Abstract
The DNA fragmentation factor 45 (DFF45) is a subunit of a heterodimeric nuclease complex critical for the induction of DNA fragmentation in vitro. To understand the in vivo role of DFF45 in programmed cell death, we generated DFF45 mutant mice. DNA fragmentation activity is completely abolished in cell extracts from DFF45 mutant tissues. In response to apoptotic stimuli, splenocytes, thymocytes, and granulocytes from DFF45 mutant mice are resistant to DNA fragmentation, and splenocytes and thymocytes are also resistant to chromatin condensation. Nevertheless, development of the immune system in the DFF45 mutant mice is normal. These results demonstrate that DFF45 is critical for the induction of DNA fragmentation and chromatin condensation in vivo, but is not required for normal immune system development.
Programmed cell death or apoptosis is a highly regulated physiological process critical in cellular differentiation and development. Insufficient or excess apoptosis may result in diseases such as neoplasia or neurodegeneration, respectively (1–4). The hallmarks of apoptosis include membrane blebbing, cytoplasmic and nuclear chromatin condensation, and cleavage of chromatin into nucleosomal fragments, i.e., DNA fragmentation (5). Genetic and biochemical analyses revealed that apoptosis is controlled by a molecular machinery that is highly conserved from nematode Caenorhabditis elegans to humans. The molecular mechanisms underlying apoptosis include regulation of the ced-9/bcl-2 family of genes, cytochrome c release, activation of caspases, chromatin condensation, DNA fragmentation and phagocytosis of dead cells, and debris by scavenger cells (6–19).
Recent in vitro biochemical studies have demonstrated that, upon induction of apoptosis, DNA fragmentation and chromatin condensation depend on a heterodimeric protein composed of DNA fragmentation factors 45 and 40 (DFF45 and DFF40, also called ICAD and CAD, respectively; refs. 20–23). Although DFF40 contains an intrinsic DNase activity, both DFF40 and DFF45 are required to generate this activity. DFF45 has been postulated to stabilize the synthesis of DFF40 (21, 23) or, alternatively, mediate correct folding and chromatin localization of DFF40 (22). Upon induction of apoptosis, DFF45 is cleaved by caspase-3 at amino acids 117 and 224 and dissociates from DFF40, resulting in the induction of DFF40 nuclease activity (20–23). Overexpression of DFF45 was shown to inhibit endogenous nuclease activity (21, 23).
DNA fragmentation in vivo may be an important step for disposal of large fragments of DNA from dying cells, which may be critical in maintaining normal tissue homeostasis (24–26). However, whether DFF45 is important for induction of DNA fragmentation in vivo, how DNA fragmentation plays a role in execution and regulation of apoptosis, and how disregulation of apoptosis at the stage of DNA fragmentation contributes to development and diseases remain unknown.
To address these issues, we have generated mutant mice carrying a deletion of the first three exons of the DFF45 gene. In vitro analyses indicate that DFF40 nuclease activity is completely abolished in tissues from the DFF45 mutant mice. Moreover, DFF45 mutant cells are resistant to DNA fragmentation and chromatin condensation in response to apoptotic stimuli. However, development of the immune cells in the DFF45 mutant mice appears normal. These findings suggest that DFF45 is essential in mediating DNA fragmentation and chromatin condensation in vivo, but is not required for normal immune system development.
MATERIALS AND METHODS
The DFF45 Gene, Targeting Construct, and ES Cell Homologous Recombinants.
Two oligonucleotide primers were used in PCRs with a mouse T cell cDNA library (CLONTECH) as the template. The sequences of these primers, which were chosen from the human DFF45 gene (20), are 5′-GGGAATTCTCGGCCTCTCAAACCGTGTCTGC and 5′-GGGGATCCCATCTGCCTCAAAGGACTCTTGG, respectively. The resulting PCR product contained part of the mouse DFF45 cDNA sequence and was used to clone the first three exons of the mouse DFF45 gene and the flanking sequences from a mouse 129 genomic library.
To make a DFF45 gene-targeting construct, four-piece DNA ligation was performed. The following DNA fragments were used: a 3.8-kb SalI-BglII fragment containing DNA from the 5′ end of the DFF45 gene, a 1.8-kb fragment containing a neo gene driven by the phosphoglycerate kinase (PGK) promoter, and an 8.0-kb EcoRI fragment containing DNA 3′ of the first three exons of the DFF45 gene and the plasmid pBluescript (Stratagene).
