Abstract
Telomere length is maintained through a dynamic balance between addition and loss of the terminal telomeric DNA. Normal telomere length regulation requires telomerase as well as a telomeric protein–DNA complex. Previous work has provided evidence that in the budding yeasts Kluyveromyces lactis and Saccharomyces cerevisiae, the telomeric double-stranded DNA binding protein Rap1p negatively regulates telomere length, in part by nucleating, by its C-terminal tail, a higher-order DNA binding protein complex that presumably limits access of telomerase to the chromosome end. Here we show that in K. lactis, truncating the Rap1p C-terminal tail (Rap1p-ΔC mutant) accelerates telomeric repeat turnover in the distal region of the telomere. In addition, combining the rap1-ΔC mutation with a telomerase template mutation (ter1-kpn), which directs the addition of mutated telomeric DNA repeats to telomeres, synergistically caused an immediate loss of telomere length regulation. Capping of the unregulated telomeres of these double mutants with functionally wild-type repeats restored telomere length control. We propose that the rate of terminal telomere turnover is controlled by Rap1p specifically through its interactions with the most distal telomeric repeats.
Telomeres are the specialized nucleoprotein structures that cap the ends of eukaryotic chromosomes, stabilizing them and protecting their integrity (1–3). Telomeric DNA of most eukaryotes is composed of tandemly repeated sequence units whose number is tightly regulated (4). This strict length control appears to be essential for telomeric function and hence for chromosomal integrity and, in many cases, for cell viability (5). A key component influencing telomere length is telomerase, which adds telomeric DNA to telomeres (1, 5). It has been proposed that telomeric repeats are constantly added by telomerase and removed by as yet unknown activity or activities, leaving mean telomeric length at a regulated average size (6). This tight length control, seen in species such as the budding yeasts S. cerevisiae and K. lactis, reflects a well regulated balance between the lengthening and shortening activities, the latter perhaps including specialized telomere shortening activities (6–11).
Telomere lengths do not always correlate directly with telomerase levels (for example, see refs. 12 and 13), indicating that additional factors are involved in telomere length control. Proteins that bind duplex telomeric DNA sequence-specifically have been shown to negatively regulate telomere length. Such proteins include the RAP1 gene product Rap1p in budding yeasts (7, 14–17), Taz1p in fission yeast (18), and TRF1 in mammalian cells (19–21). In S. cerevisiae, Rap1p nucleates a higher-order DNA–protein complex at telomeres. Assembly of this complex is mediated through interactions between the C-terminal region of Rap1p and other non-DNA-binding proteins (22, 23).
The C-terminal 30 amino acid tail of Rap1p of K. lactis is highly conserved between this budding yeast and S. cerevisiae, suggesting that the K. lactis Rap1p also interacts with other proteins through its C-terminal tail and similarly nucleates a specialized telomeric protein–DNA complex. We previously showed that in K. lactis, sequence-specific binding of the telomeric DNA at the distal end of the telomere by Rap1p is critical for telomere length regulation (17). It was proposed that telomeric DNA length is negatively controlled at two levels: through the higher-order Rap1p-nucleated DNA–protein complex at telomeres and through the interaction of Rap1p at the very terminal few repeats. This model is supported by the finding of two groups of mutations in the telomerase RNA (TER1) gene that direct the incorporation of altered telomeric DNA sequence (11). In the first group, the mutation disrupted the Rap1p binding site and telomere length control was lost immediately. The phenotypic severity of these mutations in vivo correlated directly with the degree of loss of Rap1p binding affinity to the mutated telomeric sequence in vitro (17). In addition, truncation of the C-terminal tail of Rap1p synergistically exacerbated the loss of telomere length control in these mutants, and we proposed that the telomeric complex in these double mutants is unable to limit telomerase accessibility (17). In the second group of ter1 template mutants (11), in which the Rap1p consensus binding sequence was not mutated and the in vitro binding affinity was not decreased, there was no immediate loss of telomere length control. Instead, telomere length was well controlled for an initial phase lasting hundreds of generations. During this phase, telomeres were stably maintained at shorter-than-wild-type size. However, during this phase the wild-type repeats in the inner portions of the telomeres were gradually replaced by mutant repeats. Eventually, when mutant repeats had replaced most of the wild-type telomeric repeats deep within the telomere, telomeres rapidly underwent extreme, uncontrolled lengthening (delayed runaway phenotype) (11).
