in tumor cells, endogenous production of oxygen free radicals may be a major factor in promoting instability of microsatellite sequences. This β-lactamase assay may provide a sensitive methodology for the detection and quantitation of mutations associated with the development of cancer.
It takes many years to get a cancer; ≈20 years elapse from the time of exposure to a carcinogen to the development of a clinically detectable tumor. During this time, tumors are characterized by genomic instability, resulting in phenotypic changes and the progressive accumulation of mutations (1–3). We have hypothesized that the rate of mutation in normal cells is insufficient to account for the large numbers of mutations observed in cancer cells, and therefore that tumor cells manifest a mutator phenotype (4, 5). The concept of a mutator phenotype has been supported by the demonstration of microsatellite instability (6–8), the expansion or contraction of repetitive sequences, in tumor cell DNA. The length of repetitive sequences is stable in normal cells, but increases and decreases in the number of repeats can occur in tumor cells. Instability of microsatellite sequences is most pronounced in cells from patients with hereditary nonpolyposis colon cancer, and this instability is attributed to loss of mismatch repair function. However, numerous types of sporadic cancers also display microsatellite instability, including cancer of the colon, endometrium, breast, lung (small cell and non-small cell), stomach, ovary, prostate, esophagus, bladder, glia, cervix, and pancreas (see ref. 9 for review and references). Many of these cancers have not been shown to involve mutations in mismatch repair, and thus other mechanisms may be operative to produce microsatellite instability.
To address molecular events relating to microsatellite instability, we have developed a sensitive and selective assay for the quantitative detection of frameshift mutations in repetitive nucleotide sequences. The sensitivity of this assay allows for the identification of rare frameshift events, and the versatility of this system offers considerable advantages over current methods for analysis of microsatellite instability that involve gel electrophoresis (10, 11) or screening of lacZα mutants (12–14). This assay complements other selective assays for frameshifts within repetitive sequences (15–18), without imposing limitations on the sequence or size of the repetitive sequence insert. We have utilized this assay to demonstrate that oxidative DNA damage can increase the frequency of microsatellite instability. Mutations in mismatch repair genes can reduce the efficacy of repair of reactive oxygen species damage to DNA, resulting in increased instability of repetitive sequences.
MATERIALS AND METHODS
Strains and Media.
Wild-type strain BL21 (F− ompT (lon) hsdSB (rB−mB−) was from Novagen. Mismatch repair deficient mutH strain CM1 (miniTn10Cam inserted into the mutH gene in strain CSH143) was a generous gift from Jeffrey H. Miller (University of California Los Angeles). DH5αF′ is a recA1 wild-type strain. Growth medium consisted of 2× yeast extract tryptone containing tetracycline (12.5 μg/ml) or carbenicillin (100 μg/ml).
Nonrepetitive Sequence Constructs.
The EcoRI site in plasmid pBR322 was destroyed by digesting and filling in with T4 DNA polymerase (Boehringer Mannheim) to generate plasmid pAJ2. A new unique EcoRI site was created within the coding region of β-lactamase by PCR mutagenesis with the following primers: 5′-AAATGCTTCAATAATAATGAAAAAGGAAGAGTATGAGAATTCAACATTTCCG-3′ and 5′-GTTGCCAGGGCTGCAGGCATCGTGGT-3′. The PCR product was digested with PstI and SspI (New England Biolabs), and then used to replace the PstI/SspI fragment in pAJ2 to create plasmid pAJ3. Insert DNA sequences were generated either by use of gel purified oligonucleotides (66 bp, 70 bp, and 71 bp) or by PCR amplification of the HIV reverse transcriptase gene (513 bp, 1,044 bp, 1,614 bp, and 1,618 bp). Polylinker oligonucleotides were as follows: 66 bp, 5′-CGGAGAATTCTGTACAGAGCTCTTGGTACCCCTAGGACCCGGGAAAGGCCTTCTAGAGCAGATCTTAGAATTCTATGC-3′ and 5′-GCATAGAATTCTAAGATCTGCTCTAGAAGGCCTTTCCCGGGTCCTAGGGGTACCAAGAGCTCTGTACAGAATTCTCCG-3′; 70 bp, the 66 bp construct was digested with BsrGI (New England Biolabs) and filled in with T4 DNA polymerase in the presence of all 4 dNTPs to generate a +1 frameshift and create a new unique SnaBI site; 71 bp, 5′- CGGAGAATTCTGCAGTGTACAGAGCTCTTGGTACCCCTAGGACCCGGGAAAGGCCTTCTAGAGCAGATCTTAGAATTCTATGC-3′ and 5′-GCATAGAATTCTAAGATCTGCTCTAGAAGGCCTTTCCCGGGTCCTAGGGGTACCAAGAGCTCTGTACACTGCAGAATTCTCCG-3′. Oligonucleotides were annealed, digested with EcoRI, and ligated into EcoRI-digested plasmid pAJ3 to create plasmids pAJ19, pAJ20, and pAJ21, respectively. The orientation of the polylinkers was verified by sequencing by using the upstream primer 5′-ACAATAACCCTGATAAATGC-3′.
