Abstract
Human cytomegalovirus (CMV) US28 (and the related open reading frame [ORF] US27) are G-protein-coupled receptor homologs believed to play a role in viral pathogenesis. In vitro, US28 has been shown to bind and internalize ligands, as well as activate intracellular signaling in response to certain chemokines, and to initiate the migration of smooth muscle cells to chemokine gradients. To assess the role of US28 in vivo, we examined the rhesus model and sequenced and characterized the rhesus CMV US28 locus. We found that rhesus CMV carries five tandem homologs of US28, all widely divergent from US28 and from each other. By reverse transcription-PCR and Northern analysis, we demonstrated expression of these ORFs in infected cells. With stable cell lines expressing these ORFs, we analyzed the homolog's binding and signaling characteristics across a wide range of chemokines and found one (RhUS28.5) to have a ligand binding profile similar to that of US28. In addition, we localized US28 and the rhesus CMV homolog RhUS28.5 to the envelope of infectious virions, suggesting a role in viral entry or cell tropism.
Human cytomegalovirus (HCMV) is a large double-stranded DNA herpesvirus that uses a broad array of strategies to facilitate its lifelong association with its host. In normal hosts, a complex interplay between the virus and the immune system results in an equilibrium in which neither the host nor the pathogen overpowers the other. This allows HCMV to establish an enduring latent infection within hemopoietic cells in >50% of the world's population (14) and enables transmission of periodically reactivated virus to new hosts. However, in immunocompromised individuals, such as transplant recipients, human immunodeficiency virus-seropositive patients, and developing fetuses, the balance can be tipped in favor of the virus, resulting in morbidity and mortality. For this reason, it is important to better understand viral pathogenic mechanisms to aid in the development of viral vaccines and therapeutics.
HCMV has long been known to use various mechanisms to regulate the immune environment in which it lives. US2, US3, US6, and US11 encode proteins that inhibit antigen presentation through major histocompatibility complex type I (16, 17, 54; reviewed in reference 15). UL18 and UL40 regulate the susceptibility of virus-infected cells to lysis by NK cells (23, 47). The virus also up-regulates the endogenous complement regulatory proteins CD46 and CD55 (13), CD59 and CD55 are incorporated into virions (40), and the virus induces Fc receptor expression (19, 25). Further, both HCMV (22) and rhesus CMV (RhCMV) (41) were recently shown to have biologically active homologs of interleukin-10.
In addition, HCMV expresses genes with homology to chemokines and their receptors. Chemokines are a class of cytokines important in the regulation of motility and activation of immune cells and characterized by the spacing of the their conserved cysteine motif into CC, CXC, CX3CR, and C subgroups (29). HCMV has a potent CXC chemokine homolog, UL146, as well as an uncharacterized CXC chemokine homolog, UL147 (31, 35). Additionally, HCMV expresses four genes with homology to chemokine receptors, UL33, UL78 (26), US27, and US28 (10, 30). US28 is the best characterized of these in vitro. This gene is transcribed early in infection; has been shown to bind a range of CC chemokines, including RANTES/CCL5, MCP-1/CCL2, MCP-3/CCL8, MIP1-α/CCL3, and MIP1-β/CCL4 (4) and viral chemokine homologs such as human herpesvirus 8 vMIP-2 (20); and has a strong affinity for the surface-bound chemokine fractalkine (CX3CL1) (21). Following binding of ligands, the receptor can be internalized and recycled to the surface (11) and is able to initiate intracellular signaling through coupling to G proteins (30). Other studies have suggested that US28 is able to impart constitutive signaling capacities on both infected and transfected cells, although cell type specificity and the presence of other viral or virus-induced factors may be necessary for this activity (7, 28). In in vitro assays, expression of US28 imparts chemotactic properties to vascular smooth muscle cells (44) and so may play a role in exacerbation of CMV-associated atherosclerotic disease. Finally, US28 has been associated with entry of HIV and cell-cell fusion (32, 33).
However, the role of US28 in the context of viral infection in vivo remains unclear. US28 has no known homologs in CMVs of lower species, and the species specificity of CMV prohibits examination of its function in animal models. To address this difficulty, we have sequenced the RhCMV genome and discovered and characterized US28 homologs in this virus. We have elucidated a complex locus containing five contiguous homologs of US28 and have examined their properties.
MATERIALS AND METHODS
Viruses, cells, and chemokines.
HEK293 cells (American Type Culture Collection, Manassas, Va.), low-passage (<15 passages) human dermal foreskin fibroblasts (HDF; Clonetics), or rhesus dermal fibroblasts (RhDF) (18) were cultured in Dulbecco's minimal essential medium (JRH) supplemented with 10% fetal bovine serum (HyClone) (39). HCMV strains Toledo (passage 8) (34) and TolΔ28 (48) and RhCMV strain Rh68.1 (American Type Culture Collection) (2) were propagated as previously described (39). Recombinant chemokines were obtained from R&D Systems.
Virion purification and analysis.
Virions and dense bodies from infected cell supernatants were purified over potassium tartrate-glycerol gradients as previously described (45). Virions (from the upper band) or dense bodies (from the lower band) were collected with a syringe. Purified virions or dense bodies were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue (Pierce) or transferred to nylon for Western blotting by standard techniques (37) and stained with monoclonal antibodies (MAbs) to β-actin (Sigma) or HCMV pp65 (Rumbaugh-Goodwin Institute) and then with goat anti-mouse horseradish peroxidase. The label was detected by enhanced-chemiluminescence assay (Amersham).
Genome sequencing and analysis.
RhCMV virions were purified as described above, and viral DNA was isolated by standard techniques. The resulting suspension was provided to Genome Therapeutics for sequencing. By hydroshearing, a shotgun library was created with inserts of about 3,000 bp. Individual clones were sequenced by ABI Prism BigDye terminator chemistry. The ∼220-kb genome was covered to an average accuracy of 6× sequence. Individual reads were assembled into contiguous fragments. Homologs of HCMV genes were elucidated by using the BLAST X and Clustal algorithms (1). Sequences were further analyzed with the BioNavigator bioinformatic program set (Entigen Corp.). Protein alignment was performed with the SeqVu program (Garvan Institute).
Northern blot analysis and reverse transcription (RT)-PCR.
RhDF were infected with Rh68.1 at a multiplicity of infection of 3 or mock infected, and total cellular RNA was collected at 2, 24, 48, and 72 h postinfection (hpi) with the RNeasy mini kit in accordance with the manufacturer's (Qiagen) instructions. Infected cells were also treated throughout infection with the viral polymerase inhibitor phosphonoformic acid (PFA) at 660 μM (200 μg/ml), and RNA was collected at 72 hpi. Northern blot analysis and hybridization were performed at 60oC with the NorthernMax kit in accordance with the manufacturer's (Ambion) instructions with 10 μg of RNA per lane. 32P-labeled probes spanning the whole of each open reading frame (ORF) were produced by random primed labeling of PCR products produced with the primers in Table 1 (for primer numbering, see Fig. 2A; the numbers represent 3′ positions in the sequence with GenBank accession no. AF539641). After hybridization, blots were exposed to Kodak X-Omat film.
TABLE 1.