In species such as S. cerevisiae and the malaria parasite Plasmodium berghei, the naturally variant telomeric repeat sequences allow tracking of sequence replacements in the telomeres over time. Sequence analysis of telomeres in these species has suggested that turnover occurs in the more distal portion of the telomere, with the inner region being relatively inert (24, 25). Confinement of such turnover to the distal repeats was directly demonstrated in K. lactis by monitoring incorporation of mutant repeats specified by ter1 template mutations introduced into initially wild-type telomeres (11, 26). However, the enzymatic machineries involved in telomere dynamics and repeat turnover, and the telomeric protein complex that regulates them, are only partly understood. Here we show that the Rap1p C-terminal tail is required to limit the rate of turnover of the most distal telomeric repeats in K. lactis. We also report a synthetic effect between Rap1p C-terminal truncation and a delayed runaway ter1 mutation. Together, these results identify a new role for the Rap1p-centered telomeric DNA complex, namely, controlling the rate of telomere turnover.
MATERIALS AND METHODS
Yeast Growth.
Yeast cells were grown on yeast extract/peptone/dextrose plates. For serial passages, colonies were picked and restreaked on fresh plates, as described previously (11).
Yeast Strains.
The rap1-ΔC strain was constructed by transforming K. lactis strain K7B520 with a plasmid containing the rap1-ΔC allele carrying a frameshift mutation at amino acid 635, producing KlRap1p lacking the normal C-terminal 31 amino acids. Loop-outs of the wild-type KlRap1p gene were selected following growth on plates containing 5-fluoroorotic acid. The double mutant ter1-Kpn; rap1-ΔC was constructed by transforming the rap1-ΔC strain with a plasmid containing the ter1-Kpn allele. Loop-outs of either the wild-type TER1 gene or the mutant gene (and a linked URA3 gene) were assayed. The ter-Bcl capped strains were constructed by transforming ter1-Kpn;rap1-ΔC with a plasmid carrying the mutant ter-Bcl allele.
Genomic DNA Preparation and Analysis.
Yeast DNA was prepared from colonies grown to saturation in yeast extract/peptone/dextrose liquid media and analyzed by Southern blotting, as described previously (11). Blots were probed at 50°C with an end-labeled oligomer: 5′-GGTATGTGGTGTACGGATTTGATTA-3′.
RESULTS
Turnover of Distal but Not Internal Telomeric Repeats Is Accelerated in a rap1-ΔC Mutant.
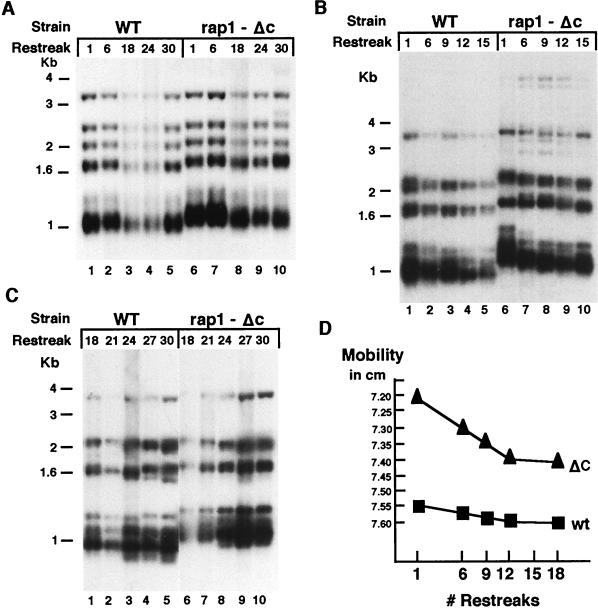
We tested whether a 30 amino acid truncation of the C-terminal tail of Rap1p (rap1-ΔC; ref. 17) affected the rate of turnover at telomere terminal domains. To monitor terminal repeat turnover, we took advantage of a phenotypically silent ter1 RNA template sequence mutation, ter1-BclI (M. J. McEachern and E.H.B., unpublished results). This mutation is located outside the Rap1p binding site of the telomeric repeat unit of K. lactis and produces no detectable short- or long-term effects on telomere mean length, length regulation, or telomere metabolism (M. J. McEachern and E.H.B., unpublished results; ref. 29). Incorporation of the BclI base change into the ends of telomeres therefore serves to mark the internalization of newly added telomeric repeats. The ter1-Bcl mutant allele was introduced into both a wild-type and a rap1-ΔC background, replacing the wild-type TER1 gene. Telomere