HIV RT PCR products were generated by using linearized template pBK8 and the following primers: forward primer, 5′-CAGCTGCAGAATTCCTATTAGTCCTATTGAA-3′; 513-bp reverse primer, 5′-CAGCTCCAGTGAATTCTAGGCTCTAAGAT-3′, 1,044-bp reverse primer, 5′-CTCAGCTGCAGTGAATTCGATATGTCCATTG-3′; and 1,614-bp reverse primer 5′-CTCAGCTGCAGTGAATTCGGATTCCAGCACT-3′. PCR products were gel purified, digested with EcoRI, and ligated into EcoRI-digested pAJ3 to generate plasmids pAJ12, pAJ13, and pAJ14, respectively. Plasmids from tetracycline-resistant colonies were isolated by the Wizard miniprep kit (Promega) and analyzed for the presence of the HIV reverse transcriptase insert by restriction digestion. Tetracycline-resistant colonies verified to contain the PCR insert were replica-plated to carbenicillin plates for analysis of β-lactamase function. Carbenicillin-resistant clones were subsequently analyzed for extent of β-lactamase activity by comparison of carbenicillin-resistant colonies to tetracycline-resistant colonies obtained from log-phase cultures. Maintenance of the HIV reverse transcriptase insert in carbenicillin-resistant colonies was verified by restriction digestion analysis of recovered plasmids.
Repetitive Sequence Constructs.
Microsatellite constructs were generated through the use of gel-purified synthetic oligonucleotides (GIBCO/BRL or Midland Certified Reagents, Midland, TX). The primers were as follows: in-frame (CA)13, 5′-GCTCGGAGAATTCTGTACAGAGCTCTTGATACCGCACACACACACACACACACACACACATCTAGAGCAGATCTTAGAATTCTATGCGAG-3′ and 5′-CACGCATAGAATTCTAAGATCTGCTCTAGATGTGTGTGTGTGTGTGTGTGTGTGTGCGGTACCAAGAGCTCTG-TACAGAATTCTCCGAGC-3′; +2 frame (CA)11, 5′-GCTCGGAGAATTCTGTACAGAGCTCTTGATACCGCACACACACACACACACACACATCTAGAGCAGATCTTAGAATTCTATGCGTG-3′ and 5′-CACGCATAGAATTCTAAGATCTGCTCTAGATGTGTGTGTGTGTGTGTGTGTGCGGTATCAAGAGCTCTGTACAGAATTCTCCG-AGC-3′; −2 frame (CA)12, 5′-GCTCGGAGAATTCTG-TACAGAGCTCTCGATACCGCACACACACACACACA-CACACACATCCAGAGCAGATCTTAGAATTCTATG-3′ and 5′-CACGCATAGAATTCTAAGATCTGCTCTGGATGTGTGTGTGTGTGTGTGTGTGTGCGGTATCGAGAGCTCTGTACAGAATTCTCC-3′; +1 frame (A)10, 5′-GCTCGGAGAATTCTGTACAGAGCTCTCGATACCGCTTCCTGTTCCAAAAAAAAACCATCCAGAGCAGATCTTA-GAATTCTATG-3′ and 5′-CACGCATAGAATTCTAAG-ATCTGCTCTGGATGGTTTTTTTTTTGGAACAGGAA-GCGGTATCGAGAGCTCTGTACAGAATTCTCC-3′; +1 frame (G)10, 5′-GCTCGGAGAATTCTGTACAGAGCTCTCGATACCGCTTCCTGTTCCGGGGGGGGGGCCATCCAGAGCAGATCTTAGAATTCTATC-3′ and 5′-CACGCATAGAATTCTAAGATCTGCTCTGGATGGCCCCCCCCCCGGAACAGGAAGCGGTATCGAGAGCTCTGTACA-GAATTCTCC-3′. Complementary oligonucleotide pairs were annealed, digested with SacI and BglII (Boehringer Mannheim), and ligated into SacI-BglII-digested pAJ19 to generate plasmids pAJ23, pAJ24, pAJ25, pAJ32, and pAJ33, respectively. Tetracycline-resistant clones were replica-plated for determination of carbenicillin resistance (in-frame sequences) or sensitivity (out-of-frame sequences), and the presence and frame of microsatellite constructs were verified by DNA sequencing of recovered plasmids with Thermosequenase kit (Amersham) by using the same sequencing primer as above.
Hydrogen Peroxide Damage.
