Abstract
Oxidation of molecular hydrogen catalyzed by [NiFe] hydrogenases is a widespread mechanism of energy generation among prokaryotes. Biosynthesis of the H2-oxidizing enzymes is a complex process subject to positive control by H2 and negative control by organic energy sources. In this report we describe a novel signal transduction system regulating hydrogenase gene (hox) expression in the proteobacterium Alcaligenes eutrophus. This multicomponent system consists of the proteins HoxB, HoxC, HoxJ*, and HoxA. HoxB and HoxC share characteristic features of dimeric [NiFe] hydrogenases and form the putative H2 receptor that interacts directly or indirectly with the histidine protein kinase HoxJ*. A single amino acid substitution (HoxJ*G422S) in a conserved C-terminal glycine-rich motif of HoxJ* resulted in a loss of H2-dependent signal transduction and a concomitant block in autophosphorylating activity, suggesting that autokinase activity is essential for the response to H2. Whereas deletions in hoxB or hoxC abolished hydrogenase synthesis almost completely, the autokinase-deficient strain maintained high-level hox gene expression, indicating that the active sensor kinase exerts a negative effect on hox gene expression in the absence of H2. Substitutions of the conserved phosphoryl acceptor residue Asp55 in the response regulator HoxA (HoxAD55E and HoxAD55N) disrupted the H2 signal-transduction chain. Unlike other NtrC-like regulators, the altered HoxA proteins still allowed high-level transcriptional activation. The data presented here suggest a model in which the nonphosphorylated form of HoxA stimulates transcription in concert with a yet unknown global energy-responsive factor.
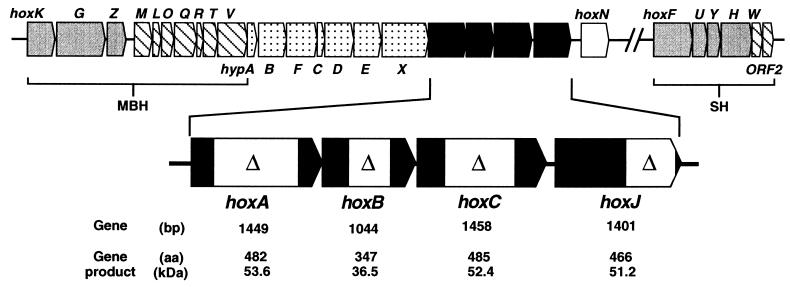
Our knowledge of the regulatory strategies of organisms capable of utilizing unusual energy sources such as molecular hydrogen, carbon monoxide, ammonia, reduced sulfur, and iron compounds is limited. The proteobacterium Alcaligenes eutrophus is one of the best-studied facultative lithoautotrophs and well adapted to the changing chemical environment. It grows on a wide range of organic substrates and alternatively assimilates CO2 via the reductive Calvin cycle, utilizing H2 as the sole energy source (1). H2 oxidation in A. eutrophus is mediated by two NiFe-containing hydrogenases: a cytoplasmic NAD-reducing flavoprotein [soluble hydrogenase (SH), ref. 2] and a cytochrome b-linked membrane-bound enzyme (MBH, ref. 3). The genes coding for the two hydrogenases are grouped together with sets of genes for auxiliary proteins involved in metal supply, metal center assembly, and enzyme maturation on the megaplasmid pHG1 (refs. 4–7; Fig. 1).
Figure 1.
The A. eutrophus hydrogenase (hox, hyp) gene cluster. The gray-colored MBH and SH structural genes together with specific accessory genes (hatched boxes) form two separate operons on megaplasmid pHG1 (4, 16). The dotted hyp genes are involved in metal center assembly of both hydrogenases (6). hoxN encodes a nickel permease (7). The hydrogenase regulatory genes are emphasized, and the location and the extent of the respective deletions are indicated by boxes. A synopsis of the data deduced from the sequence is given along the bottom of the figure.
A maturation process of this complexity needs to be regulated in a coordinate fashion. In most organisms so far studied, hydrogenase synthesis is controlled by H2 induction. A few strains, however, including A. eutrophus H16, produce hydrogenase independently of the presence of H2 under conditions of energy deprivation, e.g., during growth on poor substrates such as glycerol (4). Chemostat experiments revealed that the environmental stimulus for high-level hydrogenase expression is not limitation of the carbon source but rather starvation for electron donors (8). In the aerobic H2-oxidizing organisms, transcription of the hydrogenase genes is mediated by NtrC-type specific activator proteins (9–12). These response regulators typically are composed of three functional modules: an individual N-terminal domain implicated in signal reception, either via a sensor kinase or via interaction with a chemical signal, a central domain containing the ATPase activity essential for transcriptional activation, and a C-terminal DNA-binding domain composed of a helix–turn–helix structure (13). The latter two domains of these regulators are highly conserved (14). HoxA, the transcriptional activator of the A. eutrophus H16 hydrogenase system, binds specifically to hydrogenase promoter upstream sequences and was shown to be essential for activation of the MBH and SH promoters in vivo (15, 16).