lengths were analyzed over repeated serial passages by Southern blotting of genomic DNA digested with EcoRI, which cleaves at subtelomeric sites (Fig. 1). Over 30 serial restreaks (about 750 generations), telomeres containing Bcl repeats were stably maintained and had the mean lengths and the overall length distributions characteristic of wild-type RAP1 (15 repeats on average) or rap1-ΔC backgrounds (about 4 repeats longer) (Fig. 1A). As reported previously (17), telomeres in rap1-ΔC cells are well regulated and slightly (≈100 bp) longer than those in wild-type RAP1 cells. However, in the rap1-ΔC cells, the rate of internalization of new (Bcl-marked) telomeric repeats was higher than in RAP1 cells. This was shown by secondary digestion of the same DNA preparations as in Fig. 1A with restriction enzyme BclI (Fig. 1 B and C). BclI specifically cleaves off the marked Bcl telomeric repeats added onto the pre-existing wild-type repeats of the telomeres. Hence, the resulting telomeric fragment pattern reflects the lengths of the remaining wild-type repeat tracts located internally to the newly incorporated Bcl repeats. After replacement of the wild-type telomerase RNA gene TER1 with the ter1-BclI allele during the first nine restreaks, the remaining wild-type repeat tracts shortened faster in the rap1-ΔC strain than in wild-type RAP1 cells (Fig. 1B, compare lanes 6–8 with lanes 1–3). This result showed that the rap1-ΔC mutation increases the rate of replacement of wild-type repeats by BclI repeats in a terminal zone of the telomere. This terminal zone extended into telomeres an average of two to three 25-bp repeat units in rap1-ΔC cells and an average of about one repeat in wild-type RAP1 cells. In subsequent restreaks, the rate of BclI repeat encroachment further into the telomeres was low and similar in the two strains (Fig. 1B, compare lanes 4 and 5 with lanes 9 and 10; Fig. 1C, compare lanes 1–5 with lanes 6–10; results are graphed in Fig. 1D). Despite their greater mean length, in the telomeres in rap1-ΔC cells, the zone of increased turnover rate did not extend deeper than the extra length of these telomeres (Fig. 1D).
Figure 1.
Telomere turnover is accelerated in a rap1-ΔC background. (A) Telomeric patterns of EcoRI-digested genomic DNA from wild-type and rap1-ΔC strains followed for 30 restreaks after introduction of the ter1-Bcl telomerase RNA gene allele. The reduced signal in lanes 3 and 4 is because of their underloading. (B) Telomeric patterns of EcoRI + BclI-digested genomic DNA from wild-type and rap1-ΔC strains followed for the first 15 restreaks. (C) Telomeric patterns of EcoRI + BclI-digested genomic DNA from wild-type and rap1-ΔC strains followed for the subsequent 18 to 30 restreaks. (D) Graphic depiction of telomere turnover (as represented by the size of basal wild-type repeats remaining after BclI digestion as a function of number of restreaks) in wild-type RAP1 and rap1-ΔC strains.
In summary, these results identified two distinct zones of telomeric turnover: distal and proximal. The rap1-ΔC mutation specifically increased the turnover rate in the most distal zone of repeats. Interestingly, in rap1-ΔC telomeres, the outer boundary of the internal, more slowly turned over region was still further out than the mean length of telomeres in a wild-type RAP1 strain.
A Delayed Runaway Telomere Phenotype Is Accelerated in ter1-Kpn;rap1-ΔC Mutants.