Plasmid DNA (9 × 10−15 mol) containing either repetitive sequences or control nonrepetitive sequences within the β-lactamase gene was exposed to 1 μM or 50 μM H2O2 at room temperature for 1 h in 15 mM potassium phosphate buffer (pH 7.2). Damaged DNA was precipitated with 0.3 M sodium acetate and 2 volumes of ethanol in the presence of glycogen and purified through a microcon 30 membrane (Amicon) before electroporation of competent Escherichia coli BL21. The 1 μM H2O2 treatment corresponded to 40% survival of plasmid relative to untreated plasmid, as measured by tetracycline-resistant transformants, whereas 50 μM H2O2 treatment corresponded to 20% survival. The dose response of H2O2 with regard to DNA damage has previously been reported to be nonlinear (19).
Modulation of Hydrogen Peroxide Damage.
Plasmid containing a +2 frame (CA)11 microsatellite sequence within the β-lactamase gene was exposed to 1 μM H2O2 for 1 h at room temperature in the presence of either 1 unit catalase, 0.5 μM CuCl2, or 0.5 μM NiCl2. Damaged DNA was precipitated, purified, and transformed as described. Addition of 0.5 μM Cu2+ or Ni2+ reduced the recovery of tetracycline-resistant transformants to 20% relative to H2O2 treatment alone.
RESULTS
Design of a Positive Selection Assay for Microsatellite Instability.
The β-lactamase gene in E. coli provides resistance to cephalosporin antibiotics, such as carbenicillin (20). Using a unique restriction site in the β-lactamase gene, engineered immediately downstream of the translation start signal ATG, we cloned in-frame into the gene a series of DNA fragments of increasing lengths. Use of a modified version of pBR322, a low copy number plasmid which codes for β-lactamase as well as the tetracycline resistance gene, enabled quantitation of β-lactamase activity as the percentage of carbenicillin-resistant colonies relative to tetracycline-resistant colonies. Insertion of in-frame nonrepetitive sequences demonstrates that this region of the β-lactamase enzyme tolerates inserted DNA while maintaining functional resistance to antibiotic (Table 1). Even the insertion of a 1,614-bp fragment in-frame within β-lactamase reduced activity by only 54%. In contrast, insertion of a 1,618-bp fragment out-of-frame yielded one carbenicillin-resistant colony per 2 × 105 tetracycline-resistant colonies.
Table 1.
Tolerance and stability of DNA inserted within β-lactamase
| Insert size | Sequence | Frame | β-Lactamase activity [(Carbr/Tetr) × 100]% | Reversion frequency (Carbr/Tetr) |
|---|---|---|---|---|
| Nonrepetitive | ||||
| 66 bp | Random | In-frame | 100 | 1 |
| 70 bp | Random | +1 frame | 0.0007 | 7 × 10−6 |
| 71 bp | Random | −1 frame | 0.001 | 1 × 10−5 |
| 513 bp | HIV RT | In-frame | 95.3 | NA |
| 1,044 bp | HIV RT | In-frame | 69.2 | NA |
| 1,614 bp | HIV RT | In-frame | 43.9 | NA |
| 1,618 bp | HIV RT | +1 frame | 0.0005* | 5 × 10−6 |
| Microsatellite | ||||
| 75 bp | (CA)13 | In-frame | 100 | 1 |
| 74 bp | (CA)11 | +2 frame | 0.0045 | 4.45 ×10−5 |
| 73 bp | (CA)12 | −2 frame | 0.0036 | 3.67 × 10−5 |
| 73 bp | (A)10 | +1 frame | 0.0032 | 3.23 × 10−5 |
| 73 bp | (G)10 | +1 frame | 0.0016 | 1.59 × 10−5 |
| Mismatch repair | ||||
| 75 bp (WT strain) | (CA)11 | +2 frame | 0.0075 | 7.5 × 10−5 |
| 75 bp (mutH strain) | (CA)11 | +2 frame | 2.55 | 2.55 × 10−2 |
β-Lactamase constructs containing inserts of 1 kb and 1.6 kb were derived from and tolerated as indicated in strain NM522. The tolerance for these large inserts was much lower in strain BL21: 1 kb, 7.5%; 1.6 kb, 0%. All other inserts were stable and tolerated as indicated in BL21. NA, not applicable.
The unique restriction site was utilized for the introduction of a polycloning sequence for the directional cloning of synthetic microsatellite sequences. This polycloning sequence was constructed in each of the three coding frames, allowing for selection of microsatellite frameshift events in the form of insertions or deletions. We first determined the stability of plasmid-encoded microsatellite sequences constructed with gel-purified oligonucleotides. β-Lactamase constructs containing in-frame microsatellite sequences maintained enzyme function and rendered E. coli resistant to carbenicillin (Table 1). The frameshift frequency of out-of-frame dinucleotide (CA)11 and (CA)12 as well as mononucleotide (A)10 and (G)10 sequences in wild-type, repair-proficient E. coli BL21 was approximately 10−5 (Table 1). Mismatch repair deficiency in strain CM1, in which the mutH gene is insertionally inactivated, produced a 340-fold increase in the frequency of frameshift mutations in dinucleotide repetitive sequences (Table 1). These results are consistent with previous results demonstrating that mismatch repair function maintains the integrity of repetitive sequence length in addition to repairing single base mismatches, and that mismatch repair deficiency results in an increase in spontaneous mutations of both types.