To learn more about the molecular mechanisms that account for H2-independent and H2-responsive gene expression we selected two closely related species that differ in this regard for a comparative study. Each strain, A. eutrophus H16 and Alcaligenes hydrogenophilus, harbors the hydrogenase genes on a large plasmid. These plasmids are interchangeable and confer the regulatory phenotype of their native host (17). Sequence analysis revealed three ORFs adjacent to the response regulator gene hoxA in both A. hydrogenophilus (11) and A. eutrophus H16 (this study). The products of hoxB and hoxC are homologous to the small and large subunits of [NiFe] hydrogenases, respectively, whereas the product of hoxJ showed typical features of a sensor kinase (18, 19).
By comparing the strains and focusing the studies on A. eutrophus H16 we became aware of the function of a novel regulatory system consisting of four components. We show that HoxBC is absolutely required for H2 recognition and that this signal is transmitted to HoxJ by a yet unknown mechanism. The sensor kinase in turn modulates the response regulator HoxA to activate transcription. However, in contrast to the case for most other “two-component” systems (18, 19), our results indicate that phosphorylation of HoxA at the conserved Asp55 residue in the receiver module has a negative effect on transcriptional activation. This hypothesis is consistent with the observation that the inactive variant of the HoxJ sensor kinase, found in A. eutrophus H16, yields high-level gene expression even in the absence of H2.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions.
The A. eutrophus strains used in this study are listed in Table 1. HF strains are derivatives of the wild-type A. eutrophus H16. Alcaligenes strains were grown in mineral salts medium (20) containing 0.4% succinate (SN) or 0.2% fructose and 0.2% glycesd (FGN). H2-induction experiments were carried out in stoppered 150-ml serum flasks containing 15 ml of culture. The flasks were gassed with various amounts of H2 using gas-tight syringes. For studies on the carbon and energy-source dependence of hydrogenase synthesis cells were grown in flasks kept in desiccating jars under an atmosphere of either 50% H2/10% O2/40% N2 or 10% O2/90% N2.
Table 1.
A. eutrophus strains used in this study
| Strain | Relevant properties | Source or ref. |
|---|---|---|
| H16 | Wild type | DSM428, ATCC 17699 |
| HF409 | hoxAΔ | (11) |
| HF430 | hoxBΔ | This work |
| HF341 | hoxCΔ | This work |
| HF373 | hoxJΔ | This work |
| HF433 | hoxJ* | This work |
| HF434 | hoxJ* hoxBΔ | This work |
| HF435 | hoxJ* hoxCΔ | This work |
| HF436 | HoxJ* HoxAD55E | This work |
| HF437 | HoxJ* HoxAD55N | This work |
| HF489 | hoxJ*hoxAΔ | This work |
Plasmids.
The suicide vector pLO3 used for isolation of isogenic A. eutrophus mutants is a tetracycline-resistant derivative of plasmid pBR322 carrying the B. subtilis sacB gene, and the RP4 origin of transfer. pLO3 was constructed as follows. A Klenow-treated, 0.08-kb HindIII-EcoRI fragment containing the polylinker of pNEB193 (New England Biolabs) was inserted between the EcoRI (end-polished) and DraI sites of pBR322, resulting in pCH639. The sacB gene of pUCD800 (21) was subcloned as a 1.96-kb BamHI/NdeI, Klenow-treated fragment into StyI- (end-polished) and BsaAI-digested pCH639. The resulting plasmid, pCH640, was cut with NdeI and end-polished, and oriT from pJF145 (22) subsequently was inserted as a 0.47-kb BamHI (end-polished) fragment to give plasmid pLO3. The following plasmids were used for introducing in-frame deletions into hoxB, hoxC, and hoxJ (Fig. 1). pCH643 is derived from pLO2 (23) and carries a 0.89-kb XmnI-XhoI fragment spanning the hoxB gene but deleted for a 459-bp KpnI fragment. pCH644 is derived from pLO1 (23) and harbors a 3.3-kb PstI fragment with a copy of hoxC deleted for a 798-bp SalI (end-polished)-PmaCI fragment. pCH638 is derived from pLO1 and carries a 0.82-kb DraI-PstI fragment with a copy of hoxJ deleted for a 549-bp FspI-PvuII fragment.