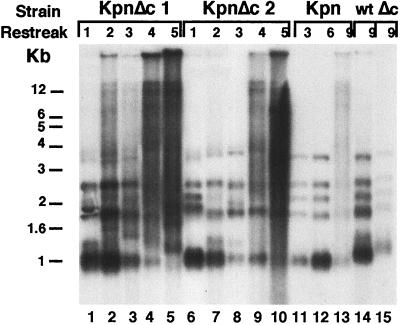
Since Rap1p plays a pivotal role in telomere length regulation, we studied its involvement in the phenotype caused by the telomerase RNA template mutation ter1-KpnI (11). As described previously, in single ter1-KpnI or single rap1-ΔC mutants, telomeres remain well controlled up to at least 15 restreaks (11, 17). In the single ter1-KpnI mutants, delayed runaway telomere elongation occurs only after several hundred cell divisions (11). As reported previously, a double-stranded DNA oligonucleotide containing the ter1-KpnI mutation binds both wild-type Rap1p and rap1p-ΔC in vitro as efficiently as the wild-type sequence oligonucleotide (17). Thus, simple loss of DNA binding affinity to the mutated telomeric repeats does not account for the loss of length regulation. Therefore, we analyzed the telomeric patterns in ter1-KpnI;rap1-ΔC double mutant cells. Interestingly, in contrast to the effect of introducing the ter1-KpnI allele into RAP1 cells, introducing it into rap1-ΔC cells caused rapid, essentially immediate loss of telomere length regulation: in seven independent ter1-Kpn;rap1-ΔC double mutant lines analyzed, by the second restreak the telomeric patterns had already deviated from the wild-type pattern, with bands becoming smeared up from the electrophoretic mobilities of the wild-type bands in neutral gels (Fig. 2, compare lanes 2 and 7 with lanes 11, 12, and 14; data not shown). This accelerated loss of length regulation in the double mutant was rapidly exacerbated on further restreaking, so that by 3–4 restreaks, telomeres had not only become very long, but also a significant fraction of the hybridization signal showed the aberrant smeared pattern characteristic of telomeres lacking length control (Fig. 2, lanes 3–5 and 8–10). These very smeared bands suggested that telomeric degradation as well as lengthening has lost regulation, which we will refer to as a deregulated telomere phenotype. Consistent with previous results (11, 17), telomeres were only slightly lengthened in the single rap1-ΔC mutant up to at least the ninth restreak (Fig. 2, lane 15) and remained slightly shortened in the single ter1-KpnI mutant cells up to the seventh restreak (Fig. 2, lanes 11 and 12; two additional independent single ter1-KpnI mutant clones gave similar results; data not shown). These results showed that the two mutations synergistically ablated telomere length control. Hence a function mediated by the Rap1p C-terminal tail at telomeres is involved in the Kpn phenotype, despite the ability of the Kpn repeat to bind Rap1p with high efficiency in vitro.
Figure 2.
Synergistic interaction between ter1-Kpn and rap1-ΔC mutations. Telomeric patterns of EcoRI-digested DNAs from two independent clones of the double mutant strains ter1-Kpn;rap1-ΔC (KpnΔC1 and KpnΔC2), a single ter1-KpnI strain (Kpn), a single rap1-ΔC strain (Δc), and a wild-type strain (wt). Strains were followed for the number of restreaks indicated above the lanes.
Internal Wild-Type Telomeric Repeat Tract Lengths Are Similar in ter1-Kpn and ter1-Kpn;rap1-ΔC Strains.