To obtain homologous recombinants, mouse J1 embryonic stem (ES) cells were transfected with 50 μg of linearized targeting construct by electroporation using a Bio-Rad Gene Pulser at 800 V and 3 μF. G418 selection was applied 24 hr after transfection at 200 μg/ml. G418-resistant colonies were picked and genomic DNA samples were isolated and digested with BglII and hybridized with a 5′ probe (Fig. 1A). Candidate homologous recombinants were confirmed by digesting their genomic DNA with EcoRV and hybridizing with a 3′ probe (Fig. 1A), and with BglII followed by hybridizing with an exon probe covering the first three exons of the DFF45 gene (Fig. 1A).
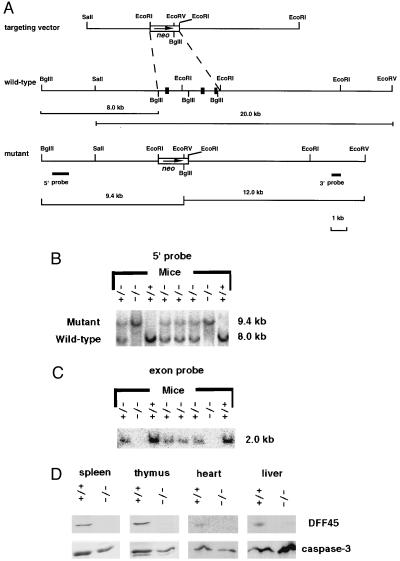
Figure 1.
Generation of DFF45 mutant mice. (A) Genomic DNA locus surrounding the first three exons (black boxes) of the DFF45 gene, the targeting vector, the mutant DFF45 gene locus, and the 5′ and 3′ hybridization probes. The exon probe contains 90, 170, and 40 bp from the first, second, and third exons of the DFF45 gene, respectively. (B and C) Identification of DFF45 mutant mice. Tail DNA samples from one litter of heterozygous intercross were digested with BglII, hybridized with a 5′ probe (B) or the exon probe (C). (D) Western blot analyses using spleen, thymus, heart, and liver extracts from wild-type (+/+) and DFF45 mutant mice (−/−) for the expression of DFF45 and caspase-3 (n = 4 mice each). Blots were probed with polyclonal antibodies against DFF45 or caspase-3, respectively.
DFF45 Mutant Mice.
ES cells amplified from the homologous recombinants were injected into blastocysts isolated from C57BL/6 female mice. The injected blastocysts were implanted back into the uteri of B6xDBA2 F1 females. The resulting male chimeric mice were bred repeatedly with C57BL/6 females, and germ-line transmission was identified initially by screening for agouti offspring. Mice heterozygous for the DFF45 gene mutation were confirmed by genomic Southern analyses of DNA isolated from their tails. Finally, mice homozygous for DFF45 gene mutation were produced by crossing heterozygous mutant animals and were identified by Southern blotting. Mutant and their wild-type control littermates 8 weeks of age were used for all subsequent analyses.
Western Blot Analysis.
Whole-cell extracts from spleen, thymus, heart, and liver were prepared by homogenizing and boiling in SDS gel loading buffer. Equal amounts of protein were analyzed by 12% acrylamide–SDS/PAGE and immunoblotted by rabbit polyclonal antibodies against human DFF45 (1:10,000), human DFF40 (1:2,000), and hamster caspase-3 (1:4,500), respectively (17, 20, 22). All immunoblots were visualized by enhanced chemiluminescence Western blotting detection reagents (Amersham).
Assay for DFF40 Nuclease Activity in Vitro.
Tissues including spleen, thymus, and testes from DFF45 mutant and wild-type mice were rinsed with 0.5 ml buffer A (20 mM Hepes⋅KOH, pH 7.5/1.5 mM MgCl2/10 mM KCl/0.5 mM EDTA/0.5 mM EGTA/1 mM DTT/1 mM phenylmethylsulfonyl fluoride) and dounced in 0.5 ml buffer A for 20 strokes in a 5-ml Wheaton douncer. The lysates were centrifuged at full speed in a microcentrifuge at 4°C for 10 min. The supernatants were centrifuged further at 100,000 × g for 30 min at 4°C in a TLA120 rotor. The supernatants were designated as S-100 fractions. An aliquot of 250 μg of S-100 fractions was incubated with 6 μl of hamster liver nuclei in the absence or presence of 150 μg caspase-3 at 37°C for 2 hr in a final volume of 60 μl buffer A. After all reactions, genomic DNA samples were extracted out, analyzed, and visualized as described previously (20).