hoxJ and hoxA mutations were generated by site-directed mutagenesis. An hoxJA1226G (hoxJ*) mutation (resulting in HoxJS422G) was constructed by the method of Chen and Przybyla (24). A 1.9-kb, hoxJ-harboring NdeI-EcoRI fragment was amplified by PCR by using the synthetic oligonucleotides 5′-GCCCAATGGCCGAGATCA-3′ and 5′-CGAAAGACcAAGCCCAGTCCCC-3′ (lowercase denotes exchanged bases) as the nonmutagenic and mutagenic primers, respectively. The 746-bp PCR product and the synthetic oligonucleotide 5′-TTCGCTTATGCTATTCCC-3′ were used as primers in a second amplification step. A 0.26-kb Tth111I-ApaI fragment of the resulting 1,136-bp PCR product subsequently was subcloned into the vector pLO3 harboring the 3′ region of hoxJ. This resulted in pCH615. The hoxA point mutations, hoxAT165G (HoxAD55E) and hoxAG163A (HoxAD55N), were constructed by amplification of the hoxA containing plasmid pCH220 (15) with the primers 5′-TGTCGTCGACAAGGAAACCCGTTCACAGG3′ and 5′-GAGGCCTGGCATGCGTTGcTCGCACAGGATCACCG-3′ (resulting in hoxAT165G) or 5′-GAGGCCTGGCATGCGTTGATtGCACAGGATCACCG-3′ (resulting in hoxAG163A). The 157-bp amplification products were cut with EcoRI and StuI, and the resulting 97-bp fragments containing the point mutations subsequently were subcloned into plasmid pCH646, a hoxA-containing pBluescript SK(+) derivative (Stratagene). This resulted in pCH647 (HoxAD55E) and pCH648 (HoxAD55N). SmaI-Ecl136II fragments (1.95 kb) from these plasmids were inserted into PmeI-digested pLO2 to give pCH649 (HoxAD55E) and pCH651 (HoxAD55N). The amplification products were checked by sequencing. Derivatives of pQE-31 (Qiagen) were used for overproduction of His-tagged HoxJ and HoxJ*. A 1.8-kb SacI-HindIII hoxJ* fragment derived from pCH622 (M. Schmelz, O.L., and B.F., unpublished data) was inserted into SacI-HindIII-digested pQE-31 to give pCH652 (His6-HoxJ* fusion). A 0.71-kb AflII-ApaI fragment from pCH652 was exchanged by the corresponding fragment derived from the hoxJ-containing plasmid pCH641 to give plasmid pCH653 (His6-HoxJ fusion). The broad-host range vectors pGE300 and pGE301 harboring the Φ(hoxF-lacZ) and Φ(hoxK-lacZ) fusions, respectively (23), were used for monitoring hydrogenase gene expression.
Isolation of Isogenic A. eutrophus Mutants.
Homogenotes carrying the defined mutations described above were introduced into A. eutrophus via an allelic-exchange procedure (23). Plasmids pCH615, pCH649, and pCH651 were used to introduce point mutations into strains H16 and HF433. hoxJΔ, hoxBΔ, hoxCΔ, and hoxAΔ mutants were isolated by using plasmids pCH638, pCH643, pCH644, and pCH553 (11), respectively (Table 1). Selected isolates were screened for the presence of the desired mutations as described previously (5).
Enzyme Assays.
SH and MBH activities were determined as described previously (2, 3). β-Galactosidase was assayed as described previously (15), and the activity (in units) was calculated according to Miller except that the cell density was measured at 436 nm (OD436).
Purification of His6-Tagged HoxJ and HoxJ* Proteins and Autophosphorylation Assays.
Escherichia coli JM109 harboring pCH652 (His6-HoxJ*) or pCH653 (His6-HoxJ) was grown in Luria–Bertani medium at 30°C. Expression of the recombinant proteins was induced by the addition of 1 mM isopropyl β-d-thiogalactoside. Four hours after induction the cells were harvested by spinning at 4,000 × g for 15 min. The pelleted cells were resuspended in buffer 1 (50 mM sodium phosphate, pH 8.0/300 mM NaCl/20 mM imidazole/1 mM phenylmethylsulfonyl fluoride) and broken by two passes through a French press at 900 psi. The crude extracts were ultracentrifuged at 60,000 × g for 45 min. The His6-tagged proteins were purified from the soluble fractions by using Ni-nitrilotriacetate Spin Columns (Qiagen) according to the manufacturer’s instructions. The eluates containing the recombinant proteins (greater than 90% purity estimated in a Coomassie-stained SDS/PAGE gel) were dialyzed against storage buffer [50 mM Tris⋅Cl, pH 8.0/100 mM KCl/1 mM DTT/20% (vol/vol) glycerol] and subsequently stored at −70°C. Autophosphorylation assays were performed in 40 mM Tris⋅Cl, pH 8.0/40 mM KCl/0.8 mM DTT containing 2.5 μg of His6-HoxJ or His6-HoxJ*. The samples were incubated for 5 min at room temperature, and the reaction was started by the addition of 0.4 mM cold ATP/0.4 mM MgCl2/10 mCi [γ-32P]ATP (5,000 Ci/mmol, Amersham) in a total reaction volume of 15 μl. The reaction was stopped by the addition of 5 μl sample buffer [125 mM Tris⋅Cl, pH 6.8/8 mM EDTA/4% (wt/vol) SDS/8% (vol/vol) 2-mercaptoethanol/20% (vol/vol) glycerol/0.02% (wt/vol) bromophenol blue] and subsequent incubation for 3 min at 55°C. Samples were separated on SDS-polyacrylamide gels, and the dried gels were scanned by using an SI 550 storage PhosphorImager (Molecular Dynamics).
RESULTS
The Regulatory Gene Region in A. eutrophus H16.