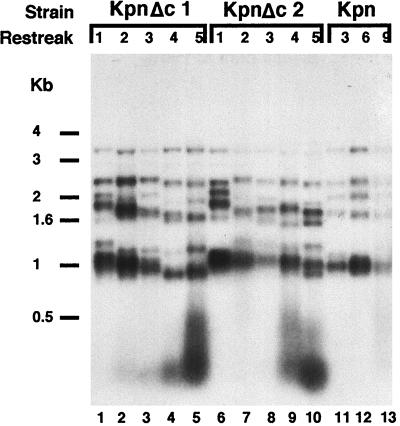
In RAP1 cells, Kpn mutant repeats impair length control only when they have replaced most of the wild-type telomeric repeats deep within the telomere (11). Therefore, it was possible that the accelerated onset of the telomere runaway phenotype in the ter1-Kpn;rap1-ΔC double mutant was a result of faster and more extensive internalization of Kpn repeats caused by the rap1-ΔC mutation. We tested this possibility by comparing the sizes and patterns of the internal tracts of remaining wild-type telomeric repeats in ter1-Kpn and ter1-Kpn;rap1-ΔC mutants, using secondary digestion with KpnI to cleave off the mutant Kpn repeats. The resulting telomeric patterns, reflecting the lengths of the basal internal tracts of wild-type repeats, were similar between ter1-Kpn single mutant strains (Fig. 3, lanes 11–13) and two independent double mutant ter1-Kpn;rap1-ΔC strains (Fig. 3, lanes 1–5 and 6–10). This similarity was especially striking in light of the very different terminal domains of the telomeres in the single and double mutants. This difference is clearly seen when the mutant repeats are not digested away by KpnI (see Fig. 2, compare lanes 5 and 10 to lane 12). In addition, there was no evidence for any large rearrangements of internal telomeric or subtelomeric domains. These results ruled out the hypothesis that the synthetic interaction between the rap1-ΔC and ter1-Kpn mutations is caused by deeper internalization of mutant Kpn repeats.
Figure 3.
Basal telomere pattern is similar in ter1-Kpn and ter1-Kpn;rap1-ΔC strains. Telomeric patterns of EcoRI + KpnI-digested genomic DNAs from two clones of the double mutant strains ter1-Kpn;rap1-ΔC (KpnΔc1 and KpnΔc2) and a single ter1-Kpn strain (Kpn). The strains are the same clonal lines shown in Fig. 2. The sizes of the telomeric restriction fragments (bands between 1- and 4-kb markers) reflect the lengths of the remaining basal internal tracts of wild-type repeats, which are not cleaved by KpnI restriction enzyme. The diffuse hybridization signal below the 0.5-kb marker is from the Kpn mutant repeat DNA cleaved off the telomeres.
Capping of Long Unregulated Telomeres with Wild-Type Repeats Restores Telomere Length Control in ter1-Kpn;rap1-ΔC Strains.
Since the rap1-ΔC protein increased the turnover rate in the terminal region of telomeres (Fig. 1), but did not cause more extensive internalization of mutant repeats into the telomere (Fig. 3), we tested whether the effect of the ter1-Kpn mutation in the rap1-ΔC background was exerted through an end effect, i. e., an interaction involving only the most distal repeats of the telomere. Previously, we used capping of mutant repeats by wild-type repeats to investigate the end effect seen in ter1;rap1-ΔC double mutants in which the mutant telomeric repeats had a lowered binding affinity to Rap1p (17). Using a similar approach, we introduced a plasmid expressing the ter1-Bcl gene into ter1-Kpn;rap1-ΔC double mutant cells, to cap the telomeres with the marked but phenotypically silent Bcl mutant repeats. The resulting telomeres were analyzed by Southern blotting after three different sets of restriction digestions. Telomeric patterns were compared with and without the introduction of the ter1-Bcl gene.