DNA Fragmentation Analysis and Visualization of Chromatin Condensation.
Splenocytes and thymocytes were prepared from DFF45 mutant and wild-type mice. Cells (2 × 106/ml) were resuspended in DMEM and were cultured in the absence or presence of actinomycin D (500 nM), etoposide (50 μM), or staurosporin (2 μM) for different lengths of time at 37°C. Bone marrow cells depleted of red blood cells were cultured in RPMI 1640 medium at 3 × 106 cells/ml in the presence or absence of cycloheximide (10 μg/ml) for 3 hr at 37°C. DNA samples then were extracted from 1 × 106 cells by overnight incubation at 56°C in 0.1 ml of lysis buffer (50 mM Tris⋅HCl, pH 7.5/10 mM EDTA/150 mM NaCl/50 μg/ml of freshly made proteinase K). All DNA samples were recovered by isopropanol precipitation, resuspended in TE-RNase, analyzed on 1.5% agarose gels, and visualized by ethidium bromide staining.
Another 1 × 106 cells from the same treatments were fixed with 3.7% formaldehyde in PBS, washed with Tris-buffered saline, and stained with 1 μg/ml 4,6-diamidino-2-phenylindole (DAPI). Two hundred cells were examined for each sample. Images of the condensation states of the chromatin were taken with a fluorescence microscope coupled to a charge-coupled device camera.
Flow Cytometric Analyses.
For immune cell development analysis, single-cell suspensions were made from thymus, spleen, mesenteric lymph nodes, Peyer’s patches, bone marrow, and blood by using standard procedures. After depletion of erythrocytes by hypotonic lysis, approximately 1 × 106 cells were analyzed for cell surface marker expression and/or cell viability. Staining was carried out in PBS containing 1% FCS at 4°C with the following mAbs (all from PharMingen): T cell receptor (TCR) α/β-fluorescein isothiocyanate (FITC), CD8α-Cychrome, CD8α-FITC, CD4-PE, B220-PE, Mac-1-PE, Gr-1-FITC, IgD-FITC, and IgM-biotin. After staining, cells were incubated with streptavidin-PE (PharMingen) where required. Flow cytometric analysis was performed at the Howard Hughes Medical Institute Flow Cytometry Core (Vanderbilt University) by using a FACSCalibur system (Becton Dickinson) and cellquest 3.1 software. Plots shown represent staining patterns of cells within the white blood cell gate based on light-scattering characteristics.
RESULTS
Cloning of Mouse DFF45 Gene and Generation of DFF45 Mutant Mice.
A 300-bp DNA fragment containing part of the first three exons of DFF45 was generated by PCR (data not shown). Genomic Southern blot analysis using this DNA fragment as the exon probe (Fig. 1A) revealed that the DFF45 gene is present as a single-copy gene with no closely related pseudogenes (data not shown). The DFF45 exon probe then was used to screen a mouse 129 genomic library to obtain part of the mouse DFF45 gene and its flanking sequences. A restriction map of the relevant mouse DFF45 gene is shown in Fig. 1A. Sequence analysis indicated that the first three exons of the mouse DFF45 gene share 79% sequence identity with their human counterparts and are 97% identical to the published mouse DFF45 sequence (refs. 20 and 21; data not shown).
The DFF45 gene-targeting construct was designed to delete the first three exons of the DFF45 gene, which encode the first 142 aa, including the first caspase-3 cleavage site (Fig. 1A). Nine ES cell clones harboring the desired homologous recombination were identified. ES cells amplified from four of the homologous recombinants were used to generate male chimeric mice, which were bred with C57BL/6 females to obtain heterozygous mutant mice. Mice homozygous for the DFF45 gene mutation were obtained by sibling-mating of heterozygotes. Genomic Southern blotting with either a 5′ probe (Fig. 1B) or the DFF45 exon probe (Fig. 1C) both identified and confirmed the desired gene mutation. Western blot analyses indicated that the DFF45 protein was absent from tissues of mutant mice, while DFF40 (data not shown) and caspase-3 (Fig. 1D) levels were largely unaffected.