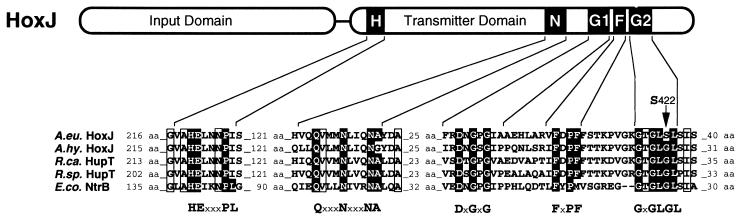
The hydrogenase regulatory gene region on megaplasmid pHG1 of A. eutrophus H16 consists of hoxA encoding the principle transcriptional activator and three newly identified ORFs, hoxB, hoxC, and hoxJ (Fig. 1). The predicted products of the A. eutrophus hoxB and hoxC are homologous (between 52 and 78% identity) to HoxB and HoxC of A. hydrogenophilus (11) and to HupU and HupV of Bradyrhizobium japonicum (25) and Rhodobacter capsulatus (26). Moreover, HoxB and HoxC share significant similarity with the small and the large subunit of standard [NiFe] hydrogenases. A comparison with the periplasmic hydrogenase from Desulfovibrio gigas whose crystal structure is known (27) showed that the cysteine and histidine residues that coordinate the three FeS centers in the small, electron-transferring subunit are well conserved in HoxB. Unlike the small subunits of standard [NiFe] hydrogenases, which are synthesized as precursors (28), the HoxB polypeptide lacks an N-terminal signal sequence, which points to a cytoplasmic location of the protein. Signatures strongly resembling sequence motifs of the active site-containing large hydrogenase subunit are present in the product of hoxC, including the two pairs of N- and C-terminal cysteine residues that provide thiol groups for nickel and iron coordination in the D. gigas enzyme (27). Unlike the majority of [NiFe] subunits, which undergo C-terminal proteolytic maturation in the course of metal center assembly (29), the HoxC protein lacks a C-terminal amino acid extension. The product of the distal gene (hoxJ) in the regulatory region (Fig. 1) shows homology to sensor kinases of bacterial two-component regulatory systems (18, 19). Sequence comparisons showed that the closest relatives are HoxJ of A. hydrogenophilus (11), HupT of R. capsulatus (30), and HupT of R. sphaeroides (31) (62.1%, 37.3%, and 37.9% identity, respectively). Fig. 2 shows an alignment of the C-terminal transmitter domain of these proteins and NtrB of E. coli (32). The typical signatures of sensor kinases (18) are almost perfectly conserved in HoxJ of A. eutrophus with the exception of the distal glycine-rich G2 motif. The A. eutrophus HoxJ bears a serine instead of a glycine residue at position 422 (Fig. 2).
Figure 2.
Partial alignment of the C-terminal transmitter domain of HoxJ of A. eutrophus with the corresponding parts of selected histidine protein kinases. Conserved amino acid residues of histidine protein kinase motifs [consensus according to Parkinson and Kofoid (18) is given in boldface letters] are emphasized by black boxes. Additional residues conserved in all sequences are boxed. A.eu., A. eutrophus; A.hy., A. hydrogenophilus; R.ca., R. capsulatus; B.ja., B. japonicum; E.co., E. coli.
To elucidate the function of the newly identified ORFs in A. eutrophus, in-frame deletions were constructed and recombined into the genome to yield isogenic mutants. Whereas disruption of the response regulator gene hoxA is accompanied by a complete failure to express the two hydrogenases (9, 15), strains bearing deletions in hoxB (HF430), hoxC (HF431), and hoxJ (HF373) were barely affected in lithoautotrophic growth and accordingly contained almost wild-type levels of SH and MBH activity (Table 2). That a mutation in hoxJ had no effect on hydrogenase synthesis in A. eutrophus H16 was in marked contrast to observations in A. hydrogenophilus (11) and R. capsulatus (30), raising the question of whether the A. eutrophus hoxJ product had lost its regulatory function in the course of evolution.
Table 2.
Hydrogenase activities of A. eutrophus mutants
| Strain | Genotype | Hydrogenase activity*
|
|
|---|---|---|---|
| SH | MBH | ||
| H16 | Wild type | 4.4 | 1.8 |
| HF409 | hoxAΔ | <0.1 | <0.1 |
| HF430 | hoxBΔ | 3.8 | 1.4 |
| HF341 | hoxCΔ | 4.7 | 2.0 |
| HF373 | hoxJΔ | 4.1 | 2.2 |
Strains were grown in FGN medium to an OD436 of 8. Values [units ·(mg protein)−1] give the average of at least two independent determinations.
HoxJ* Mediates H2-Responsive Hydrogenase Regulation.
