Abstract
We obtained mice deficient for major histocompatibility complex (MHC) molecules encoded by the H-2K and H-2D genes. H-2 KbDb −/− mice express no detectable classical MHC class I-region associated (Ia) heavy chains, although β2-microglobulin and the nonclassical class Ib proteins examined are expressed normally. KbDb −/− mice have greatly reduced numbers of mature CD8+ T cells, indicating that selection of the vast majority (>90%) of CD8+ T cells cannot be compensated for by β2-microglobulin-associated molecules other than classical H-2K and D locus products. In accord with the greatly reduced number of CD8+ T cells, spleen cells from KbDb −/− mice do not generate cytotoxic responses in primary mixed-lymphocyte cultures against MHC-disparate (allogeneic) cells. However, in vivo priming of KbDb −/− mice with allogeneic cells resulted in strong CD8+ MHC class Ia-specific allogeneic responses. Thus, a minor population of functionally competent peripheral CD8+ T cells capable of strong cytotoxic activity arises in the complete absence of classical MHC class Ia molecules. KbDb −/− animals also have natural killer cells that retain their cytotoxic potential.
Major histocompatibility complex (MHC) class I molecules, expressed on almost all nucleated cells of the body, are heterodimeric type I membrane proteins, composed of an MHC-encoded class I heavy chain, a β2-microglobulin (β2m) light chain, and a short peptide 8–10 aa in length, all of which are essential for the formation of a stable MHC class I protein (1). Those MHC class I proteins whose heavy chains are encoded by highly polymorphic genes (H-2K, D, and L in mice) are termed “classical” or class I-region associated (Ia) MHC. Cell-surface MHC class Ia proteins are essential for the thymic development of CD8+ T cells (2) and for providing protection against lysis by natural killer (NK) cells (3). Recent work suggests that cell-surface MHC class Ia proteins are also involved critically in maintaining the levels of memory CD8+ T cells in the periphery (4, 5). For studies of these processes, mice that are totally deficient in the classical class I MHC proteins would be valuable.
Mice deficient for β2m or the MHC-encoded peptide transporter (TAP), a complex involved in loading peptides onto newly synthesized MHC class I proteins, do not express normal levels of cell-surface MHC class I proteins and consequently are defective in MHC class I-restricted antigen presentation and in thymic selection of CD8+ T cells (6–8). These TAP −/− and β2m −/− mice have been used widely as MHC class I-deficient models, but this deficiency is not complete. Thus, a sizable fraction of MHC class I heavy chains apparently folds normally and is transported to the cell surface even in the absence of β2m (in particular, the H-2Db gene product) or in the absence of TAP-dependent peptides (8–12). In addition, “empty” class I molecules are expressed on the cell surface of TAP −/− and β2m −/− animals, so that total MHC I cell-surface levels are decreased at most 10-fold (12, 13). Although such empty class I molecules decay rather rapidly in ex vivo cultured cells, nothing is known about their half-life in vivo. TAP −/− and β2m −/− animals also possess a limited repertoire of self-MHC class I-restricted CD8+ T cells, which can be explained by their selection on the remaining low levels of MHC class I (14–17). These animals reject allogeneic skin grafts and tumors and give rise to viral and allogeneic CD8+ T cell-mediated responses (14, 17–21), consistent with the continued presence of functional MHC class I proteins.
In addition to the classical H-2K and H-2D MHC class I loci, there are fewer polymorphic MHC class I-encoded proteins (encoded by genes in the Q, T and M regions; refs. 22 and 23). Moreover, there are additional genes, clearly related more closely to MHC class I genes than to any other gene family, but not linked to the MHC (CD1, ZAG, FcRn, and HFE; refs. 22–27). Both MHC-linked and MHC-unlinked class I genes of this type are called MHC class Ib genes, and the molecules they encode (MHC class Ib proteins) are structurally similar to the classical MHC class Ia proteins. Many of the class Ib molecules require β2m and possibly TAP-dependent peptides for their folding and transport to the cell surface (28). In some cases, presentation of peptides by class Ib proteins to αβ or γδ T cells has been shown (28).