Primers used for PCR
| Primer sequence | Designation | Map nucleotide | ORF |
|---|---|---|---|
| GCTGAAGCGGCCGCTATGAATAACACATCTTGCAACTTC | 1 | 3598 | RhUS28.1 |
| CTCGCGAATTCCACACAGACCACATGTAC | 2 | 4630 | RhUS28.1 |
| GCTGAAGCGGCCGCATTCAACATGACCAACGCCGG | 3 | 2514 | RhUS28.2 |
| CTCGAGAATTCGCATTTCCGTGGATTCG | 4 | 3539 | RhUS28.2 |
| GCTGAAGCGGCCGCCATGACCAACACTAAC | 5 | 1366 | RhUS28.3 |
| ATCGCGAATTCGAGTCTTTTGTGAGCC | 6 | 2401 | RhUS28.3 |
| GCTGAAGCGGCCGCTATGAATTCGAGCCAGCAC | 7 | 37 | RhUS28.4 |
| CTCGAGAATTCGTACGCGACTAAGACAGAG | 8 | 1034 | RhUS28.4 |
| GCTGAAGCGGCCGCAAAGATGACTACCACCAC | 9 | 4752 | RhUS28.5 |
| CTCGAGAATTCATAACCTAGCACCTCCCC | 10 | 6255 | RhUS28.5 |
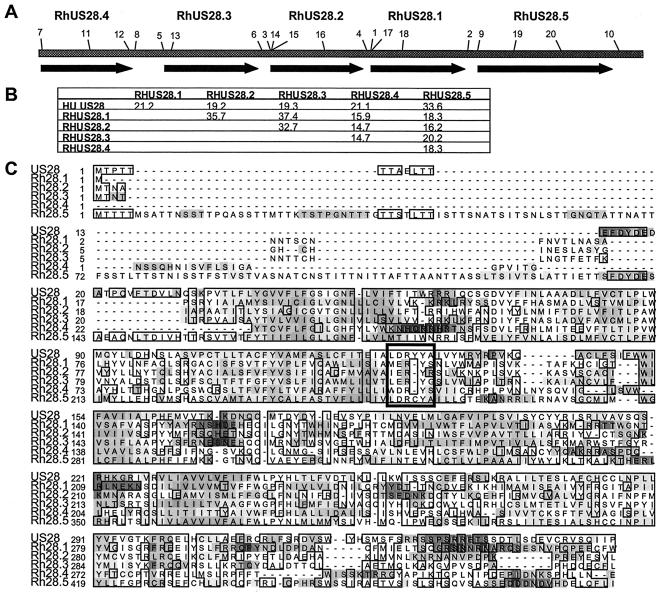
FIG. 2.
(A) Scale diagram showing RhUS28 ORFs. Primers used in this study are numbered. (B) Percent identity comparisons of RhCMV and HCMV US28 homologs by Clustal analysis. (C) Alignment of RhCMV and HCMV US28 homologs. Shaded regions indicate regions of hydrophobicity and hydrophilicity. Boxes indicate regions of amino acid similarity. The putative DRY box motif is indicated by the heavy box.
RT-PCR was performed on the above RNA template, including the PFA-treated sample, with the GeneAmp RNA PCR core kit in accordance with the manufacturer's (Roche) instructions, with amplification for 30 cycles and an annealing temperature of 55°C. RNA was primed with a dTTT primer, and cDNA was primed with the intragenic primers in Table 2 (see Fig. 2A). A control reaction mixture was prepared in which reverse transcriptase was included or omitted and run with primers to the rhesus major immediate-early region. PCR products were separated on a 2% agarose gel.
TABLE 2.
Intragenic primers used for RT-PCR
| Primer sequence | Designation | Map nucleotide |
|---|---|---|
| AGTGCACCTGTGCTCCTTC | 11 | 547 |
| GGAGCGATATTCCAACTTA | 12 | 977 |
| CAGCGCCTACACTGTACTC | 13 | 1429 |
| GTGACAGTGTCCGGCGTT | 14 | 2507 |
| CAACGCCGGACACTGTCA | 15 | 2506 |
| TGGCTATTAACAAGGTCGT | 16 | 3077 |
| ATGTACAGCATTGTTATCT | 17 | 3647 |
| CTAAGTTGGAATATCGCTC | 18 | 3914 |
| TACCATTGAAACCACATC | 19 | 5128 |
| TGGTTCATACTCTGCTAC | 20 | 5639 |
| TCGGAGGAGCACCATAGAAGGTACC | MIE UP | |
| AAGGCCCAGTAGAAGGTCCTCCAGAAGTCC | MIE LOW |
DNA cloning and cell lines.
All restriction endonucleases were obtained from New England Biolabs. PCR was performed with an annealing temperature of 55°C and Taq master mix in accordance with the manufacturer's (Qiagen) instructions. ORFs Rh28.1 to -5 were individually cloned into pIRES-puro (Clontech), which was modified to express an in-frame N-terminal FLAG epitope tag (pPF IRES-puro), by restriction endonuclease digestion of PCR products and vector and cloned by standard techniques. HCMV US28 cloned into the pCDNA3 vector (Invitrogen) was also used (30). Primers for each ORF (as detailed in the description of Northern blot analysis) were designed to encode 5′ NotI and 3′ EcoRI restriction sites (underlined). Inserts were sequenced after insertion into all vectors (Biotech Core) to ensure sequence identity and fidelity.
To make stable cell lines expressing these ORFs, HEK293 cells were transfected by electroporation, cultured with 3 μg of puromycin per ml (800 μg of G-418 per ml for the HCMV US28 line), and isolated by limiting dilution. Lines were expanded and then stained with anti-FLAG M1 antibody (Sigma) or an isotype control, followed by goat anti-mouse-phycoerythrin, and analyzed by flow cytometry. High-expression lines were amplified and maintained under selection. For transient transfections, cells were harvested 48 h after transfection by electroporation and analyzed for FLAG expression as described above.
Binding analyses and internalization assays.
Virions, infected fibroblasts, or transfected HEK293 cells were incubated at 4°C for 3 h with ∼0.08 nM 125I-labeled fractalkine (Pharmacia Biotech) alone or with unlabeled fractalkine in binding buffer (25 mM HEPES, 140 mM NaCl, 1 mM CaCl2, 5 mM MgCl2, 0.2% bovine serum albumin, pH 7.1). Cells were aspirated onto polyethyleneimine-treated GF/C glass fiber filters (Packard) with a cell harvester (Packard) and washed twice with wash buffer (25 mM HEPES, 500 mM NaCl, 1 mM CaCl2, 5 mM MgCl2, pH 7.1). Scintillant was added to the dried filters, and counts per minute were measured on a Packard Topcount scintillation counter. For Displacemax (6), cells incubated in the presence of 125I-labeled fractalkine were interrogated in parallel as described above with a broad array of unlabeled chemokine competitors (2 μg/ml) to assess their ability to displace radiolabeled fractalkine.
For internalization assays, HEK293 transfectant cells were harvested, resuspended in binding buffer, and seeded to 96-well plates at 100,000 cells per well on ice in the presence or absence of an inhibitor of US28 and RhUS28.5 (C0232391). Tracer (∼0.08 nM 125I-labeled fractalkine plus 1 nM unlabeled fractalkine) was added to wells, and parallel plates were incubated at 4 or 37°C and then harvested at indicated time points and counted as described above.
RESULTS
HCMV and RhCMV express fractalkine binding receptors.