The DFF45 mutant mice appeared healthy and did not show any obvious growth abnormalities compared with their normal or heterozygous littermates. The genotype distribution of the first 66 offspring produced by heterozygous breeding was 19 wild-type mice, 32 heterozygous mutants, and 15 homozygous mutants, indicating no segregation distortion. Both male and female DFF45 mutant mice were fertile when crossed with wild-type mice, and normal numbers of pups were obtained with no obvious sex bias. The DFF45 mutant females nursed their offspring to adulthood.
Lack of DFF40 Nuclease Activity in Extracts from DFF45 Mutant Tissues.
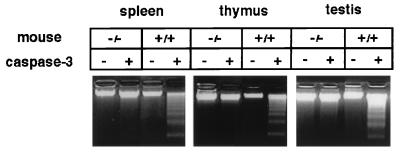
In normal cells, DFF45 is associated with DFF40. Upon induction of apoptosis, DFF45 is cleaved away by caspase-3, resulting in the induction of DFF40 nuclease activity (21, 22). To investigate the effects of DFF45 deficiency on DFF40 nuclease activity in vivo, we isolated extracts from spleen, thymus, and testes from DFF45 mutant and wild-type control mice (n = 3 each). We then assayed for DFF40 nuclease activity in vitro in the presence or absence of activated caspase-3, as described previously (20). As shown in Fig. 2, there was abundant DFF40 nuclease activity in tissues from control mice in the presence of caspase-3 based on DNA laddering. In contrast, this activity was completely absent in tissue extracts prepared from mutant spleen, thymus, and testes. Thus, mutating the DFF45 gene led to the inactivation of DFF40 nuclease activity, as judged by this assay, indicating that DFF45 is essential for normal expression of DFF40 nuclease activity in these tissues in vivo.
Figure 2.
Lack of DFF40 nuclease activity in DFF45 mutant mice. Cell extracts from spleen, thymus, and testes from both DFF45 mutant (−/−) and control (+/+) mice (n = 3 each) were isolated. DFF40 nuclease activity in these extracts was assayed by using hamster liver nuclei in the presence or absence of activated caspase-3. Genomic DNA samples were extracted, separated on 1.5% agarose gels, and visualized by ethidium bromide staining.
Resistance to DNA Fragmentation and Chromatin Condensation in DFF45 Mutant Cells.
To understand the consequences of DFF45 gene mutation on apoptosis, we measured the extent of DNA fragmentation and chromatin condensation in response to various apoptotic stimuli by using cells from DFF45 mutant mice. We first isolated DNA from multiple tissues, including brain, liver, kidney, heart, testis, ovary, uterus, and skin and measured steady-state levels of spontaneous DNA fragmentation in both DFF45 mutant and control mice by gel electrophoresis (n = 5 each). No significant spontaneous DNA fragmentation was observed in any tissues from DFF45 mutant mice, which is similar to that found for normal control mice (data not shown).
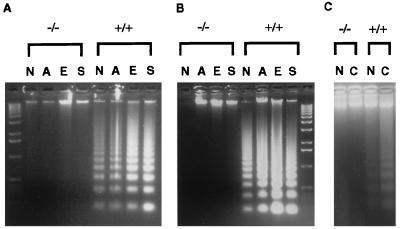
We next isolated cells from spleen and thymus from mutant and wild-type mice and exposed these cells to actinomycin D, etoposide, and staurosporine for different time periods. Samples were then divided into two parts and analyzed for DNA fragmentation or for chromatin condensation. To assay for DNA fragmentation, DNA samples were isolated from one-half of the cells and analyzed on agarose gels and compared with DNA samples from untreated cells. After 10 hr of drug treatment, the DNA samples from splenocytes and thymocytes from DFF45 mutant mice were devoid of DNA fragmentation, similar to those from the untreated cells. In contrast, the DNA samples from wild-type control cells exhibited standard DNA laddering under these experimental conditions (Fig. 3 A and B). After 24 hr of treatment, DNA samples from thymocytes were still intact and those from the splenocytes started undergoing some DNA fragmentation, but at a much more reduced level compared with the wild-type cells (data not shown). These results suggest that, upon induction of apoptosis, DFF45 is required for DNA fragmentation in splenocytes and thymocytes in vivo.
Figure 3.
Resistance to DNA fragmentation in cells from DFF45 mutant mice. Genomic DNA samples were isolated from splenocytes (A) and thymocytes (B) from both mutant (−/−) and control (+/+) mice with or without actinomycin D, etoposide, and staurosporine treatment (n = 5 mice each), and from granulocytes with or without cycloheximide treatment (C, n = 3 mice each). All DNA samples were extracted, separated on 1.5% agarose gels, and visualized by ethidium bromide staining. N, no treatment; A, actinomycin D; E, etoposide; S, staurosporine; C, cycloheximide.