Induction of Microsatellite Instability by Oxidative Damage.
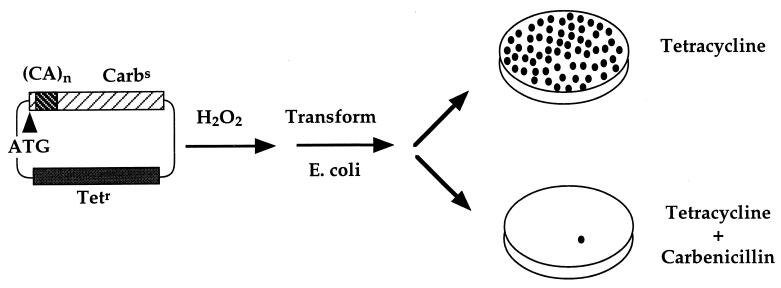
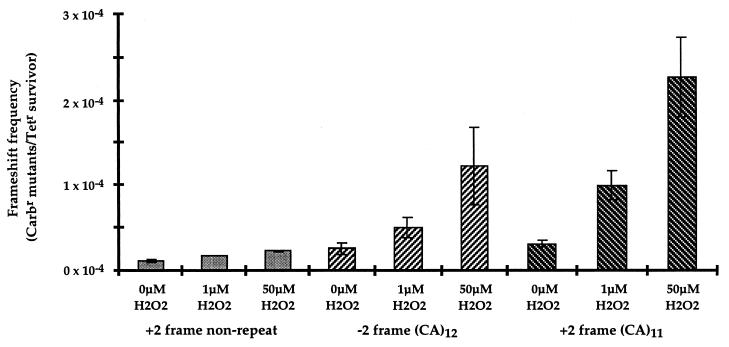
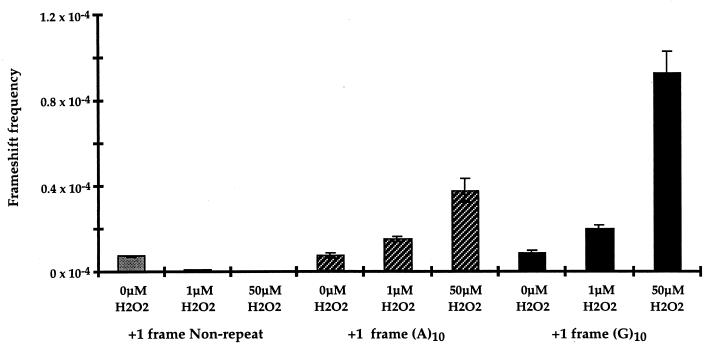
Oxidative DNA damage is postulated to contribute to somatic mutations (21, 22). In addition, microsatellite instability has been demonstrated to be associated with the non-neoplastic but inflammatory settings of pancreatitis (23) and ulcerative colitis (24). We therefore utilized our selection assay to determine whether microsatellite instability can result from reactive oxygen species-induced DNA damage. Plasmids with β-lactamase constructs containing out-of-frame microsatellite sequences that disrupt the production of a functional β-lactamase were the substrate for H2O2 damage in vitro. Following transformation of wild-type E. coli, growth on plates containing carbenicillin could indicate a compensating frameshift to restore β-lactamase function (Fig. 1). The damage-induced mutation frequency was quantitated as carbenicillin mutants recovered per tetracycline-resistant survivor to correct for plasmid loss. The ratio of carbenicillin-resistant colonies to tetracycline-resistant colonies was observed to increase with increasing concentrations of H2O2 by using plasmid constructs containing either dinucleotide (Fig. 2) or mononucleotide (Fig. 3) repetitive sequences. The frequency of 2-bp CA (Fig. 2) and 1-bp G (Fig. 3) frameshifts were enhanced 10-fold after incubation with 50 μM H2O2. In contrast, there was no detected increase in mutation frequency with control plasmids containing nonrepetitive sequences of the same length and reading frame, and present in the same location (Figs. 2 and 3). The induction of microsatellite instability by hydrogen peroxide appears to be independent of the SOS response, as a 17-fold increase in frameshift mutation of the (CA)11 construct occurs in the recA1 strain DH5αF′. The dose of H2O2 in these experiments is critical, as lower doses do not saturate the repair capacity of the cell, and higher doses result in extensive damage and plasmid loss. Although the concentration of H2O2 in our experiment (50 μM) is higher than the steady-state level in E. coli cells (0.15 μM) (25), the amount of plasmid DNA in our experiment (8.7 × 10−15 mol) is also considerably higher than the amount of DNA in a bacterial cell (4.5 × 10−24 mol). The levels of hydrogen peroxide utilized in our in vitro experiments are comparable to or lower than the levels of hydrogen peroxide reported to occur in several human tumor cell lines (26).