Nonclassical MHC molecules are functionally diverse. For example, the FcRn receptor transports IgG from ingested milk across the intestinal epithelium of neonatal mice and rescues circulating IgG from degradation (24). In addition, CD1 plays a crucial role in the function of NK1.1 T cells and in their positive selection (26, 29). H-2 M3a presents N-formylated peptides derived from certain bacterial and mitochondria-encoded proteins, whereas MHC class Ia molecules do not seem to bind such peptides (25, 30, 31). Expression of H-2 Qa-2 has been correlated with the expression of an early embryo gene, Ped, that maps in the H-2 Qa-2 region and controls the rate of preimplantation embryonic development (32). Though the functional role of Qa-2 in the immune response is not known, it binds a wide variety of peptides, and its expression depends on TAP (32). Because of the requirement of β2m and TAP in the expression of nonclassical MHC molecules (22, 33), many class Ib molecules fail to be expressed on cells in animals that lack β2m or TAP. Combined, these findings suggest that neither β2m −/− nor TAP −/− mice can be considered ideal models for mice whose lack of MHC class Ia molecules is complete and limited to only the classical MHC class Ia molecules.
Besides their well established role as restriction elements for T cell recognition, MHC class I molecules are also involved in providing protection from lysis by NK cells (3, 34–36). Little is known about the development of NK cells, but a number of reports support the existence of an educational process for NK-cell tolerance, and it has been proposed that NK cells calibrate to endogenous levels of self-MHC class I during development. For example, β2m −/− mice possess functional but self-tolerant NK cells, whereas β2m −/− Con A-stimulated blasts are killed by NK cells from β2m-expressing parental mice (37, 38). The mechanism of this self-tolerance is not well understood but includes the regulation of NK-cell inhibitory receptors. In addition, recent reports show that NK-cell reactivity may also depend on MHC class Ib molecules (39, 40), which are not expressed normally in either β2m −/− or TAP −/− mice. This further motivates the use of mice that are selectively deficient for MHC class Ia in such studies.
To obtain MHC class Ia deficient animals, we took advantage of mice of the 129 strain with defective K or D genes, which account for all of the classical MHC class Ia heavy chains in mice of the H-2bc MHC haplotype (4, 41). The genetic distance between these genes is only 0.2 centimorgan (42). We obtained H-2 KbDb −/− mice by crossing H-2Kb −/− with H-2Db −/− animals and screening progeny for the rare intra-H-2 recombinant that carries both targeted genes on the same chromosome. Here, we report characteristics of these mice.
MATERIALS AND METHODS
Mice and Cell Lines.
Kb −/− and Db −/− single-knockout mice are reported elsewhere (4, 41). They are generated from E14TG2a embryonic stem cells derived from the 129/Ola mouse strain of the H-2bc haplotype. The H-2K and H-2D locus products of the H-2bc haplotype are serologically indistinguishable from H-2Kb and H-2Db, but the H-2bc haplotype differs from H-2b in the H-2T region that contains genes encoding class Ib molecules (22). C57BL/6, BALB/c, and β2m −/− strains were purchased from The Jackson Laboratory. P815 is an H-2d-expressing mouse mastocytoma derived from the DBA strain. EL4 is a T cell lymphoma derived from C57BL/6. T2-Kb is an H-2Kb transfectant of T2 that has an antigen processing mutation caused by a genomic deletion in the MHC class II region (43). C4.4–25− is a β2m-defective mutant of EL-4 and carries a stop codon in the sixth codon of the β2m gene (ref. 44; R.G. and K. Magnusson, unpublished work). RMA-S is an NK-sensitive TAP-2 defective lymphoma of RMA (3), both derived from C57BL/6. YAC-1 is an NK-sensitive lymphoma derived from the A/Sn strain, commonly used to detect NK-cell cytotoxic activity.
Antibodies.
Anti-H-2Kb coupled to fluorescein isothiocyanate (FITC), anti-H-2Db coupled to FITC, anti-H-2Kb coupled to phycoerythrin (PE), anti-H-2Dd, anti-CD4 coupled to PE, anti-CD8 coupled to FITC, anti-T cell receptor (TCR) coupled to PE, goat anti-mouse IgG coupled to horseradish peroxidase, goat anti-rabbit IgG coupled to horseradish peroxidase, anti-Qa-2, and biotinylated anti-Qa-2 antibodies were purchased from PharMingen. Rabbit anti-MHC I free heavy chain (RAFHC) and antiexon 8 of Kb antibodies have been described (45).
Metabolic Labeling and Immunoprecipitations.
Spleen cells were incubated for 30 min in methionine and cysteine-free DMEM, labeled with 1 mCi of methionine/[35S]cysteine for 45 min, and chased for ≈2 h in RPMI medium 1640 containing 10% fetal calf serum. Cells were lysed in Nonidet P-40 lysis mix (0.5% Nonidet P-40/50 mM Tris, pH 7.5/5 mM MgCl2/1 mM phenylmethylsulfonyl fluoride/2 μg/ml leupeptin/1 μg/ml aprotinin). Lysates were precleared twice with normal rabbit or mouse serum and formalin-fixed Staphylococcus aureus, followed by the indicated MHC class Ia or Ib specific reagents. Immunoprecipitates, adjusted for the total amount of incorporation as measured by trichloroacetic acid precipitation, were run on SDS/12.5% PAGE. Gels were developed with dimethyl sulfoxide/2,5-diphenyloxazole and exposed to Kodak X-Omat AR film.