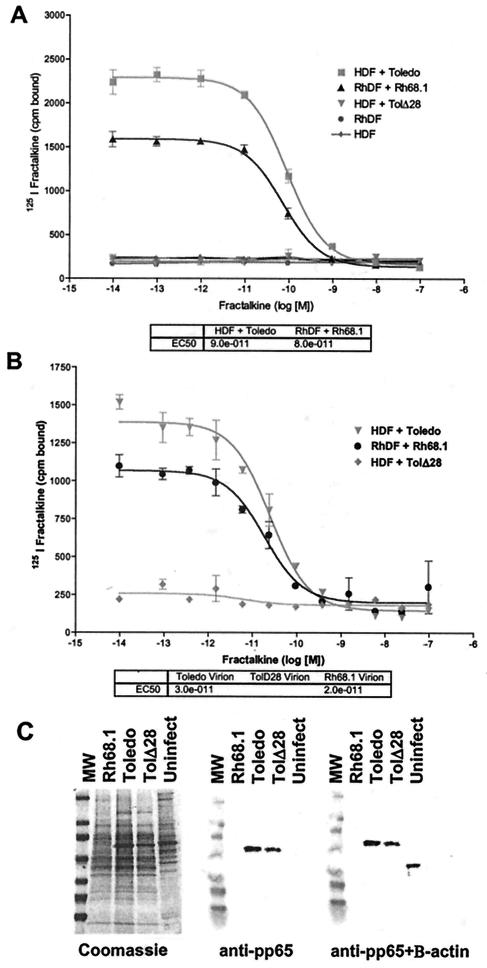
To determine if RhCMV expresses a fractalkine binding receptor with properties similar to those of HCMV US28, RhCMV-infected cells and purified virions were assessed in competitive radiolabeled ligand binding assays. HDF were infected at an multiplicity of infection of 3 with HCMV strain Toledo or TolΔ28 (48), and RhDF were infected with RhCMV strain Rh68.1 and then harvested at 72 hpi. Infected and mock-infected cells were incubated with 125I-labeled fractalkine in the presence of increasing amounts of unlabeled fractalkine competitor, and then the free label was washed away and the cell- or virion-associated label was evaluated. Strain Toledo-infected HDF strongly bound the labeled fractalkine with a 50% infective concentration (IC50) of 90 pM, while strain TolΔ28, with US28 deleted, and mock-infected cells showed no binding to this label. Similarly, Rh68.1-infected RhDF showed strong binding to the labeled fractalkine, with an IC50 of 80 pM (Fig. 1A). This suggests that RhCMV has a fractalkine binding homolog of HCMV US28 that is expressed on the surface of infected cells.
FIG. 1.
Competitive binding analysis of infected fibroblasts (A) or purified virions (B). Mock-infected or infected cells or virions were contacted with 125I-labeled fractalkine in the presence of increasing amounts of unlabeled fractalkine in triplicate. The results shown are representative of at least three assays. Error bars indicate 2 standard deviations. IC50s calculated by single-site competitive analysis are shown in the tables. (C) Purified virions or uninfected cell lysate separated by SDS-4 to 20% PAGE and stained with Coomassie blue (left). Protein was transferred to nitrocellulose and stained with an anti-HCMV pp65 MAb (center). Blots were subsequently stained with an anti-β actin MAb. MW, molecular weight markers.
Next, virions and dense bodies were purified from supernatants of infected cultures at 96 hpi by potassium tartrate-glycerol density gradient centrifugation. To confirm that purified virions were free of contaminating cellular proteins, we performed SDS-PAGE. Virions separated by SDS-PAGE were stained with Coomassie blue or analyzed by Western blotting. Rh68.1, Toledo, and TolΔ28 virions showed patterns characteristic of CMV, although the pp65 band prominent in HCMV virions was reduced in RhCMV (Fig. 1C, left). When stained with antibody to HCMV pp65, strains Toledo and TolΔ28 had a reactive band at about 65 kDa, but Rh68.1 did not, presumably because of failure of the anti-HCMV pp65 antibody to recognize the rhesus counterpart (Fig. 1C, center). Analysis of the RhCMV genome at this locus (GenBank accession numbers: rhesus, AAL08224; human, NP040018) shows only around 40% sequence identity at this locus, so the failure of the HCMV-directed antibody to recognize this protein is not surprising. Further staining of the blot with an antibody to the β-actin protein abundantly found in fibroblasts reacted only with the uninfected cell lysate, demonstrating an absence of contaminating cellular protein in the virion preparations (Fig. 1C, right). Virions were processed for binding analysis as described above and showed binding similar to that of infected cells. Strain Toledo virions bound the labeled fractalkine with an IC50 of 30 pM, while the virions with US28 deleted showed no binding to this label. Strain Rh68.1 virions bound fractalkine with an IC50 of 20 pM (Fig. 1B). Dense bodies isolated from the lower band of potassium tartrate-glycerol density gradients showed binding characteristics similar to those of the virions (data not shown). This is the first demonstration of virion localization of US28 and suggests a role for US28 in binding or other early events in cell infection.
Analysis of the RhCMV US28 locus.
In order to further characterize the rhesus US28 homolog, the rhesus viral genome was sequenced as described in Materials and Methods. Analysis of the genome revealed that the RhCMV genome is highly homologous to the HCMV genome. The genomes are colinear and have positional and sequence homologs of most ORFs. In addition, the structural arrangements of the viruses are similar, with coding regions divided into short and long regions. This is dissimilar to the rodent CMV genomes, which have a single contiguous coding region and no known homologs of US28 (36), suggesting a comparatively greater phylogenetic separation of these genomes from HCMV.
BLAST analysis revealed a series of five juxtaposed ORFs with positional and sequence homology to HCMV US28, designated RhUS28.1, RhUS28.2, RhUS28.3, RhUS28.4, and RhUS28.5 (Fig. 2A). Clustal analysis showed wide divergence in the sequences of these ORFs from each other, as well as from HCMV US28 (Fig. 2B). However, all had the characteristics of seven-transmembrane proteins, including conserved alternating regions of hydrophobicity and hydrophilicity, and showed significant sequence similarity (Fig. 2C). All of the ORFs also contained a recognizable “DRY box” (boxed), a motif important to efficient interactions of G-protein alpha subunits with G-coupled protein receptors (GPCRs) (53). However, the cysteine insertion in the DRY motif of RhUS28.5 is not normally associated with functional GPCRs and may underlie the failure of this ORF to facilitate Ca2+ mobilization in our analyses. Additionally, the RhUS28.5 ORF had an extended N terminus, adding approximately 130 amino acids compared to US28 and most known chemokine receptors. To assess whether any of the putative transcripts were spliced, we sequenced PCR-amplified cDNAs generated by poly(T)-primed RT-PCR of whole-cell RNA from infected cells at 48 hpi. All transcripts were demonstrated to be unspliced (data not shown).
Kinetics of expression of RhUS28 genes.