Figure 1.
Assay for the selection of oxidative damage-induced frameshift mutations in repetitive sequences. A modified version of the low copy number plasmid pBR322 was constructed to contain the tetracycline resistance gene and a carbenicillin resistance gene containing an out-of-frame microsatellite sequence. This construct renders bacteria tetracycline-resistant and carbenicillin-sensitive. Following treatment with H2O2, damaged plasmid was transformed into wild-type BL21 E. coli. Colony growth on tetracycline-containing plates was indicative of plasmid survival, whereas growth on tetracycline plus carbenicillin was indicative of a compensating frameshift mutation to restore the reading frame to generate functional β-lactamase. Mutation frequency is calculated as the number of carbenicillin-resistant mutants recovered per tetracycline-resistant survivor.
Figure 2.
Induction of dinucleotide frameshift mutations by hydrogen peroxide. Plasmid DNA (9 × 10−15 mol) containing either repetitive sequences or control nonrepetitive sequences within the β-lactamase gene was exposed to 1 μM or 50 μM H2O2. The 1 μM H2O2 treatment corresponded to a 40% survival of plasmid relative to untreated plasmid as measured by tetracycline-resistant transformants, whereas 50 μM H2O2 treatment corresponded to 20% survival. Dinucleotide (CA)11 or (CA)12 repeats and control nonrepetitive polylinker in +2 frame are shown.
Figure 3.
Induction of mononucleotide frameshift mutations by hydrogen peroxide. Treatment conditions were identical to those for dinucleotide repeats in Fig. 2. Mononucleotide (A)10 or (G)10 repeats and control nonrepetitive polylinker in +1 frame are shown.
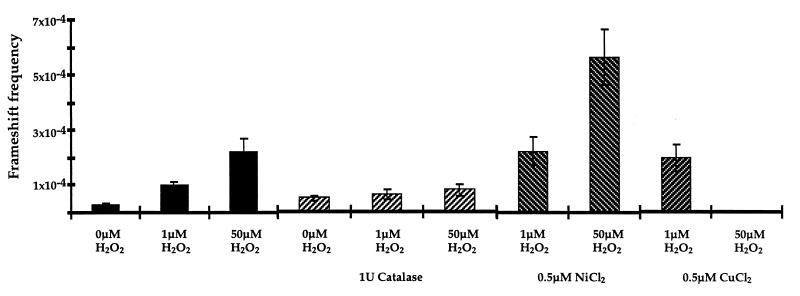
The frequency of induced frameshift mutations reflected the extent of exposure to agents that generate oxygen-free radicals. The presence of catalase, a hydrogen peroxide scavenger (27), during incubation with H2O2 reduced the frequency of induced mutations (Fig. 4). The addition of Cu2+ and Ni2+, which catalyze the reduction of H2O2 to hydroxyl radical (28), further increased the frameshift mutation frequency (Fig. 4). These results provide supporting evidence that the frameshift mutations resulted from the generation of reactive oxygen species. We have not observed any induction of frameshift mutations in repetitive sequences following exposure to other DNA damaging agents, such as UV radiation or N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) (unpublished results). Oxidatively damaged DNA was analyzed in the mismatch repair-deficient stain CM1. No increase in microsatellite instability relative to undamaged DNA was observed, but the interpretation of this experiment is not straightforward. The spontaneous mutation frequency in this strain is 2.55 × 10−2, potentially precluding the detection of further instability. In addition, doses of hydrogen peroxide that produced significant increases in microsatellite frameshift frequency in wild-type cells were lethal in the mismatch repair-defective cells, possibly due to an overwhelming mutational load.
Figure 4.
Modulation of frameshift frequency by exposure to oxygen radicals. Plasmid containing a +2 frame (CA)11 microsatellite sequence within the β-lactamase gene was exposed to 1 μM H2O2 or 50 μM H2O2 for 1 h at room temperature in the presence of either 1 unit catalase, 0.5 μM CuCl2, or 0.5 μM NiCl2. Addition of 0.5 μM Cu2+ or Ni2+ in the presence of 1 μM H2O2 reduced the recovery of tetracycline-resistant transformants to 30% relative to H2O2 treatment alone. In the presence of 50 μM H2O2 and 0.5 μM Cu2+, no survivors were detected.
Sequence Analysis of Frameshift Events Selected by Antibiotic Resistance.