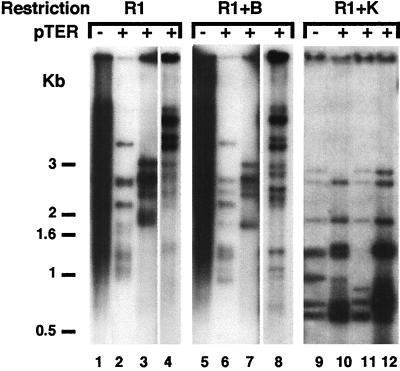
First, genomic DNA was digested with EcoRI alone (Fig. 4, lanes 1–4). EcoRI has only subtelomeric recognition sites and therefore the pattern observed reflects the whole telomeric DNA regardless of mutations in the telomere repeats. In contrast to the control double mutant, in which the smear characteristic of deregulated telomeres was seen (Fig. 4, lane 1), on introduction of the ter1-Bcl gene (pTER lanes) the telomeric pattern immediately exhibited several new discrete bands (Fig. 4, lanes 2–4). This result indicated that on capping of deregulated telomeres in ter1-Kpn;rap1-ΔC strains by ter1-Bcl repeats, telomere length control was resumed. Notably, these newly controlled telomeres were still longer than the wild type. Second, genomic DNA was double digested with EcoRI and BclI (Fig. 4, lanes 5–8), to determine the length of the tracts containing wild-type plus Kpn repeats, minus the terminally added Bcl repeats. These EcoRI and BclI double digest telomeric bands were slightly shorter and sharper than those bands seen after digestion with EcoRI alone (Fig. 4, lanes 5–8). The slight shortening showed that very few Bcl repeats had been incorporated onto the termini, and the sharpness of these bands indicated that telomere length control was regained. Third, genomic DNA was double digested with EcoRI and KpnI (Fig. 4, lanes 9–12), to cleave off the Kpn mutant telomeric repeats incorporated before capping. Therefore, the resulting telomeric patterns indicated the lengths of basal wild-type repeats internal to the Kpn repeats. These internal wild-type tracts in the three independent transformed strains (Fig. 4, lanes 10–12) were similar to those in the nontransformed strain (Fig. 4, lane 9). In addition, in all three transformants the telomeric restriction fragment patterns were similar overall. Hence, no major rearrangement events had occurred in the internal region of the telomeres, or in the subtelomeric regions, that could account for the difference in telomere length control.
Figure 4.
Capping of long telomeres in ter1-Kpn;rap1-ΔC strains by very few Bcl repeats. Telomeric patterns of three independent ter1-Kpn;rap1-ΔC transformants (+) that carry the ter-BclI gene on a plasmid (pTER) and untransformed controls (−). Each set was subjected to three different restriction digestion protocols: EcoRI only (R1), reflecting total telomeric pattern, EcoRI + BclI (R1 + B), reflecting telomeric pattern except the Bcl cap, and EcoRI+KpnI (R1+K), reflecting the basal wild-type telomeric pattern.
In summary, telomere length control was restored in a ter1-Kpn;rap1-ΔC background by capping of the deregulated telomeres by a few terminal Bcl repeats. Notably, this control was regained despite the presence of extensive tracts of mutant Kpn repeats adjacent to the cap. Furthermore, these newly capped telomeres were now controlled at longer-than-wild-type average lengths that were often similar to their average length before capping.
DISCUSSION
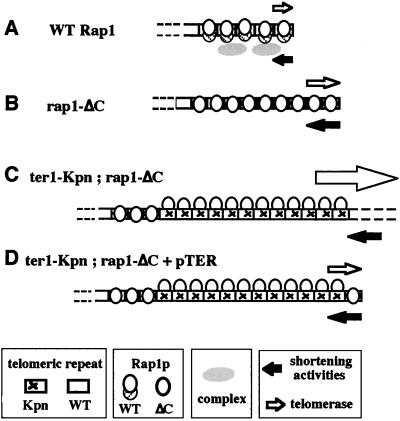
Here we have shown that a C-terminal truncation mutant of Rap1p (rap1-ΔC) accelerates telomeric turnover. This acceleration was specific to the most terminal telomeric domain. Turnover of the telomeric ends is normally balanced so that telomeric length remains close to a given average size. The length distribution of telomeres thus reflects the balance between the addition of telomeric repeats by telomerase and their depletion caused by normal DNA replication and possibly other telomere shortening activities (Fig. 5A). We and others have previously suggested that the telomeric DNA–protein complex nucleated by Rap1p affects the accessibility of the telomere to telomerase (17, 27). We suggest that the increase in the dynamics of terminal telomeric metabolism caused by the rap1-ΔC mutation involves more frequent and/or prolonged actions of telomerase at the telomere, as well as a correspondingly increased rate of action of telomere shortening activity(ies) (Fig. 5B).
Figure 5.