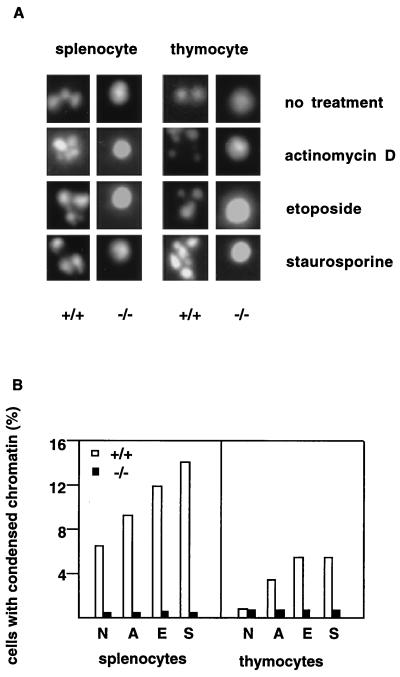
The second half of the cells was used to assess chromatin condensation. After 10 hr of drug treatment, chromatin condensation in mutant cells was severely impaired as shown by DAPI staining of the nuclear DNA (Fig. 4 A and B). Only after prolonged incubation of the DFF45-deficient cells in culture medium (24 hr) was some chromatin condensation observable, but at a much reduced level compared with the wild-type cells (data not shown). Splenocytes and thymocytes from three mutant and three control mice all gave consistent results, respectively, suggesting that DFF45 is required for chromatin condensation in splenocytes and thymocytes in vivo.
Figure 4.
Resistance to chromatin condensation in cells from DFF45 mutant mice. Splenocytes and thymocytes from both DFF45 mutant (−/−) and wild-type control (+/+) mice (n = 3 each) were cultured in the absence or presence of actinomycin D, etoposide, and staurosporine. DAPI stainings of these cells were visualized under a fluorescence microscope, and 200 cells for each treatment condition were observed (A). Percentage of cells exhibiting chromatin condensation was counted and plotted (B). N, no treatment; A, actinomycin D; E, etoposide; S, staurosporine.
Inflammatory responses are regulated in part by elimination and clearance of immune effector cells through apoptosis (27). Since granulocytes represent a major subset of inflammatory mediators, we investigated the possible role of DFF45 in granulocyte apoptosis. Since cycloheximide treatment of bone marrow granulocytes rapidly induces apoptosis (28), we examined cycloheximide-induced apoptosis of bone marrow cells from DFF45 mutant mice and wild-type littermates. We treated bone marrow granulocytes (GR-1-positive cells) with cycloheximide for 3 hr as described (29). DNA samples were harvested and analyzed by agarose gel electrophoresis. As shown in Fig. 3C, the DFF45 mutant granulocytes consistently showed less DNA fragmentation than the wild-type control granulocytes. Granulocytes from three mutant and three control mice gave consistent results, respectively. This result parallels those from the splenocytes and thymocytes, suggesting that DFF45 is required for DNA fragmentation in granulocytes in vivo.
No Overt Alterations in the Development of the Immune System in DFF45 Mutant Mice.
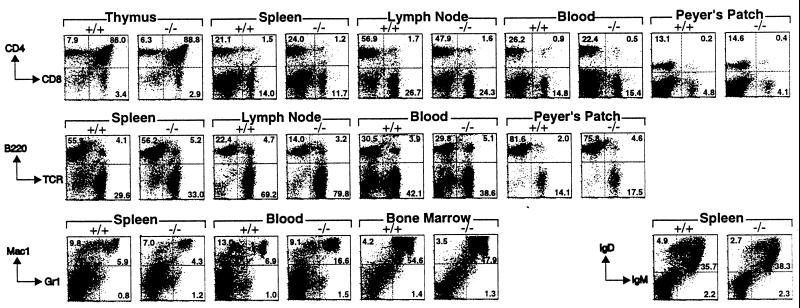
Since programmed cell death plays a prominent role during the development of the immune system (30), we analyzed the effects of DFF45 deficiency on immune system development in more detail. Lymphoid tissues including thymus, spleen, lymph nodes, and Peyer’s patches from the mutant mice were similar in size to those from normal littermate controls. Flow cytometric analyses of thymus, spleen, lymph nodes, Peyer’s patches, blood, and bone marrow from mutant and wild-type mice at 8 weeks of age then were performed. Total numbers of immune cells recovered from DFF45 mutant animals were not significantly different from those of the wild-type animals (data not shown). White blood cells derived from these tissues were analyzed for the expression of selective cell surface markers (Fig. 5). The distribution of T and B lineage cells in all lymphoid compartments tested appeared similar in mutant and wild-type animals. Similarly, myeloid populations also were largely unaffected, except for a small increase of granulocytes (Mac-1+/Gr-1+) in the blood of DFF45 mutant animals. Although this modest increase was seen in two independent experiments, more animals will need to be analyzed to assess the significance of this finding. Thus, it appears that the presence of DFF45 is not obligatory for the development of the immune system.