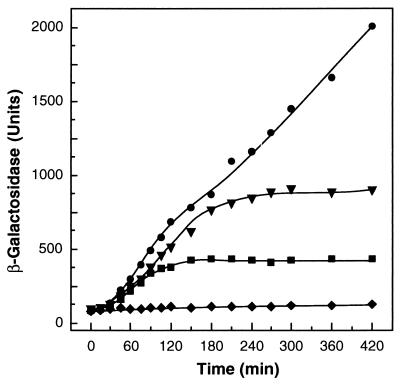
Two observations drew our attention to the C-terminal domain of the A. eutrophus HoxJ. (i) Cells of A. eutrophus H16 that bear a HoxJ hybrid protein in which the C-terminal part of the indigenous HoxJ protein had been replaced by the A. hydrogenophilus counterpart exhibited H2-dependent hydrogenase gene expression (data not shown). (ii) The exchanged protein segment contributed the G2 motif with a perfect consensus (Fig. 2). This prompted us to modify the native HoxJ protein of A. eutrophus H16 by substituting Gly for Ser422. The altered allele (hoxJ*) was introduced into A. eutrophus H16 by double-recombination, yielding the isogenic strain HF433, which was examined for its regulatory properties by monitoring β-galactosidase activity from vector-based Φ(hoxF-lacZ) and Φ(hoxK-lacZ) translational fusions. The results are documented representatively for hoxK expression (Fig. 3). Cultures were grown heterotrophically on fructose and glycerol at 36°C for 24 h to an optical density of 7.0 at 436 nm. Under these conditions hydrogenase gene expression is completely blocked because of the temperature sensitivity of the transcriptional activator HoxA (15). At zero time the cells were shifted to 30°C and H2 was added at various concentrations. A. eutrophus HF433 (hoxJ*) expressed Φ(hoxK-lacZ) only when H2 was added, and the final level of β-galactosidase activity correlated well with the concentration of H2 supplied (Fig. 3), indicating that SH and MBH gene expression ceased when H2 was consumed by the cells. Similar regulatory responses were obtained with the Φ(hoxF-lacZ) fusion (data not shown), confirming previous observations that the SH and MBH are expressed coordinately (4, 16).
Figure 3.
Induction of MBH gene expression by H2. A. eutrophus HF433 cells harboring the plasmid-based Φ(hoxK-lacZ) fusion were grown under hydrogenase-repressing conditions and subsequently induced with H2 as described in the text. •, 10% (vol/vol) H2; ▾, 1% (vol/vol) H2; ■, 0.1% (vol/vol) H2; ♦, without H2.
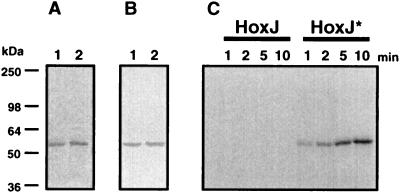
The highly conserved glycine residue at position 315 in the G2 motif of NtrB from E. coli is essential for the positive function of this sensor kinase. An altered NtrB in which Gly315 had been replaced by Ala was incapable of autophosphorylation and hence could not activate its cognate response regulator NtrC (33). To examine whether the corresponding Gly422 in HoxJ* affects its phosphorylation capacity, His6-HoxJ* and His6-HoxJ fusion proteins were isolated from E. coli. The resulting proteins were homogenous on the basis of SDS/PAGE and immunochemical analysis (Fig. 4 A and B). Phosphorylation experiments with [γ-32P]ATP as the substrate showed that the His6-HoxJ protein did not mediate autophosphorylation (Fig. 4C) whereas restoration of the consensus motif in HoxJ* (Fig. 2) led to recovery of its autophosphorylation activity (Fig. 4C). Phosphorylated His6-HoxJ* was stable in sodium hydroxide at pH 13 but acid labile at pH 1.0 (data not shown) as reported for other histidine protein kinases (19). Moreover, competition studies revealed that ATP and not GTP served as the phosphoryl donor for HoxJ* (data not shown). These results are compatible with the notion that a histidyl phosphate is formed at the conserved His220, which is the likely phosphoryl acceptor of HoxJ* (Fig. 2).
Figure 4.
Purification and autophosphorylation of His6-tagged HoxJ and HoxJ* proteins. The fusion proteins were purified as described in Materials and Methods. A portion (2.5 μg) of each protein was separated by SDS/PAGE and either Coomassie-stained (A) or blotted onto a nitrocellulose membrane and visualized with an alkaline phosphatase-coupled Ni-nitrilotriacetate conjugate (B). Lane 1, His6-HoxJ; lane 2, His6-HoxJ*. Time-dependent autophosphorylation (C) of the fusion proteins was performed as specified in Materials and Methods. Approximate molecular masses are given along the left side of the figure.
HoxB and HoxC Are Essential for H2 Sensing.
The results thus far showed that HoxJ* acts in monitoring H2 as an environmental stimulus, leaving open the question of whether the sensor kinase acts as transmitter or direct receptor of the gaseous substrate. Despite the fact that the hydrogenase-related genes hoxB and hoxC were dispensable for SH and MBH gene expression in the parent strain H16 (Table 2), we reexamined their possible regulatory function in the HoxJ* background. In-frame deletion alleles of hoxB and hoxC were introduced into the recipient HF433. The resulting isogenic mutants HF434 (hoxBΔ) and HF435 (hoxCΔ) failed to grow autotrophically on H2 and contained only 20% and less than 10%, respectively, of the reference activities for SH and MBH. Transfer of plasmid-borne hoxB and hoxC wild-type alleles restored lithotrophic growth and hydrogenase activities (data not shown). With the aid of Φ(hoxF-lacZ) and Φ(hoxK-lacZ) fusions it was demonstrated that the products of hoxB and hoxC are active on the transcriptional level, as illustrated representatively for hoxK-lacZ expression in Fig. 5. These data provide evidence that HoxB and HoxC are absolutely necessary for H2 sensing in the HoxJ* strain.
Figure 5.