A model for telomere turnover in wild-type and mutant strains. Telomere length reflects a balance between elongation by telomerase (open right-pointing arrows) and an unknown activity (or activities) acting to shorten the telomere (solid left-pointing arrows). The sizes of arrows reflect the extent of activity. (A) Wild-type cells. Rap1p binds telomeric repeats and nucleates a specialized telomeric DNA–protein complex. A balance between telomerase action and limited telomere shortening activity (or activities) sets mean telomere length and the distribution of lengths within a narrow range. (B) rap1-ΔC cells. In the terminal zone of the telomere, accessibility to both telomerase and shortening activities (telomere turnover) is enhanced compared with wild-type RAP1 cells. (C) Terminally located Kpn mutant telomeric repeats (specified by ter1-Kpn telomerase), in the presence of rap1-ΔC, greatly enhance access to both telomerase and telomere degradation activities. Telomeres elongate and are degraded uncontrollably (deregulated telomeres). (D) Capping of previously deregulated telomeres by a functionally wild-type telomeric repeat restores normal cap function at the end of the telomere. This function is sufficient to reduce telomerase accessibility and cause both telomere lengthening and telomere shortening to become controlled, even though mean overall telomere length is markedly longer than wild type.
The association of telomeric turnover primarily with the distal domain of the telomere is intriguing in light of a previously suggested two-domain hypothesis for telomeric structure in S. cerevisiae. This model divides the telomere into two distinct functional domains, with the distal domain involving interactions of the C-terminal tail of Rap1p with Rif1p and Rif2p proteins (27, 28). Here we have shown that in K. lactis, terminal turnover is specifically accelerated when Rap1p lacks its evolutionarily conserved C-terminal tail. The simplest interpretation of these two experimental lines of evidence for a terminal telomeric zone, both of which implicate functions mediated by the C-terminal tail of Rap1p, is that they are manifestations of the same terminal zone.
In vitro, the truncated Rap1p-ΔC protein binds Kpn and wild-type telomeric repeat sequences indistinguishably and also binds both repeats indistinguishably from wild-type Rap1p (ref. 17 and data not shown). Although cells containing a single rap1-ΔC or ter1-Kpn mutation can control telomeres tightly for long periods, combining these mutations resulted in rapid instead of delayed deregulation of telomere length. Hence, the ter1-Kpn mutation causes short-term control of telomere length to become dependent on an intact C-terminal tail of Rap1p. Furthermore, we found no apparent changes in the length of the basal internal wild-type tracts in ter1-Kpn;rap1-ΔC double mutants compared with the single ter1-Kpn mutant, suggesting that the synthetic effect of rap1-ΔC with ter1-kpn may specifically involve interactions at the most terminal region of the telomere (Fig. 5C). A direct test of this hypothesis showed that capping the ends of the long, deregulated telomeres of ter1-Kpn;rap1-ΔC double mutants with only a very few functional Rap1p binding sites (the Bcl repeats) was sufficient to restore cap function to these otherwise dramatically altered telomeres (Fig. 5D). Significantly, the newly controlled telomeres were now set at a longer size.
The deregulated telomeres in the double ter1-Kpn;rap1-ΔC mutant contained large numbers of mutant Kpn repeats that, despite their ability to bind Rap1p as efficiently as wild-type repeats in vitro (17), were apparently invisible to the length control system. It was proposed previously that the number of bound Rap1 C-terminal domains at a telomere determine its length (28). However, the results reported here show that with Kpn repeats simply the number of binding sites for C-terminal-containing Rap1p molecules is insufficient to regulate telomere length. We therefore propose that monitoring of telomere length also requires the correct assembly of the complex nucleated by Rap1p on telomeric DNA. According to this model, Kpn mutant repeats are subtly defective in such complex assembly (although the nature of the defect remains unidentified), and telomere length becomes deregulated only when a second event occurs. In the single ter1-Kpn mutant, the second event is proposed to be the eventual loss of a critical minimal length of wild-type repeat tract and hence loss of a correctly assembled complex of sufficient size (11). In the double ter1-Kpn;rap1-ΔC mutant, we propose that the second event leading to loss of length control is the truncation of the C-terminal tail of Rap1p, causing assembly of a defective Rap1p-nucleated complex and consequent higher turnover at telomeric ends (Fig. 5C). Together with previous findings, these results suggest that Rap1p C-terminal interactions with proteins at the distal domain of the telomere, together with sequence-specific telomeric DNA binding by Rap1p, control the rate of telomere repeat turnover.