Figure 5.
Normal immune system development in DFF45 mutant mice. Cells were harvested from lymphoid organs from wild-type (+/+) and DFF45 mutant (−/−) mice and analyzed for the expression of the indicated cell surface markers. Each plot shown is representative of results from three DFF45 mutant and three control mice, except for Peyer’s Patch, which is representative of results from two DFF45 mutant and two control mice. Numbers indicate the percentage of cells in each quadrant.
Peripheral blood cell counts from DFF45 mutant mice also appeared normal. Cell types surveyed included total white blood cells, red blood cells, and platelets. Among the white blood cells, percentages of neutrophils, lymphocytes, monocytes, eosinophils, and basophils all fell within normal range (data not shown). A preliminary histological survey of all the major internal organs in DFF45 mutant mice revealed no significant alterations, including hypertrophy, when compared with those from the control mice (G. Boivin and M.X., unpublished results). Thus, all the major organ systems in DFF45 mutant mice appeared to have developed normally despite defects in DNA fragmentation and chromatin condensation in response to apoptotic stimuli.
DISCUSSION
To understand the in vivo function of DFF45 in programmed cell death, we generated DFF45 mutant mice by using targeted homologous recombination in ES cells. Analysis of the mutant mice indicated that cell extracts from DFF45 mutant mice lack in vitro DNA fragmentation activity. DFF45 mutant cells are strikingly resistant to DNA fragmentation and chromatin condensation in response to apoptotic stimuli. Despite this inability to undergo DNA fragmentation, development of the immune system appears normal. These results indicate that DFF45 is essential for the induction of DNA fragmentation and chromatin condensation in vivo, but is not required for the immune system development.
DFF45 in DNA Fragmentation, Chromatin Condensation, and Apoptosis.
Our findings that DFF45 mutant cells are resistant to DNA fragmentation and chromatin condensation in response to several apoptotic stimuli strongly suggest that DFF45 plays a critical role in vivo in internucleosomal cleavage of DNA and in the induction of chromatin condensation during apoptosis. Moreover, these findings indicate that DFF45 function cannot be fully compensated by other mechanisms, such as the existence of DFF45 homologs or other nuclease systems, in DFF45 mutant mice.
Previous studies suggested that DFF45 may function both as a specific inhibitor and a molecular chaperone for the DFF40 nuclease (21, 22, 31). On the one hand, the inhibitory activity of DFF45 may ensure that no DFF40 nuclease would be active until an apoptotic signal activates caspase-3, which results in cleavage and dissociation of DFF45. On the other hand, DFF45 may be required for DFF40 synthesis, correct folding, or recruitment to proper chromosomal locations (21, 22, 31). Our results suggest that this latter molecular chaperone function of DFF45 is more dominant in vivo, since we did not detect any DFF40 nuclease activity in DFF45 mutant cell extracts. How DFF45 fulfills these functions will require further biochemical investigations.
At present, we do not know whether DFF45 also plays a role in the feedback regulation of programmed cell death. A previous study showed that an intact cell nucleus and DNA fragmentation are not required for apoptosis under certain conditions (32). Consistent with this view, we have noted early plasma membrane changes and activation of caspase-3 in the absence of DFF45 (J.Z. and M.X., unpublished results). Together, these findings suggest that upstream events for apoptosis in DFF45 mutant cells could occur in response to certain apoptotic stimuli. Nevertheless, it remains to be determined whether these upstream events occur at a reduced level in DFF45 mutant cells.
DFF45 Function in Development.