H2-dependent hydrogenase gene expression in HF433-derived hoxB and hoxC deletion strains. Strains harboring the Φ(hoxK-lacZ) fusion were grown in fructose glycerol mineral medium until they reached the glycerol growth phase (OD436 = 7), and hoxK expression subsequently was induced by adding 10% (vol/vol) H2. ○, HF433 (hoxJ*) without H2; •, HF433 (hoxJ*) +H2; ▵, HF434 (hoxJ*hoxBΔ) +H2; □, HF435 (hoxJ*hoxCΔ) +H2.
HoxA Is the Target for H2 Sensing.
The discovery of an H2 sensor consisting of at least the H2 receptor HoxBC and the histidine protein kinase HoxJ* raised the question of whether and how these components interact with the response regulator HoxA. The first approach to attack this problem experimentally aimed at mutating hoxA by exchanging the nucleotides that determine the conserved aspartate residue, the general target for phosphorylation of response regulators (18, 19). Site-directed mutagenesis of hoxA was done in vitro, yielding two different mutant alleles corresponding to Asp55-to-Glu (HoxAD55E) and Asp55-to-Asn (HoxAD55N), respectively. The mutant alleles were introduced by double-recombination into the HoxJ* derivative HF433, and the resulting isogenic mutants were characterized phenotypically and compared with a hoxAΔ strain. To our great surprise exchange of the Asp55 showed no effect on lithoautotrophic growth (data not shown) and yielded high levels of hydrogenase gene expression. By contrast, disruption of hoxA by an in-frame deletion completely disrupted growth on H2 and prevented gene transcription, confirming the important role of the regulator. Thus, Asp55 of HoxA is not essential for hydrogenase gene transcription but rather appears to function negatively (Table 3).
Table 3.
MBH and SH gene expression in hoxA mutants of A. eutrophus
| Strain | Characteristic | Hydrogenase activity*
|
β-Galactosidase activity†
|
||
|---|---|---|---|---|---|
| MBH | SH | Φ(hoxK-lacZ) | Φ(hoxF-lacZ) | ||
| H16 | HoxJ | 1.41 | 4.8 | 7,523 ± 995 | 6,484 ± 762 |
| HF409 | hoxAΔ | <0.01 | <0.1 | 71 ± 1 | 27 ± 4 |
| HF433 | HoxJ* | <0.01 | 0.1 | 119 ± 26 | 970 ± 80 |
| HF436 | HoxJ* HoxAD55E | 2.00 | 4.8 | 7,797 ± 538 | 8,094 ± 688 |
| HF437 | HoxJ* HoxAD55N | 1.97 | 6.0 | 6,840 ± 740 | 7,294 ± 1,292 |
| HF489 | hoxJ*hoxAΔ | <0.01 | <0.1 | 83 ± 6 | 28 ± 1 |
Strains were grown in FGN medium to an OD436 of 8. Values [units ⋅ (mg protein)−1] give the average of at least two independent determinations.
Strains were grown in FGN medium to an OD436 of 8. Values [units ⋅ (OD436)−1] give the average of at least three independent determinations ± SD.
H2 Induction in the HoxJ* Strain Is Affected by the Carbon and Energy Source.
Batch culture and chemostat experiments have shown that the two hydrogenases of A. eutrophus H16 are expressed at extremely low levels in rapidly growing cells, e.g., during cultivation on succinate. Both enzymes reach intermediate levels on substrates such as fructose and are strongly derepressed when growth rate is limited by the energy source, e.g., on glycerol (8, 16). To determine whether organic substrates could interfere with the H2-mediated induction of the system in the hoxJ* strain, the reporter activity of plasmid-borne SH and MBH gene fusions was monitored under mixotrophic conditions (Table 4). The results show that neither hydrogenase operon was induced by H2 when a preferentially utilized substrate such as succinate was supplied. However, induction by H2 occurred when a poor substrate such as glycerol was provided.
Table 4.
Energy-responsive control of hydrogenase gene expression in A. eutrophus HF433
| Activities | Succinate*
|
Fructose-glycerol*
|
||
|---|---|---|---|---|
| −H2 | +H2 | −H2 | +H2 | |
| MBH† | <0.01 | <0.01 | <0.01 | 0.23 |
| SH† | <0.01 | <0.01 | 0.05 | 0.70 |
| Φ(hoxK-lacZ)‡ | 7 ± 1 | 5 ± 1 | 119 ± 26 | 1,626 ± 44 |
| Φ(hoxF-lacZ)‡ | 6 ± 1 | 7 ± 1 | 970 ± 80 | 2,929 ± 154 |
Strains were grown in SN medium to an OD436 of 3 and in FGN medium to an OD436 of 8.
Values [hydrogenase activity in units·(mg protein)−1] give the average of at least two independent determinations.
Values (β-galactosidase activity in units) give the average of at least four independent determinations ±SD.
DISCUSSION
This study reports a novel bacterial signal transduction chain capable of sensing molecular hydrogen, the smallest molecule on earth. The system, discovered in the facultative lithoautotroph A. eutrophus, consists of the putative H2 receptor protein HoxBC, the histidine protein kinase HoxJ*, and the response regulator HoxA, which is essential for transcriptional activation under all conditions (Fig. 6).