To verify that the enhanced frequency of mutations resulting from oxidative damage was the result of frameshifts induced by processing of the damaged DNA in E. coli, we sequenced the microsatellite insert and adjacent 100 nucleotides. Because the entire plasmid was exposed to H2O2, and therefore damage to DNA was nontargeted, the compensating frameshift mutation could have occurred anywhere from the ATG to the first stop codon in β-lactamase. Sequencing verified that the carbenicillin-resistant phenotype resulted from compensating frameshift mutations in β-lactamase and indicated that all frameshifts occurred within microsatellite sequences (Table 2). In total, 68 constructs were sequenced, and all 68 contained mutations within the microsatellite insert that restored the reading frame to β-lactamase. One clone also contained a single base substitution of C·G → T·A within the repetitive sequence. Therefore, despite the fact that the oxidative DNA damage was nontargeted, all frameshift mutations recovered were located in the microsatellite sequence. The preference for frameshifts in microsatellite sequences compared with nonrepetitive sequences was calculated as the total mutation frequency divided by the number of nucleotides in the target region for each construct, and was >70-fold.
Table 2.
Sequence analysis of H2O2-induced carbenicillin-resistant mutants
| Repeat | Damage | Frameshift, bp | Inside repeat | Outside repeat |
|---|---|---|---|---|
| (CA)11 | H2O2 | +2 | 18/18 | 0/18 |
| (+2 frame) | −4 | |||
| (CA)12 | H2O2 | +2 | 7/10 | 0/10 |
| (−2 frame) | −4 | 3/10 | ||
| (CA)11 | H2O2/Cu2+ | −2 | 10/10 | 0/10 |
| (+2 frame) | +4 | |||
| (CA)11 | H2O2/Ni2+ | −2 | 10/10 | 0/10 |
| (+2 frame) | +4 | |||
| (A)10 | H2O2 | −1 | 10/10 | 0/10 |
| (+1 frame) | +2 | |||
| (G)10 | H2O2 | −1 | 10/10 | 0/10 |
| (+1 frame) | +2 |
DISCUSSION
A potential source of the large number of mutations produced during tumor progression is damage to DNA by endogenous reactive chemicals, and in particular, damage by reactive oxygen species (29–32). Endogenous oxidative damage to DNA is extensive and has been estimated by Ames and coworkers (32 and references therein) to account for 20,000 hits per cell per day in humans. Oxidative adducts appear to occur at a frequency that is one or more orders of magnitude higher than nonoxidative adducts (30–33). Cerutti (29) has argued that prooxidant states, in which the intracellular concentration of activated forms of oxygen is increased, contribute to tumor progression due either to deficient defense against oxidative damage or overproduction of reactive oxygen species. In accordance with this reasoning, oxygen radical-induced DNA damage has been implicated in the progression of human breast cancers (34). In addition, tumor cell lines corresponding to melanomas, neuroblastomas, colon carcinoma, and ovarian carcinoma have been shown to constitutively overproduce hydrogen peroxide at rates up to 0.5 nmol/104 cells/h, and accumulate hydrogen peroxide at levels as large as those produced by stimulated polymorphonuclear lymphocytes (26). We have investigated the possibility that oxidative damage is causally associated with microsatellite instability through the use of a selection-based assay.
Detection of frameshifts within repetitive sequences, and identification of the agents that produce them, is an issue of considerable interest, and several assays have been developed for this purpose. Cupples et. al. (15) have developed a series of five frameshift tester strains in which defined mononucleotide A or G, or dinucleotide CG repeats inactivate the lacZ gene of E. coli. Following treatment of E. coli with mutagens, or in the absence of mismatch repair, frameshift mutations occurring within the repetitive sequences restore lacZ reading frame, enabling selection for Lac+ phenotype. Two plasmid-based systems have been described for the detection of repetitive sequence instability in yeast (13, 14). In one system, the lacZ gene has been disrupted by GT repeats, and in another, the URA3 gene has been altered to contain in-frame GT repeats. Frameshift mutations are detected either by screening for blue yeast colonies on 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) plates for reversion to lacZ reading frame, or by forward mutation of URA3 to 5-fluoro-orotic acid resistance. Both of these assays have been used to demonstrate that microsatellite instability can result from mutations in mismatch repair genes, or, to a lesser extent, from mutations in pol3, the polymerase δ homolog (14). Tran et. al. (18) have inserted mononucleotide A repeats of various lengths within the LYS2 gene in the genome of Saccharomyces cerevisiae to produce a Lys− phenotype. In the absence of mismatch repair, or in the presence of pol3 mutations, frameshift mutations occur in longer repeats to restore a Lys+ phenotype. Fuchs and coworkers (17, 35) have created a (GC)3 run within the tetracycline-resistance gene on plasmid pBR322 of E. coli and demonstrated that frameshift mutations can be induced within the GC repeat to restore tetracycline resistance by treatment of either cells or plasmid DNA with frameshift mutagens. These assays have provided valuable insight into some of the factors that influence microsatellite instability.