We found that development of the immune system appears normal in DFF45 mutant mice. Moreover, these mutant mice stayed healthy for up to at least 8 weeks of age and none of them developed any apparent abnormalities. Our preliminary histological analyses suggest that DFF45 is dispensable for the development of all the major organ systems. Together, these observations indicate that resistance to DNA fragmentation and chromatin condensation has not produced detectable detrimental effects to the development of the immune system and likely other systems up to this age in DFF45 mutant mice. This raises the critical question of whether the endogenous system for DNA removal after apoptosis is critical for normal development and maintenance of tissue homeostasis, or whether such DNA removal system is equally effective for both small (fragmented) and large (unfragmented) DNA. A systematic analysis of development in DFF45 mutant mice at different ages will be essential to resolve these issues.
Alternatively, there may be compensatory mechanisms or redundant pathways for DNA fragmentation and chromatin condensation that allow normal development of the immune system and possibly other systems in DFF45 mutant mice. Such developmental compensation or redundancy has been observed previously in many mutant mice lacking apoptotic executioner molecules, including caspases-1 and 3 (29, 33–36). Possible compensatory or redundant mechanisms for DNA fragmentation may involve the existence of other nucleases that produce DNA fragmentation in apoptosis (37–39). Alternatively, redundant DFF45 activity may exist to partially compensate for the loss of DFF45 in the mutant mice. The recent identification of CIDE-A and CIDE-B indicates that DFF45 homologs do exist (40). Whether there is an up-regulation of CIDE activity in DFF45 mutant mice and whether CIDEs directly interact with DFF40 to regulate DFF40 nuclease activity remain to be investigated.
Despite these unresolved issues, the DFF45 mutant mice provide valuable new tools for studying the role of defective DNA fragmentation in various biological and pathological processes, including normal development, neurodegeneration triggered by excitatory signals or free radicals, normal and autoimmune responses, genomic instability in various tissues induced by adverse conditions, and tumorigenesis.
Acknowledgments
We are grateful to J. Duffy and M. Yin and the ES cell-gene-altered mouse service core for blastocyst injections. We thank K. Fukasawa, D. Ballard, E. Oltz, and G. Hicks for sharing reagents; S. Pixley and N. Koster for helping with microscopy; and D. McFarland for help with flow cytometry. We thank P. Stambrook and K. Fukasawa for advice and stimulating discussions and P. Stambrook for critical comments on the manuscript. J.Z. is an Effie D. Beeman National Alliance for Research on Schizophrenia and Depression investigator and is supported by National Institute on Drug Abuse (DA11284). X.W. is supported in part by grants from the American Cancer Society (RE258) and National Institutes of Health (GMRO1–55942). M.X. is a National Alliance for Research on Schizophrenia and Depression investigator and is supported by a startup fund from the University of Cincinnati College of Medicine and by the National Institute on Drug Abuse (DA11005).
ABBREVIATIONS
- DFF45
45-kDa subunit of DNA fragmentation factor
- ES
embryonic stem
Footnotes
A Commentary on this article begins on page 12077.
References
- 1. Raff M C. Nature (London) 1992;356:397–400. doi: 10.1038/356397a0. [DOI] [PubMed] [Google Scholar]
- 2.Thompson C B. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 3.Wyllie A H. Curr Opin Genet Dev. 1995;5:97–104. doi: 10.1016/s0959-437x(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 4.White E. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Wyllie A H. Nature (London) 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- 6.Yuan J-Y, Horvitz H R. Dev Biol. 1990;138:33–41. doi: 10.1016/0012-1606(90)90174-h. [DOI] [PubMed] [Google Scholar]
- 7.Yuan J-Y, Horvitz H R. Development. 1992;116:309–320. doi: 10.1242/dev.116.2.309. [DOI] [PubMed] [Google Scholar]