Figure 6.
Model of the H2-sensing signal transduction chain that controls hydrogenase gene expression in Alcaligenes eutrophus.
The H2 receptor HoxBC resembles typical features of [NiFe] hydrogenases (34). Homologous regulatory proteins in the phototroph R. capsulatus, the nitrogen-fixing B. japonicum, and the hydrogen bacterium A. hydrogenophilus (11, 25, 26) point to the existence of a new class of H2-interacting proteins that differ from the typical energy-generating hydrogenases (27, 34) in two major respects: the lack of a C-terminal proteolytically cleavable extension in the large subunit, which generally is considered to be necessary for metal center assembly (29), and the absence of an export-triggering, N-terminal leader peptide in the small subunit (28), suggesting a cytoplasmic location of the H2 receptor proteins. Recently, we succeeded in demonstrating that HoxBC of A. eutrophus is a nickel-containing protein that mediates H2 oxidation with redox dyes as electron acceptors (L. Kleihues, O.L., and B.F., unpublished data). Binding of H2 to the HoxBC homologue HupUV was shown recently in R. capsulatus (35); however, H2 oxidation by this protein was not demonstrated.
Our results clearly show that HoxB and HoxC are absolutely required for lithoautotrophic growth in an H2-responsive strain of A. eutrophus. This observation is in marked contrast to the behavior of HupUV-deficient mutants of R. capsulatus, which showed high-level hydrogenase gene expression pointing to a different regulatory mechanism in the phototroph (26).
That HoxB and HoxC are essential for H2 sensing support the notion that the hydrogenase-like protein rather than the kinase HoxJ* is the primary receptor of H2. This conclusion raises the question of how signal transduction proceeds from the receptor protein to the sensor kinase. The input domain of HoxJ* does not contain any obvious signatures suggestive of a binding site for a redox active cofactor. Provided no additional components are involved we favor the hypothesis that the regulatory interaction of HoxBC and HoxJ* is guided through an H2-mediated conformational change in HoxBC (Fig. 6).
The lack of autophosphorylation activity associated with the A. eutrophus H16 HoxJ protein was shown to be based on the presence of a serine in the place of glycine at position 422 within the G2 motif of the C-terminal transmitter module. The HoxJ* form that has glycine at position 422 acts as an autokinase. Unlike the native HoxJ protein, the HoxJ* form exerts a negative effect on hydrogenase gene transcription in the absence of H2 (Fig. 6). This observation suggests that HoxJ*-mediated phosphorylation of HoxA prevents transcriptional activation. The hypothesis that phosphorylated HoxA fails to activate transcription is supported further by the behavior of mutants in which the Asp55 phosphoryl acceptor site of HoxA (D55E and D55N) was modified. As with the HoxJ strain of A. eutrophus, these mutants also showed H2-independent, high-level hydrogenase gene expression. Although the phosphoryl transfer from the HoxJ* kinase to its cognate response regulator HoxA awaits experimental proof, the data presented here indicate the existence of an uncommon signal transduction system that distinguishes the HoxA-HoxJ* pair from prototypic two-component systems (18, 19). Normally, amino acid substitutions at the phosphoryl acceptor residue corresponding to Asp55 in HoxA lead to a dramatic decrease in transcriptional activation by response regulators, including the well documented NtrC (36). Interestingly the AlgB regulator of Pseudomonas aeruginosa appears to be another exception to this rule since corresponding mutant forms of the response regulator mediate high-level gene expression. In the case of AlgB, however, the conserved Asp residue had no regulatory impact at all (37).
The experiments reported here show that Asp55 of HoxA is crucial for H2-mediated signal transduction. The absence of HoxJ* or the presence of an Asp55-modified HoxA protein restricts the system to global control in response to electron donor availability. The mechanism(s) underlying this energy-related control, which affects transcription activation by HoxA, is unknown. We conclude from this study that global control enables the cell to grow mixotrophically by using H2 as an additional energy source as long as organic substrates are growth rate-limiting.
Acknowledgments
We are grateful to Karin Marin for her excellent assistance in nucleotide sequence determination and to Edward Schwartz for valuable suggestions and critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft and by the Fonds der Chemischen Industrie.
ABBREVIATIONS
- MBH
membrane-bound hydrogenase
- SH
soluble hydrogenase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the EMBL/GenBank/DDBJ databases (accession no. U82564).