We have sought to complement these sensitive and powerful selective assays through the introduction of an additional level of versatility. The assay used in this paper is similar to that described by Fuchs (17, 35). We have developed a selective assay for microsatellite instability that allows us to monitor large pieces of DNA in a plasmid for frameshift mutations. The demonstrated tolerance for large DNA inserts within the β-lactamase gene in this study, together with the introduction of a polycloning sequence, provides a facile method for studying repetitive and nonrepetitive sequences of differing size and sequence simply by ligation of oligonucleotides containing the desired sequence within the polylinker. Thus this system affords a flexibility not available with other methods. Furthermore, this systems offers the ability to directly clone microsatellite sequences amplified from cell lines or tumor samples into a selection-based assay for microsatellite instability. The majority of microsatellite loci analyzed in instability studies are on the order to 100 to 300 bp in length, which will be readily accommodated by our cloning system. The plasmid can subsequently be easily recovered for sequence analysis of microsatellite frameshift events. We have utilized this β-lactamase assay to study induction of frameshifts in repetitive sequences. Previous studies have established that agents which perturb the DNA duplex through intercalation or induction of bulky adducts can induce frameshifts in repetitive DNA sequences (15–17, 35–38). We have demonstrated with this assay that oxidative damage to DNA by hydrogen peroxide, which is not a classical frameshift mutagen, is sufficient to promote instability of repetitive sequences in the presence of wild-type DNA repair.
The results with H2O2 damage suggest that oxygen-induced DNA lesions can affect the faithful maintenance of microsatellite sequences. We therefore postulate that endogenous production of oxygen-free radicals in tumor cells may be a major factor in the observed instability of microsatellites in the absence of mismatch repair deficiency. The oxygen damage-induced microsatellite instability could occur as an early step in the development of cancer, because microsatellite instability has been associated with inflammatory but non-neoplastic settings. Reactive oxygen species induce damage to DNA in the form of base alterations, abasic sites, and strand breaks, any of which could contribute to the observed instability of repetitive sequences. The mechanism by which oxygen-induced DNA alteration promotes microsatellite instability needs to be identified. One mechanism could be pausing of DNA polymerase at sites of oxidative base damage to provide the opportunity for strand displacement misalignment of the growing strand at a different position on the template. However, we have observed oxygen-induced frameshifting with mononucleotide and dinucleotide microsatellites of different sequences, suggesting that a common lesion, such as a strand break, is more likely than the alteration of a specific nucleotide to contribute to instability. Within the context of a microsatellite sequence, strand scission by reactive oxygen species to produce a free DNA end could increase the probability of stand misalignment, leading to the generation of insertion or deletion loops. Thus, damage to DNA by reactive oxygen species could increase the formation of slipped-strand intermediates through the generation of single strand breaks in repetitive sequences.
Strand dissociation and realignment in nonrepetitive sequences are likely to be highly accurate, precluding the formation of slipped-strand intermediates that promote frameshifts. The observed preference for damage-induced frameshifts within microsatellite sequences could also result if the repair of lesions within repetitive sequences is less efficient than that in nonrepetitive sequences. There is some evidence that repetitive sequences may adopt a non-B DNA form in vivo (39). As a consequence of this altered structure, lesions within repetitive sequences may be less efficient substrates for DNA repair enzymes. The possible persistence of oxidative lesions within repetitive sequences could contribute to the increased frameshift frequency.
In conclusion, we have developed a selective and sensitive assay for frameshift mutations that has provided evidence that microsatellite instability can result from damage to DNA by oxygen free radicals. Such a finding may suggest that antioxidant therapy would be appropriate to delay the progression of tumors that exhibit microsatellite instability, particularly those that lack mutations in mismatch repair genes. Sensitive detection of microsatellite instability will make possible the identification of additional molecular processes and endogenous sources of DNA damage that influence microsatellite instability. This assay should have wide applicability to diagnostic and mechanistic questions relating to microsatellite instability in cancer development.