Acknowledgments
We thank M. McEachern and J. Roy for ter1 constructs and Sandy Johnson and Chris Smith for critical reading of the manuscript. A.K. was supported by a European Molecular Biology Organization Fellowship and by a postdoctoral training grant from the National Institutes of Health. This work was supported by Grants GM26259 and DE11356 from the National Institutes of Health to E.H.B.
References
- 1. Blackburn E H. Cell. 1994;77:621–623. doi: 10.1016/0092-8674(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E H. Annu Rev Biochem. 1984;53:163–194. doi: 10.1146/annurev.bi.53.070184.001115. [DOI] [PubMed] [Google Scholar]
- 3.Sandell L L, Zakian V A. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 4.Zakian V A. Annu Rev Genet. 1989;23:579–604. doi: 10.1146/annurev.ge.23.120189.003051. [DOI] [PubMed] [Google Scholar]
- 5.Greider C W. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 6.Shampay J, Blackburn E H. Proc Natl Acad Sci USA. 1988;85:534–538. doi: 10.1073/pnas.85.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li B, Lustig A J. Genes Dev. 1996;10:1310–1326. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- 8.Wellinger R J, Ethier K, Labrecque P, Zakian V A. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- 9.Zakian V A. Annu Rev Genet. 1996;30:141–172. doi: 10.1146/annurev.genet.30.1.141. [DOI] [PubMed] [Google Scholar]
- 10.Blackburn E H. Trends Biochem Sci. 1991;16:378–381. doi: 10.1016/0968-0004(91)90155-o. [DOI] [PubMed] [Google Scholar]
- 11.McEachern M J, Blackburn E H. Nature (London) 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 12.Nilsson P, Mehle C, Remes K, Roos G. Oncogene. 1994;9:3043–3048. [PubMed] [Google Scholar]
- 13.Bryan T M, Englezou A, Gupta J, Bacchetti S, Reddel R R. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berman J, Tachibana C Y, Tye B K. Proc Natl Acad Sci USA. 1986;83:3713–3717. doi: 10.1073/pnas.83.11.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shore D, Nasmyth K. Cell. 1989;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 16.Larson G P, Castanotto D, Rossi J J, Malafa M P. Gene. 1994;150:35–41. doi: 10.1016/0378-1119(94)90854-0. [DOI] [PubMed] [Google Scholar]
- 17.Krauskopf A, Blackburn E H. Nature (London) 1996;383:354–357. doi: 10.1038/383354a0. [DOI] [PubMed] [Google Scholar]
- 18.Cooper J P, Nimmo E R, Allshire R C, Cech T R. Nature (London) 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 19.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 20.Broccoli D, Chong L, Oelmann S, Fernald A A, Marziliano N, van Steensel B, Kipling D, Le Beau M M, de Lange T. Hum Mol Genet. 1997;6:69–76. doi: 10.1093/hmg/6.1.69. [DOI] [PubMed] [Google Scholar]
- 21.Bilaud T, Koering C E, Binet-Brasselet E, Ancelin K, Pollice A, Gasser S M, Gilson E. Nucleic Acids Res. 1996;24:1294–1303. doi: 10.1093/nar/24.7.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig A J, Gasser S M. J Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 24.Wang S S, Zakian V A. Mol Cell Biol. 1990;10:4415–4419. doi: 10.1128/mcb.10.8.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponzi M, Pace T, Dore E, Picci L, Pizzi E, Frontali C. Nucleic Acids Res. 1992;20:4491–4497. doi: 10.1093/nar/20.17.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McEachern M J, Blackburn E H. Pezcoller Found Symp. 1997;8:111–127. [Google Scholar]
- 27.Shore D. Trends Biochem Sci. 1997;22:233–235. doi: 10.1016/s0968-0004(97)01082-7. [DOI] [PubMed] [Google Scholar]
- 28.Marcand S, Gilson E, Shore D. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 29.Roy J, Boswell Fulton T, Blackburn E H. Genes Dev. 1998;12(20):3286–3300. doi: 10.1101/gad.12.20.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]