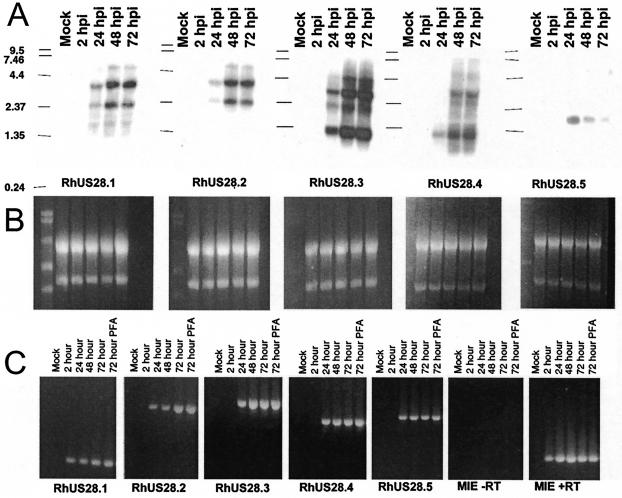
To determine which, if any, of the RhUS28 ORFs are expressed in infected cells, Northern analysis was performed. RNA was extracted from infected RhDF at 2, 24, 48, and 72 hpi, as well as from mock-infected cells. RNA was separated on formaldehyde gels and transferred to nylon and then hybridized with probes specific for each of the five RhUS28 ORFs. Loading consistency was confirmed by ethidium bromide imaging of rRNA (Fig. 3B). None of the probes hybridized to RNA from mock-infected cells, but the RhUS28.3 and RhUS28.4 probes hybridized predominantly to two bands of approximately 1.3 and 3.0 kb (Fig. 3A). These bands appeared to be of the same size, suggesting that these ORFs are expressed in a single transcription unit. However, sequence analysis revealed an abundance of poly(A)-like sequences in this region, so that overlapping transcripts may account for this banding pattern. Isolation of these transcripts, as well as 5′ mapping, is needed to more fully understand the complex transcription patterns within this locus. The bands were observed at 24, 48, and 72 hpi, suggesting early transcription. RT-PCR confirmed these results (Fig. 3C). Similarly, the RhUS28.1 and RhUS28.2 probes hybridized predominantly to transcripts of 2.4 and 3.5 kb with kinetics similar to those of RhUS28.3 and RhUS28.4 by Northern analysis and RT-PCR (Fig. 3A and C). The RhUS28.5 probe hybridized to a single transcript of 1.8 kb detected at 24 hpi by Northern analysis and RT-PCR that apparently declines in abundance at later times (Fig. 3A and C). All transcripts showed normal expression by RT-PCR in the presence of the viral DNA polymerase inhibitor PFA and so may be classified as early or immediate-early transcripts (Fig. 3C). In addition, RNA cycled in the presence or absence of reverse transcriptase was annealed with primers in different exons of the RhCMV major immediate-early region and confirmed the absence of contaminating DNA (Fig. 3C). Despite the apparent redundancy of the RhCMV US28 locus compared to the HCMV US28 locus, all ORFs were shown to be transcribed at early or immediate-early times, suggesting a role for each in viral replication or pathogenesis. The complexity of transcription in this region necessitates further detailed study to more fully understand the transcription unit.
FIG. 3.
(A) Northern blot analysis of infected or mock-infected cell RNA from the indicated times after infection. Blots were hybridized with 32P-labeled probes to the indicated ORF. (B) Normalization of RNA loading was confirmed by visualization of rRNA with ethidium bromide. (C) For RT-PCR, RNA was poly(dT) primed and reverse transcribed and then cDNA was amplified with ORF-specific primers. RNA collected from cells infected in the presence of PFA is included. Reverse transcriptase-negative control reaction mixtures were amplified with primers to the RhCMV major immediate-early region. The values on the left of panel A are molecular sizes in kilobase pairs.
Functional analysis of the RhUS28 locus.
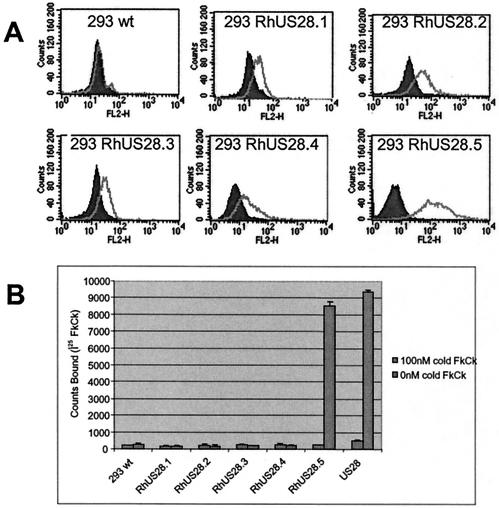
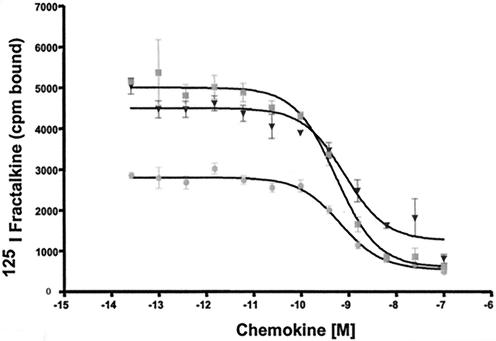
To determine whether these ORFs have functional homology to US28, we examined the ability of cells expressing the ORFs to bind the CX3CL1 chemokine fractalkine, a hallmark of US28 activity. Stable HEK293 cell lines expressing FLAG-tagged RhUS28.1, RhUS28.2, and RhUS28.5 were produced by selection of transfectants with puromycin. HEK293 cells transiently expressing RhUS28.3 and RhUS28.4 were examined, as extended propagation of these cells in the presence of puromycin selected for populations with decreased surface expression of the ORFs. Surface receptor presence on all cells was assessed on the day of assay by flow cytometry with anti-FLAG antibodies and was shown to be robust (Fig. 4A). Cells were incubated with radiolabeled fractalkine in the presence or absence of 100 nM unlabeled fractalkine competitor. Only RhUS28.5, but not the other US28 homologs, was able to bind the fractalkine label. As expected, a control line expressing US28 also showed robust binding to fractalkine (Fig. 4B). RhUS28.5 or US28 transfected HEK293 cells were next subjected to a competitive binding analysis in the presence of increasing concentrations of unlabeled fractalkine or vMIPII, a chemokine produced by human herpesvirus 8 that also binds strongly to US28 (20). Single-site competitive analysis of the results gave an IC50 for RhUS28.5-transfected HEK293 of 600 pM for fractalkine and 800 pM for vMIPII, while US28-transfected HEK293 cells had a similar affinity for fractalkine of 700 pM (Fig. 5). The 10-fold difference in binding affinity between the infected cells or virions and the transfected cells may be a result of conformational changes in the receptor induced by the cellular environment or the presence of other viral ORFs in the infected cells. Alternatively, it is possible that the N-terminal FLAG tag may affect the receptor characteristics. However, in our laboratory, we have observed no differences in the characteristics of tagged and untagged US28 and other chemokine receptor-expressing cell lines (data not shown).
FIG. 4.
(A) Stable or transiently transfected HEK293 cells expressing FLAG-tagged RhUS28 ORFs were stained with an anti-FLAG MAb, and expression was analyzed by flow cytometry. (B) Transfected cells were incubated with 125I-labeled fractalkine (FkCk) in the presence or absence of 100 nM unlabeled fractalkine. Error bars indicate 2 standard deviations. The results shown are representative of at least three assays. wt, wild type.
FIG. 5.
Stably transfected HEK293 cells expressing RhUS28.5 or US28 were incubated with 125I-labeled fractalkine in the presence of increasing amounts of unlabeled fractalkine or vMIPII (▪, RhUS28.5 plus fractalkine; ▾, RhUS28.5 plus vMIPII; •, US28 plus fractalkine). IC50s calculated by single-site competitive analysis are indicated in the table at the bottom. Error bars indicate 2 standard deviations. The values shown are representative of at least three assays.