- 8.Hengartner M O, Ellis R E, Horvitz H R. Nature (London) 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- 9.Yuan J-Y, Shaham S, Ledoux S, Ellis M H, Horvitz H R. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 10.Fernandes-Alnemri T, Litwack G, Alnemri E S. J Biol Chem. 1994;269:30761–30764. [PubMed] [Google Scholar]
- 11.Hengartner M O, Horvitz H R. Cell. 1994;76:665–676. doi: 10.1016/0092-8674(94)90506-1. [DOI] [PubMed] [Google Scholar]
- 12.Kumar S, Kinoshita M, Noda M, Copeland N G, Jenkens N A. Genes Dev. 1994;8:1613–1626. doi: 10.1101/gad.8.14.1613. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Miuru M, Bergeron L, Zhu H, Yuan J. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson W E, Ali A, Thornberry N A, Vailancourt J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A, et al. Nature (London) 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Kim C N, Yang J, Jemmerson R, Wang X. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 16.Schlegel J, Peters I, Orrenius S, Miller D K, Thornberry N A, Yamin T T, Nicholson D W. J Biol Chem. 1996;271:1841–1844. doi: 10.1074/jbc.271.4.1841. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Zelenski N G, Yang J, Sakai J, Brown M S, Goldstein J L. EMBO J. 1996;15:1012–1020. [PMC free article] [PubMed] [Google Scholar]
- 18.Vaux D L. Cell. 1997;90:389–390. doi: 10.1016/s0092-8674(00)80497-3. [DOI] [PubMed] [Google Scholar]
- 19.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Zou H, Slaughter C, Wang X. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 21.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. Nature (London) 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 22.Liu, X., Li, P., Widlak, P., Zou, H., Luo, X., Garrard, W. T. & Wang, X. (1998) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 23.Sakahira H, Enari M, Nagata S. Nature (London) 1998;391:96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- 24.Peitsch M C, Mannherz H G, Tschopp J. Trends Cell Biol. 1994;4:37–41. doi: 10.1016/0962-8924(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 25.Bortner C D, Oldenburg N B, Cidlowski J A. Trends Cell Biol. 1995;5:21–27. doi: 10.1016/s0962-8924(00)88932-1. [DOI] [PubMed] [Google Scholar]
- 26.Wyllie A H. Nature (London) 1998;391:20–21. doi: 10.1038/34040. [DOI] [PubMed] [Google Scholar]
- 27.Homburg C H, Roos D. Curr Opin Hematol. 1996;3:94–99. doi: 10.1097/00062752-199603010-00014. [DOI] [PubMed] [Google Scholar]
- 28.Tsuchida H, Takeda Y, Takei H, Shinzawa H, Takahashi T, Sendo F. J Immunol. 1995;154:2403–2412. [PubMed] [Google Scholar]
- 29.Woo M, Hakem R, Soengas M S, Duncan G S, Shahinian A, Kagi D, Hakem A, McCurrach M, Khoo W, Kaufman S A, et al. Genes Dev. 1998;12:806–819. doi: 10.1101/gad.12.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osborne B A. Curr Opin Immunol. 1996;8:245–254. doi: 10.1016/s0952-7915(96)80063-x. [DOI] [PubMed] [Google Scholar]
- 31.Halenbeck R, MacDonald H, Roulston A, Chen T T, Conroy L, Williams L T. Curr Biol. 1998;8:537–540. doi: 10.1016/s0960-9822(98)79298-x. [DOI] [PubMed] [Google Scholar]
- 32.Schulze-Osthoff K, Walczak H, Droge W, Krammer P H. J Cell Biol. 1994;127:15–20. doi: 10.1083/jcb.127.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuida K, Lippke J A, Ku G, Harding M W, Livingston D J, Su M S-S, Flavell R A. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 34.Li P, Allen H, Banerjee S, Franklin S, Herzog L, Johnston C, McDowell J, Paskind M, Rodman L, Salfeld J, et al. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 35.Zheng H, Fletcher D, Kozak W, Jiang M, Hofmann K J, Conn C A, Soszynski D, Grabiec C, Trumbauer M E, Shaw A, et al. Immunity. 1995;3:9–19. doi: 10.1016/1074-7613(95)90154-x. [DOI] [PubMed] [Google Scholar]
- 36.Kuida K, Zheng T S, Na S, Kuan C-y, Yang D, Karasuyama H, Rakic P, Flavell R A. Nature (London) 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- 37.Gaido M L, Cidlowski J A. J Biol Chem. 1991;269:30761–30764. [Google Scholar]
- 38.Nikonova L V, Beletsky I P, Umansky S R. Eur J Biochem. 1993;215:893–901. doi: 10.1111/j.1432-1033.1993.tb18107.x. [DOI] [PubMed] [Google Scholar]
- 39.Peitsch M C, Polar B, Stephan H, Crompton T, MacDonald H R, Mannherz H G, Tschopp J. EMBO J. 1993;12:371–377. doi: 10.1002/j.1460-2075.1993.tb05666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inohara N, Koseki T, Chen S, Wu X, Nunez G. EMBO J. 1998;17:2526–2533. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]