References
- 1. Bowien B, Schlegel H G. Annu Rev Microbiol. 1981;35:405–452. doi: 10.1146/annurev.mi.35.100181.002201. [DOI] [PubMed] [Google Scholar]
- 2.Schneider K, Schlegel H G. Biochim Biophys Acta. 1976;452:66–80. doi: 10.1016/0005-2744(76)90058-9. [DOI] [PubMed] [Google Scholar]
- 3.Schink B, Schlegel H G. Biochem Biophys Acta. 1979;567:315–324. doi: 10.1016/0005-2744(79)90117-7. [DOI] [PubMed] [Google Scholar]
- 4.Friedrich B, Schwartz E. Annu Rev Microbiol. 1993;47:351–383. doi: 10.1146/annurev.mi.47.100193.002031. [DOI] [PubMed] [Google Scholar]
- 5.Bernhard M, Schwartz E, Rietdorf J, Friedrich B. J Bacteriol. 1996;178:4522–4529. doi: 10.1128/jb.178.15.4522-4529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dernedde J, Eitinger T, Patenge N, Friedrich B. Eur J Biochem. 1996;235:351–358. doi: 10.1111/j.1432-1033.1996.00351.x. [DOI] [PubMed] [Google Scholar]
- 7.Eitinger T, Wolfram L, Degen O, Anthon C. J Biol Chem. 1997;272:17139–17144. doi: 10.1074/jbc.272.27.17139. [DOI] [PubMed] [Google Scholar]
- 8.Friedrich C G. J Bacteriol. 1982;149:203–210. doi: 10.1128/jb.149.1.203-210.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eberz G, Friedrich B. J Bacteriol. 1991;173:1845–1854. doi: 10.1128/jb.173.6.1845-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Soom C, Verreth C, Sampaio M J, Vanderleyden J. Mol Gen Genet. 1993;239:235–240. doi: 10.1007/BF00281623. [DOI] [PubMed] [Google Scholar]
- 11.Lenz O, Strack A, Tran-Betcke A, Friedrich B. J Bacteriol. 1997;179:1655–1663. doi: 10.1128/jb.179.5.1655-1663.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richaud P, Colbeau A, Toussaint B, Vignais P M. J Bacteriol. 1991;173:5928–5932. doi: 10.1128/jb.173.18.5928-5932.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morett E, Segovia L. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.North A K, Klose K E, Stedman K M, Kustu S. J Bacteriol. 1993;175:4267–4273. doi: 10.1128/jb.175.14.4267-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmer D, Schwartz E, Tran-Betcke A, Gewinner P, Friedrich B. J Bacteriol. 1995;177:2373–2380. doi: 10.1128/jb.177.9.2373-2380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz E, Gerischer U, Friedrich B. J Bacteriol. 1998;180:3197–3204. doi: 10.1128/jb.180.12.3197-3204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedrich B, Friedrich C G, Meyer M, Schlegel H G. J Bacteriol. 1984;158:331–333. doi: 10.1128/jb.158.1.331-333.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkinson J S, Kofoid E C. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 19.Stock J B, Surette M G, Levit M, Park P. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 25–51. [Google Scholar]
- 20.Eberz G, Eitinger T, Friedrich B. J Bacteriol. 1989;171:1340–1345. doi: 10.1128/jb.171.3.1340-1345.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado C I. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fürste J P, Pansegrau W, Ziegelin G, Kröger M, Lanka E. Proc Natl Acad Sci USA. 1979;86:1771–1775. doi: 10.1073/pnas.86.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenz O, Schwartz E, Dernedde J, Eitinger M, Friedrich B. J Bacteriol. 1994;176:4385–4393. doi: 10.1128/jb.176.14.4385-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen B, Przybyla A E. BioTechniques. 1994;17:657–659. [PubMed] [Google Scholar]
- 25.Black L K, Fu C, Maier R J. J Bacteriol. 1994;176:7102–7106. doi: 10.1128/jb.176.22.7102-7106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elsen S, Colbeau A, Chabert J, Vignais P M. J Bacteriol. 1996;178:5174–5181. doi: 10.1128/jb.178.17.5174-5181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volbeda A, Charon M H, Piras C, Hatchikan E C, Frey M, Fontecilla-Camps J C. Nature (London) 1995;373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- 28.Berks B C. Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 29.Maier T, Böck A. In: Advances in Inorganic Biochemistry, Mechanisms of Metallocenter Assembly. Hausinger R P, Eichhorn G L, Marzilli L G, editors. New York: VHC; 1996. pp. 173–192. [Google Scholar]
- 30.Elsen S, Richaud P, Colbeau A, Vignais P M. J Bacteriol. 1993;175:7404–7412. doi: 10.1128/jb.175.22.7404-7412.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomelsky M, Kaplan S. Microbiology. 1995;141:1805–1814. doi: 10.1099/13500872-141-8-1805. [DOI] [PubMed] [Google Scholar]
- 32.Ninfa A J, Magasanik B. Proc Natl Acad Sci USA. 1986;83:5909–5911. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atkinson M R, Ninfa A. J Bacteriol. 1993;175:7016–7023. doi: 10.1128/jb.175.21.7016-7023.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L-F, Mandrand M A. FEMS Microbiol Rev. 1993;104:243–270. doi: 10.1111/j.1574-6968.1993.tb05870.x. [DOI] [PubMed] [Google Scholar]
- 35.Vignais P M, Dimon B, Zorin N A, Colbeau A, Elsen S. J Bacteriol. 1997;179:290–292. doi: 10.1128/jb.179.1.290-292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klose K E, Weiss D S, Kustu S. J Mol Biol. 1993;232:67–78. doi: 10.1006/jmbi.1993.1370. [DOI] [PubMed] [Google Scholar]
- 37.Ma S, Selvaraj U, Ohman D E, Quarless R, Hassett D J, Wozniak D J. J Bacteriol. 1998;180:956–968. doi: 10.1128/jb.180.4.956-968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]