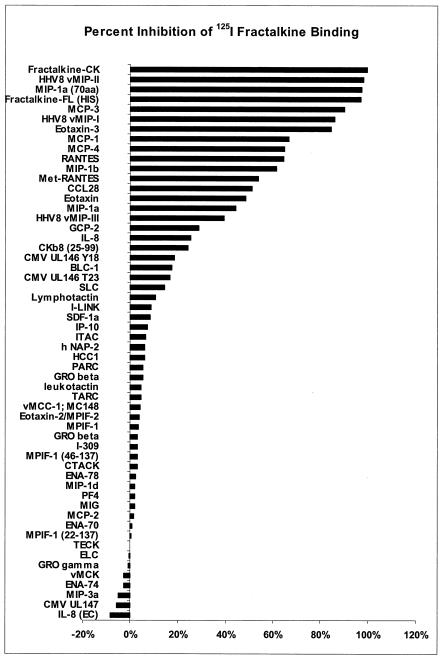
To determine a comprehensive binding profile of RhUS28.5, a stable cell line expressing this ORF was incubated with 125I-labeled fractalkine in the presence of a wide range of human and viral chemokine elements at ∼200 nM. Results are expressed as percent inhibition of binding of 125I-labeled fractalkine compared to that with no competitor (Fig. 6). RhUS28.5 was determined to be a promiscuous receptor able to bind a range of CC chemokines, as well as the CX3CL1 molecule, fractalkine, and certain viral chemokines. The binding profile of the rhesus homolog RhUS28.5 was essentially identical to that of US28 (data not shown), further evidencing its similarity to its HCMV counterpart. A preponderance of the most active competitors are from the family of ligands that bind the proinflammatory receptors CCR1, -2, and -5, and US28 shows the strongest homology to CCR1. However, the ORFs also bound ligands to CCR3, CX3CR, and a number of viral chemokines with strong affinity but were generally unreactive with CXC chemokines.
FIG. 6.
Stably transfected HEK293 cells expressing RhUS28.5 were incubated with 125I-labeled fractalkine in the presence of 56 unlabeled human and viral chemokine elements at ∼200 nM. Results are expressed as percent inhibition of labeled fractalkine binding. Each bar represents one chemokine element. The values shown are representative of at least two assays in triplicate.
Internalization of ligand by RhUS28.5.
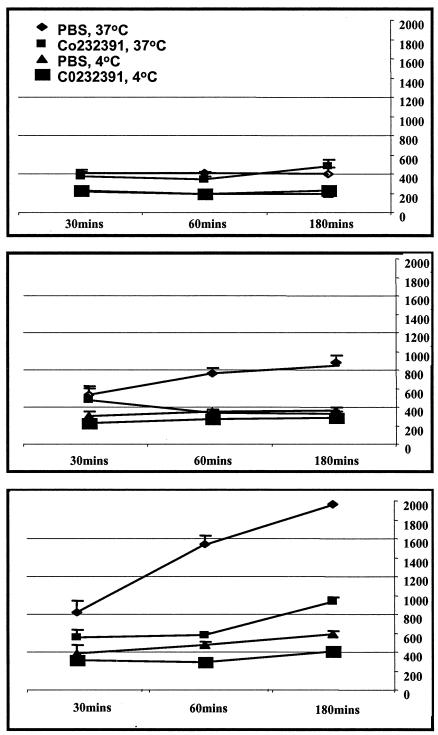
To determine whether RhUS28.5 has further functional similarities to US28, HEK293 cells stably transfected with RhUS28.5 were next analyzed for mobilization of intracellular calcium. Despite high surface expression of the receptor and binding to fractalkine, these cells were not induced to mobilize calcium in response to known ligands (e.g., fractalkine and vMIPII; results not shown). Control HEK293 cells stably transfected with US28 showed strong signaling in these experiments. To determine if RhUS28.5 induces an intracellular signal through pathways we were not able to detect in our assay, we next examined internalization of the receptor in response to ligand binding, a surrogate for functional stimulation of a receptor. Cells were incubated with radiolabeled fractalkine at 37 or 4°C, and the amount of label associated with the cells over time was assayed. Wild-type HEK293 cells showed little interaction with the labeled fractalkine at either temperature. US28-transfected HEK293 cells showed robust time-dependent association with the label at 37°C, while only background levels of surface-bound ligand were observed at 4°C, a temperature inhibitory to internalization. This indicates that label bound to surface receptors was carried into the cell cytoplasm as receptors were internalized at 37°C and recycled to the surface over time (Fig. 7). This is in agreement with previously published results (4, 21). The phenotype of the RhUS28.5-transfected HEK293 cells was intermediate in repeated assays. To demonstrate that internalization was mediated by US28 or RhUS28.5, cells were assayed in the presence of an inhibitor of fractalkine binding to RhUS28.5 and US28, C0232391, which has a fractalkine binding IC50 of 140 nM for RhUS28.5 and 300 nM for US28 and was added to a final concentration of 750 nM in these experiments. In the presence of this inhibitor, incorporated label was held to background levels for all cells and temperatures (Fig. 7). These results suggest that while binding of fractalkine to US28 induces receptor internalization and actively modulates intracellular signaling pathways, RhUS28.5 has a more passive phenotype in these assays.
FIG. 7.
Wild-type or stably transfected HEK293 cells expressing RhUS28.5 or US28 were incubated with 125I-labeled fractalkine at 4 or 37°C in the presence or absence of an inhibitor of RhUS28-US28-fractalkine binding (C0232391) for the periods indicated. The top panel shows wild-type HEK293, the middle panel shows HEK293 cells expressing RhUS28.5, and the bottom panel shows HEK293 cells expressing US28. The values shown are representative of at least four assays. The x axis represents time in minutes, and the y axis represents counts per minute of internalized radioactivity. PBS, phosphate-buffered saline.
DISCUSSION
The herpesviruses can infect a broad range of hosts, ranging from mollusks, fish, poultry, and livestock to higher mammals and humans. Among these are the cytomegaloviruses, ubiquitous pathogens with representatives characterized in an array of mammals including mice, rats, guinea pigs, nonhuman primates, and humans. In humans, CMV is involved in numerous pathologies in the immune compromised but has its greatest impact on human health through congenital infection of neonates resulting in neurological malformation, deafness, learning disabilities, and death, making control of congenital infection a top priority according to the Institute of Medicine (42). However, the study of the molecular mechanisms of pathogenesis presents a quandary. The double-stranded DNA genome has one of the largest viral coding capacities known and can only be propagated for study in a narrow cell range in vitro. Many genes have been shown to be dispensable in culture and are thought to be involved in cell tropism, pathogenesis, or immune interaction in the natural environment of the host. Hence, to more fully understand the importance of such genes and their potential as targets for manipulation in vaccines and therapeutics, a relevant in vivo model is essential. However, the extreme species specificity of CMVs hampers the study of HCMV in lower mammals. Likewise, genome divergence and differences in the life cycles of rodent CMVs in their hosts limit the utility of these as models for the study of HCMV pathogenesis. As a solution to this problem, recent work has focused on the use of nonhuman primates as models for HCMV. Aspects of pathogenesis are broadly conserved between humans and rhesus macaques (24, 46), and recent sequencing of the chimpanzee CMV (accession no. NC003521), as well as our sequencing of the RhCMV genome, has revealed good conservation of both individual genes and genome structure between HCMV and nonhuman primate CMVs. To this end, we have begun to characterize the RhCMV US28 locus in order to facilitate elucidation of its role in viral persistence and disease.