Acknowledgments
This paper is dedicated to the memory of Earl Benditt, whose unique insights into mutations and disease encourage our efforts. We thank Roel Schaaper, Michael Fry, Terry Newcomb, and Baek Kim for critical discussions. This work was supported by grants from the National Institute of Health (OIG CA39903-14), National Institutes of Health/National Institute on Aging Grant AG 01751-18 (to L.A.L.), National Institutes of Health Grant CA69732-02 (to A.L.J.). R.C. was supported by National Institutes of Health Grant R01 CA68124-04.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Loeb L A, Springgate C F, Battula N. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- 2.Nowell P C. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 3.Richards B, Zhang H, Phear G, Meuth M. Science. 1997;277:1523–1526. doi: 10.1126/science.277.5331.1523. [DOI] [PubMed] [Google Scholar]
- 4.Loeb L A. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 5.Loeb L A. Cancer Res. 1994;54:5059–5063. [PubMed] [Google Scholar]
- 6.Perucho M. J. Biol Chem. 1996;377:675–684. [PubMed] [Google Scholar]
- 7.Aaltonen L A, Peltomaki P, Leach F S, Sistonen P, Pylkkanen L, Mecklin J-P, Jarvinen H, Powell S M, Jin J, Hamilton S R, et al. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 8.Peinado M A, Malkhosyan S, Velazquez A, Perucho M. Proc Natl Acad Sci USA. 1992;89:10065–10069. doi: 10.1073/pnas.89.21.10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson A L, Loeb L A. Genetics. 1998;148:483–490. doi: 10.1093/genetics/148.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ionov Y, Peinado M A, Malkhosyan S, Shibata D, Perucho M. Nature (London) 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 11.Peltomaki P, Lothe R A, Aaltonen L A, Pylkkanen L, Nystrom-Lahti M, Seruca R, David L, Holm R, Ryberg D, Haugen A, et al. Cancer Res. 1993;53:5853–5855. [PubMed] [Google Scholar]
- 12.Levinson G, Gutman G A. Nucleic Acids Res. 1987;15:5323–5359. doi: 10.1093/nar/15.13.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson S T, Petes T D. Mol Cell Biol. 1992;12:2749–2757. doi: 10.1128/mcb.12.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strand M, Prolla T A, Liskay R M, Petes T D. Nature (London) 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 15.Cupples C G, Cabrera M, Cruz C, Miller J H. Genetics. 1990;125:275–280. doi: 10.1093/genetics/125.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hour T-C, Lee C-C, Lin J-K. Mutagenesis. 1995;10:433–438. doi: 10.1093/mutage/10.5.433. [DOI] [PubMed] [Google Scholar]
- 17.Maenhaut-Michel G, Janel-Bintz R, Samuel N, Fuchs R P P. Mol Gen Genet. 1997;253:634–641. doi: 10.1007/s004380050366. [DOI] [PubMed] [Google Scholar]
- 18.Tran H T, Keen J D, Kricker M, Resnick M A, Gordenin D A. Mol Cell Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imlay J A, Chin S M, Linn S. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 20.Bush K. Antimicrob Agents Chemother. 1989;33:259–263. doi: 10.1128/aac.33.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutchinson F. Prog Nucleic Acid Res Mol Biol. 1985;32:115–154. doi: 10.1016/s0079-6603(08)60347-5. [DOI] [PubMed] [Google Scholar]
- 22.Newcomb T G, Loeb L A. In: Oxidative DNA Damage and Mutagenesis. Jickoloff J A, Hoekstra M F, editors. Vol. 1. Totowa, NJ: Humana; 1997. pp. 1–18. [Google Scholar]
- 23.Brentnall T A, Chen R, Lee J G, Kimmey M B, Bronner M P, Haggitt R C, Kowdley K V, Hecker L M, Byrd D R. Cancer Res. 1995;55:4264–4267. [PubMed] [Google Scholar]
- 24.Brentnall T A, Crispin D A, Bronner M P, Cherian S P, Hueffed M, Rabinovitch P S, Rubin C E, Haggitt R C, Boland C R. Cancer Res. 1996;56:1237–1240. [PubMed] [Google Scholar]
- 25.Gonzalez-Flecha B, Demple B. J Biol Chem. 1995;270:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 26.Satrowski T P, Nathan C F. Cancer Res. 1991;51:794–798. [PubMed] [Google Scholar]
- 27.Halliwell B. Annu Rev Nutr. 1996;16:33–50. doi: 10.1146/annurev.nu.16.070196.000341. [DOI] [PubMed] [Google Scholar]
- 28.Fenton H J H. J Chem Soc. 1894;65:899–910. [Google Scholar]
- 29.Cerutti P A. Science. 1985;227:375–381. doi: 10.1126/science.2981433. [DOI] [PubMed] [Google Scholar]
- 30.Feig D I, Reid T M, Loeb L A. Cancer Res. 1994;54:1890s–1894s. [PubMed] [Google Scholar]
- 31.Ames B N, Shigenaga M K, Hagen T M. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beckman K B, Ames B N. J Biol Chem. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 33.Ames B N, Gold L S, Willet W C. Proc Natl Acad Sci USA. 1995;92:5258–5265. doi: 10.1073/pnas.92.12.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malins D C, Polissar N L, Gunselman S J. Proc Natl Acad Sci USA. 1996;93:2557–2563. doi: 10.1073/pnas.93.6.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuchs R P P, Bintz R. Environ Health Perspect. 1990;88:83–87. doi: 10.1289/ehp.908883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shearman C W, Loeb L A. J Biol Chem. 1983;258:4477–4484. [PubMed] [Google Scholar]
- 37.Shelton M L, DeMarini D M. Mutat Res. 1995;327:75–86. doi: 10.1016/0027-5107(94)00186-9. [DOI] [PubMed] [Google Scholar]
- 38.Malia S A, Vyas R R, Basu A K. Biochemistry. 1996;35:4568–4577. doi: 10.1021/bi9525132. [DOI] [PubMed] [Google Scholar]
- 39.Aranda A, P’erez-Ort’in J E, Behnam C J, Del-Olmo M L. Yeast. 1997;13:313–326. doi: 10.1002/(SICI)1097-0061(19970330)13:4<313::AID-YEA93>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]