As HCMV US28 has been shown to strongly bind fractalkine (21), we first assessed fractalkine binding by RhCMV-infected cells. Unlike rat and murine CMV-infected cells, RhCMV-infected cells demonstrated strong binding to fractalkine, suggesting the presence of a US28 homolog in the RhCMV genome. Additionally, we demonstrated US28-mediated fractalkine binding by both virions and dense bodies of these viruses. This is the first demonstration of localization of US28 to viral particles, which may mediate its delivery to the membranes of infected cells at the earliest times in viral infection. It has been suggested that US28 may initiate intracellular signaling events through coupling to G proteins, and such early modulation of signaling events may be crucial in the establishment of an environment favorable for viral replication. Alternatively, virion US28 may modify cell tropism by binding to cell surface proteins such as fractalkine, thereby facilitating viral absorption and entry. Cytokine-activated endothelial cells have been shown to support viral replication (38, 51), and surface expression of fractalkine in these cells is also up regulated in response to cytokines (12). Binding of plasma-borne virions or infected cells to fractalkine on activated endothelial cells lining the vasculature may play a role in dissemination from initial sites of mucosal infection and provide a molecular basis for observations of CMV's role in cardiovascular disease (8, 43) and transplant-related vascular disease (9).
Sequencing of the RhCMV genome revealed a complex locus with five potential US28 homologs. While this gene is absent in rat and murine CMVs (36, 49) and has not been detected in guinea pig CMV, HCMV has two adjacent GPCR homologs, US27 and US28, speculated to have resulted from a gene duplication event. This arrangement was recently shown to be identical in the chimpanzee CMV genome (accession no. NC003521). The significance of the apparent redundancy in the RhCMV US28 locus is unclear. Previous work by Martin (27) revealed five adjacent GPCR homologs similar to US28 and the RhCMV ORFs in the so-called stealth virus genome. This virus is widely believed to be an isolate of African green monkey CMV.
Transcription analysis by RT-PCR showed that all five RhCMV ORFs were expressed in viral infection from 24 hpi on. These results were confirmed by Northern blotting of transcripts with probes specific to each ORF. Sequencing of cDNAs from cells infected for 48 h demonstrated that all transcripts were unspliced. RhUS28.1 and RhUS28.2 appeared to be expressed on common transcripts, and transcription increased at 48 hpi. Similarly, RhUS28.3 and RhUS28.4 appeared to be coexpressed through a separate transcription unit, again increasing in intensity from 48 hpi on. The RhUS28.5 ORF was expressed through a single transcript that peaked at 24 hpi, and expression was reduced at later times of infection. These data suggest that, rather than being untranscribed pseudogenes, all of the ORFs are actively transcribed in infected cells and hence may all play a role in the biology of infection.
Further analysis of the functions of these genes was performed by expression in HEK293 cell lines. RhUS28.5 showed clear functional homology to HCMV US28, binding strongly to fractalkine, vMIPII, and other ligands for US28, even in the absence of other viral factors. We were unable to demonstrate binding of RhUS28.1 to RhUS28.4 to our radiolabeled fractalkine tracer. Additional experiments (data not shown) were performed in which intracellular calcium flux was measured by high-throughput FLIPR technology. ORFs 1 to 5 were not reactive in this assay with any of the approximately 35 chemokines tested, although stromal cell-derived factor was shown to signal through the endogenous CXCR4 receptor found on HEK293 cells. The IC50 of fractalkine binding to transfected RhUS28.5 was virtually identical to that seen for transfected US28. Similarly, the IC50 of fractalkine binding to infected rhesus or human cells was the same. However, the IC50 observed for transfected cells was approximately 10-fold lower than that in infected cells. The affinity of fractalkine for virion-bound RhUS28.5 or US28 was somewhat higher than that for infected cells. This suggests that viral or cellular factors can influence the conformation, G-protein coupling, or other characteristics of these receptors, leading to increased ligand avidity in the context of viral infection. Heterogeneity of receptor-G-protein coupling in different cell types, cell-specific posttranslational modifications, and lipid rafting have all been suggested to affect receptor-ligand avidity in other model systems (5, 52).
As HCMV US28 has previously been shown to bind multiple ligands, we decided to more fully characterize the binding profiles of US28 and the RhUS28.5 homolog. Competitive displacement of a signature chemokine from US28 or RhUS28.5 in either infected (data not shown) or stably transfected cells demonstrated a broad and diverse set of potential ligands for these ORFs. More than a dozen chemokines, CC, CX3C, and viral, were able to bind with IC50s in the low nanomolar to subnanomolar range. The binding profiles of US28 and RhUS28.5 were virtually indistinguishable (data not shown). Our unpublished work with other chemokine receptors has demonstrated a variety of binding profiles. Some receptors have one or a few ligands, while others may have multiple ligands. However, US28 displays the broadest binding capacity of any receptor we have observed within the chemokine system. This may suggest that US28 is a promiscuous receptor facilitating signal transduction through an array of host stimuli. Alternatively, recent publications have suggested that US28 signals constitutively or in response to endogenously produced ligands (7, 28, 50). These studies appear to have some dependency on the cell type or the presence of other viral factors. We attempted to demonstrate intracellular signaling by CRE, N-FAT, SRE, and NF-κB reporter-mediated assays in the presence and absence of G-protein inhibitors such as pertussis toxin (results not shown). We were unable to demonstrate either inducible or constitutive activity in these assays in the HEK293 transfectants. However, given the level of controversy surrounding the nature of the signaling capacity of human US28, further studies of the rhesus receptors appear warranted. Alternatively, US28 may act to modulate the environment of infected cells by sequestering chemokines from the medium without subsequent signaling events (4, 11). In our study, transfected US28 was clearly shown to mediate internalization of ligand from the medium, and we and others have previously demonstrated US28-dependent calcium mobilization in response to ligand (3, 48). RhUS28.5 demonstrated low but repeatedly detectable levels of ligand internalization, but we were unable to demonstrate RhUS28.5-dependent calcium mobilization in HEK293 cells.
The apparent redundancy at the rhesus US28 locus invites the question of the relatedness of the human and rhesus viruses. The human US28 and rhesus RhUS28.5 genes show clear similarities in ligand binding and internalization, and differences in signaling capacity in our assays may be explained by the populations of G proteins and other cofactors available in our cellular systems. As noted previously, HCMV US27 has been proposed to have arisen from a gene duplication event and has not been demonstrated to have a clear function in vitro, but it should not be assumed that this ORF is without biological relevance. Similarly, although no function was attributed to RhUS28.1 to RhUS28.4 in our assays, it seems likely that one or more of these ORFs is a surrogate for US27. The conservation of the locus expansion in the related virus African green monkey CMV, which also has five copies of a US28-ike ORF (27), further suggests the biological importance of this locus. It is possible that the alternate expansion of this locus in humans represents a different but not unrelated evolutionary divergence between rhesus monkey and human CMVs.
To further expand our understanding of US28, it is necessary to study its role in vivo. To this end, we have enabled the use of the rhesus model by our characterization of the RhUS28 locus. Further, we have constructed recombinant viruses in which RhUS28.5 or the RhUS28.1 to RhUS28.4 regions are deleted and will use these viruses to analyze the role of these genes in viral pathogenesis in this model. Preliminary results (not shown) have confirmed that a virus with RhUS28.1 to RhUS28.4 deleted has normal fractalkine binding activity, while deletion of the RhUS28.5 gene ablates the fractalkine binding capacity of this virus. By in vivo testing of the hypotheses developed by in vitro experimentation over the past decade, we hope to shed new light on the true role of this chemokine receptor homolog in viral biology.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Asher, D. M., C. J. Gibbs, D. J. Lang, D. C. Gajdusek, and R. M. Chanock. 1974. Persistent shedding of cytomegalovirus in the urine of healthy rhesus monkeys. Proc. Soc. Exp. Biol. Med. 145:794-801. [DOI] [PubMed] [Google Scholar]
- 3.Billstrom, M. A., G. L. Johnson, N. J. Avdi, and G. S. Worthen. 1998. Intracellular signaling by the chemokine receptor US28 during human cytomegalovirus infection. J. Virol. 72:5535-5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodaghi, B., T. R. Jones, D. Zipeto, C. Vita, L. Sun, L. Laurent, F. Arenzana-Seisdedos, J. L. Virelizier, and S. Michelson. 1998. Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells. J. Exp. Med. 188:855-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourne, H. R. 1997. How receptors talk to trimeric G proteins. Curr. Opin. Cell Biol. 9:134-142. [DOI] [PubMed] [Google Scholar]
- 6.Burns, J. M., D. J. Dairaghi, M. Deitz, M. Tsang, and T. J. Schall. 2002. Comprehensive mapping of poxvirus vCCI chemokine-binding protein: expanded range of ligand interactions and unusual dissociation kinetics. J. Biol. Chem. 277:2785-2789. [DOI] [PubMed] [Google Scholar]
- 7.Casarosa, P., R. A. Bakker, D. Verzijl, M. Navis, H. Timmerman, R. Leurs, and M. J. Smit. 2001. Constitutive signaling of the human cytomegalovirus-encoded chemokine receptor US28. J. Biol. Chem. 276:1133-1137. [DOI] [PubMed] [Google Scholar]
- 8.Cines, D. B., E. S. Pollak, C. A. Buck, J. Loscalzo, G. A. Zimmerman, R. P. McEver, J. S. Pober, T. M. Wick, B. A. Konkle, B. S. Schwartz, E. S. Barnathan, K. R. McCrae, B. A. Hug, A. M. Schmidt, and D. M. Stern. 1998. Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood 91:3527-3561. [PubMed] [Google Scholar]
- 9.De La Melena, V. T., C. N. Kreklywich, D. N. Streblow, Q. Yin, J. W. Cook, C. Soderberg-Naucler, C. A. Bruggeman, J. A. Nelson, and S. L. Orloff. 2001. Kinetics and development of CMV-accelerated transplant vascular sclerosis in rat cardiac allografts is linked to early increase in chemokine expression and presence of virus. Transplant. Proc. 33:1822-1823. [DOI] [PubMed] [Google Scholar]
- 10.Federsppiel, B., I. G. Melhado, A. M. Duncan, A. Delaney, K. Schappert, I. Clark-Lewis, and F. R. Jirik. 1993. Molecular cloning of the cDNA and chromosomal localization of the gene for a putative seven-transmembrane segment (7-TMS) receptor isolated from human spleen. Genomics 16:707-712. [DOI] [PubMed] [Google Scholar]
- 11.Fraile-Ramos, A., T. N. Kledal, A. Pelchen-Matthews, K. Bowers, T. W. Schwartz, and M. Marsh. 2001. The human cytomegalovirus US28 protein is located in endocytic vesicles and undergoes constitutive endocytosis and recycling. Mol. Biol. Cell 12:1737-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goda, S., T. Imai, O. Yoshie, O. Yoneda, H. Inoue, Y. Nagano, T. Okazaki, H. Imai, E. T. Bloom, N. Domae, and H. Umehara. 2000. CX3C-chemokine, fractalkine-enhanced adhesion of THP-1 cells to endothelial cells through integrin-dependent and -independent mechanisms. J. Immunol. 164:4313-4320. [DOI] [PubMed] [Google Scholar]
- 13.Gordon, D. L., T. A. Sadlon, S. L. Wesselingh, S. M. Russell, R. W. Johnstone, and D. F. Purcell. 1992. Human astrocytes express membrane cofactor protein (CD46), a regulator of complement activation. J. Neuroimmunol. 36:199-208. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths, P. D. E., and V. C. Emery. 1997. Cytomegalovirus, p. 445-470. In D. D. Richman, R. J. Whitley, and F. G. Hayden (ed.), Clinical virology. Churchill Livingstone, Secaucus, N.J.
- 15.Gruhler, A., and K. Fruh. 2000. Control of MHC class I traffic from the endoplasmic reticulum by cellular chaperones and viral anti-chaperones. Traffic 1:306-311. [DOI] [PubMed] [Google Scholar]
- 16.Hengel, H., J. O. Koopmann, T. Flohr, W. Muranyi, E. Goulmy, G. J. Hammerling, U. H. Koszinowski, and F. Momburg. 1997. A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity 6:623-632. [DOI] [PubMed] [Google Scholar]
- 17.Jones, T. R., E. J. Wiertz, L. Sun, K. N. Fish, J. A. Nelson, and H. L. Ploegh. 1996. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc. Natl. Acad. Sci. USA 93:11327-11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaur, A., M. D. Daniel, D. Hempel, D. Lee-Parritz, M. S. Hirsch, and R. P. Johnson. 1996. Cytotoxic T-lymphocyte responses to cytomegalovirus in normal and simian immunodeficiency virus-infected rhesus macaques. J. Virol. 70:7725-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keller, R., R. Peitchel, J. N. Goldman, and M. Goldman. 1976. An IgG-Fc receptor induced in cytomegalovirus-infected human fibroblasts. J. Immunol. 116:772-777. [PubMed] [Google Scholar]
- 20.Kledal, T. N., M. M. Rosenkilde, F. Coulin, G. Simmons, A. H. Johnsen, S. Alouani, C. A. Power, H. R. Luttichau, J. Gerstoft, P. R. Clapham, I. Clark-Lewis, T. Wells, and T. W. Schwartz. 1997. A broad-spectrum chemokine antagonist encoded by Kaposi's sarcoma-associated herpesvirus. Science 277:1656-1659. [DOI] [PubMed] [Google Scholar]
- 21.Kledal, T. N., M. M. Rosenkilde, and T. W. Schwartz. 1998. Selective recognition of the membrane-bound CX3C chemokine, fractalkine, by the human cytomegalovirus-encoded broad-spectrum receptor US28. FEBS Lett. 441:209-214. [DOI] [PubMed] [Google Scholar]
- 22.Kotenko, S. V., S. Saccani, L. S. Izotova, O. V. Mirochnitchenko, and S. Pestka. 2000. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc. Natl. Acad. Sci. USA 97:1695-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leong, C. C., T. L. Chapman, P. J. Bjorkman, D. Formankova, E. S. Mocarski, J. H. Phillips, and L. L. Lanier. 1998. Modulation of natural killer cell cytotoxicity in human cytomegalovirus infection: the role of endogenous class I major histocompatibility complex and a viral class I homolog. J. Exp. Med. 187:1681-1687. (Erratum, 188: 614, 1998.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockridge, K. M., G. Sequar, S. S. Zhou, Y. Yue, C. P. Mandell, and P. A. Barry. 1999. Pathogenesis of experimental rhesus cytomegalovirus infection. J. Virol. 73:9576-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackowiak, P. A., and M. Marling-Cason. 1987. Immunoreactivity of cytomegalovirus-induced Fc receptors. Microbiol. Immunol. 31:427-434. [DOI] [PubMed] [Google Scholar]
- 26.Margulies, B. J., H. Browne, and W. Gibson. 1996. Identification of the human cytomegalovirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology 225:111-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, W. J. 2000. Chemokine receptor-related genetic sequences in an African green monkey simian cytomegalovirus-derived stealth virus. Exp. Mol. Pathol. 69:10-16. [DOI] [PubMed] [Google Scholar]
- 28.Minisini, R., C. Tulone, A. Lüske, D. Michel, T. Mertens, P. Gierschik, and B. Moepps. 2003. Constitutive inositol phosphate formation in cytomegalovirus-infected human fibroblasts is due to expression of the chemokine receptor homologue pUS28. J. Virol. 77:4489-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy, P. M. 2002. International Union of Pharmacology. XXX. Update on chemokine receptor nomenclature. Pharmacol. Rev. 54:227-229. [DOI] [PubMed] [Google Scholar]
- 30.Neote, K., D. DiGregorio, J. Y. Mak, R. Horuk, and T. J. Schall. 1993. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell 72:415-425. [DOI] [PubMed] [Google Scholar]
- 31.Penfold, M. E., D. J. Dairaghi, G. M. Duke, N. Saederup, E. S. Mocarski, G. W. Kemble, and T. J. Schall. 1999. Cytomegalovirus encodes a potent alpha chemokine. Proc. Natl. Acad. Sci. USA 96:9839-9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pleskoff, O., C. Tréboute, and M. Alizon. 1998. The cytomegalovirus-encoded chemokine receptor US28 can enhance cell-cell fusion mediated by different viral proteins. J. Virol. 72:6389-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pleskoff, O., C. Treboute, A. Brelot, N. Heveker, M. Seman, and M. Alizon. 1997. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science 276:1874-1878. [DOI] [PubMed] [Google Scholar]
- 34.Plotkin, S. A., S. E. Starr, H. M. Friedman, E. Gonczol, and R. E. Weibel. 1989. Protective effects of Towne cytomegalovirus vaccine against low-passage cytomegalovirus administered as a challenge. J. Infect. Dis. 159:860-865. [DOI] [PubMed] [Google Scholar]
- 35.Prichard, M. N., M. E. Penfold, G. M. Duke, R. R. Spaete, and G. W. Kemble. 2001. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev. Med. Virol. 11:191-200. [DOI] [PubMed] [Google Scholar]
- 36.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Smiley, M. L., E. C. Mar, and E. S. Huang. 1988. Cytomegalovirus infection and viral-induced transformation of human endothelial cells. J. Med. Virol. 25:213-226. [DOI] [PubMed] [Google Scholar]
- 39.Spaete, R. R., and E. S. Mocarski. 1985. Regulation of cytomegalovirus gene expression: α and β promoters are trans activated by viral functions in permissive human fibroblasts. J. Virol. 56:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spear, G. T., N. S. Lurain, C. J. Parker, M. Ghassemi, G. H. Payne, and M. Saifuddin. 1995. Host cell-derived complement control proteins CD55 and CD59 are incorporated into the virions of two unrelated enveloped viruses, human T-cell leukemia/lymphoma virus type I (HTLV-I) and human cytomegalovirus (HCMV). J. Immunol. 155:4376-4381. [PubMed] [Google Scholar]
- 41.Spencer, J. V., K. M. Lockridge, P. A. Barry, G. Lin, M. Tasng, M. E. T. Penfold, and T. J. Schall. 2002. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J. Virol. 76:1285-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stratton, K. R., J. S. Durch, and R. S. Lawrence (ed.). 1999. Vaccines for the 21st century: a tool for decisionmaking. Committee to Study Priorities for Vaccine Development. Division of Health Promotion and Disease Prevention, Institute of Medicine, National Academy of Sciences, Washington, D.C.
- 43.Streblow, D. N., S. L. Orloff, and J. A. Nelson. 2001. Do pathogens accelerate atherosclerosis? J. Nutr. 131:2798S-2804S. [DOI] [PubMed] [Google Scholar]
- 44.Streblow, D. N., C. Soderberg-Naucler, J. Vieira, P. Smith, E. Wakabayashi, F. Ruchti, K. Mattison, Y. Altschuler, and J. A. Nelson. 1999. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell 99:511-520. [DOI] [PubMed] [Google Scholar]
- 45.Talbot, P., and J. D. Almeida. 1977. Human cytomegalovirus: purification of enveloped virions and dense bodies. J. Gen. Virol. 36:345-349. [DOI] [PubMed] [Google Scholar]
- 46.Tarantal, A. F., M. S. Salamat, W. J. Britt, P. A. Luciw, A. G. Hendrickx, and P. A. Barry. 1998. Neuropathogenesis induced by rhesus cytomegalovirus in fetal rhesus monkeys (Macaca mulatta). J. Infect. Dis. 177:446-450. [DOI] [PubMed] [Google Scholar]
- 47.Tomasec, P., V. M. Braud, C. Rickards, M. B. Powell, B. P. McSharry, S. Gadola, V. Cerundolo, L. K. Borysiewicz, A. J. McMichael, and G. W. Wilkinson. 2000. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287:1031. [DOI] [PubMed] [Google Scholar]
- 48.Vieira, J., T. J. Schall, L. Corey, and A. P. Geballe. 1998. Functional analysis of the human cytomegalovirus US28 gene by insertion mutagenesis with the green fluorescent protein gene. J. Virol. 72:8158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vink, C., E. Beuken, and C. A. Bruggeman. 2000. Complete DNA sequence of the rat cytomegalovirus genome. J. Virol. 74:7656-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waldhoer, M., T. N. Kledal, H. Farrell, and T. W. Schwartz. 2002. Murine cytomegalovirus (CMV) M33 and human CMV US28 receptors exhibit similar constitutive signaling activities. J. Virol. 76:8161-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waldman, W. J., J. M. Sneddon, R. E. Stephens, and W. H. Roberts. 1989. Enhanced endothelial cytopathogenicity induced by a cytomegalovirus strain propagated in endothelial cells. J. Med. Virol. 28:223-230. [DOI] [PubMed] [Google Scholar]
- 52.Wedegaertner, P. B., P. T. Wilson, and H. R. Bourne. 1995. Lipid modifications of trimeric G proteins. J. Biol. Chem. 270:503-506. [DOI] [PubMed] [Google Scholar]
- 53.Wess, J. 1993. Molecular basis of muscarinic acetylcholine receptor function. Trends Pharmacol. Sci. 14:308-313. [DOI] [PubMed] [Google Scholar]
- 54.Wiertz, E. J., D. Tortorella, M. Bogyo, J. Yu, W. Mothes, T. R. Jones, T. A. Rapoport, and H. L. Ploegh. 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384:432-438. [DOI] [PubMed] [Google Scholar]