Peptide N-Glycosidase Treatment and Immunoblotting.
Tissues from different organs were homogenized in 0.5–2 ml of Nonidet P-40 lysis mix and incubated at 4°C for 1 h. Lysates were spun for 30 min at 14,000 rpm in an Eppendorf centrifuge and diluted in lysis mix to 2 mg/ml total protein. Lysates were then heat-denatured in 0.6 M 2-mercaptoethanol and 2.4% SDS at 95°C and treated for 4 h at 30°C with peptide N-glycosidase in PBS containing 25 mM EDTA, 1% Nonidet P-40, 0.4% SDS, and 0.1 M 2-mercaptoethanol. Lysates were then separated by SDS/PAGE and transferred to nitrocellulose. The filters were incubated with the indicated first antibodies (RAFHC 1:1000 and anti-exon 8 of Kb antibodies 1:2000), followed by horseradish-peroxidase-coupled goat anti-mouse or anti-rabbit IgG. The immune complexes were visualized by chemiluminescence.
Flow-Cytometry Evaluation of MHC I, CD4, CD8, and TCR Surface Expression.
Screening for KbDb −/− mice was done by collecting ≈100 μl of tail blood into 50 mM EDTA in PBS. Red blood cells were removed by osmotic lysis. Splenocyte and thymocyte samples from KbDb −/− and control mice were prepared by making single-cell suspensions in RPMI medium 1640. Cells were then stained with FITC or with PE-labeled antibodies in PBS containing 25% fetal calf serum and 1% normal mouse serum. Expression of surface markers was analyzed in a fluorescence-activated cell sorter (FACScan or FACStar, Becton Dickinson) with cellquest software. We collected ≈2,500 cells per blood sample and 10,000–50,000 cells per spleen or thymus sample.
Cytotoxic T Lymphocyte (CTL) Generation and Complement-Mediated Depletion of T Cells.
KbDb −/−, β2m −/−, BALB/c, or C57BL/6 mice were injected twice (primed) with either C57BL/6 or BALB/c splenocytes (≈107 cells per 300 μl of PBS, injected i.p.) with a 10-day interval between injections. CTL were obtained by 5- to 7-day mixed-lymphocyte cultures (MLCs) with C57BL/6 or BALB/c irradiated spleen cells used as stimulators and spleen cells from either naive or primed mice as responders. For MLC, 25 × 106 responder spleen cells were mixed with 2–5 × 106 stimulator cells and were used as effector cells after 5–7 days. Complement-mediated depletion of CD8+ or CD4+ T cells was done by incubating ≈105 responder cells with 10 μg of the appropriate antibody for 30 min at 4°C, followed by wash and incubation with 10 μg of complement for 1 h at 37°C in 6% CO2.
Generation of Con A Lymphoblasts and NK Cells.
Spleen cells (5 × 107), from which erythrocytes had been depleted, were placed in 10 ml of complete medium with 5 μg/ml of Con A (Sigma). After 2 days the cells were harvested, and viable cells were isolated by centrifugation with Lymphoprep (Nycomed, Oslo). For cytotoxicity assays with tumor target cells, NK effector cells were generated from erythrocyte-depleted splenocytes from mice injected 1 day earlier with tilorone (Sigma). For a cytotoxicity assay with Con A lymphoblasts, NK effector cells were obtained by culturing 25 × 106 spleen cells in 10 ml of complete medium and 2000 units/ml recombinant interleukin 2 at 37°C in 6% CO2. After 5 days both nonadherent and adherent cells were removed to obtain lymphokine-activated killer cells.
Cytotoxic 51Cr Release Assay.
The effector cells (see above) were tested at the indicated effector:target ratios, in RPMI medium 1640 supplemented with 10% fetal calf serum, against 5 × 103 51Cr-labeled target cells per well (96-well plate). Each effector:target ratio was assayed in triplicate, and assays were incubated for 3.5–4.5 h.
RESULTS
Generation of KbDb −/− Mice.
To obtain KbDb −/− mice we made use of single Kb −/− and Db −/− knockout animals generated on the H-2bc haplotype in the 129 mouse strain (4, 41). These animals were crossed, and their offspring were screened for an intra-H-2 recombinant that carried both targeted loci on the same chromosome with the following breeding scheme.
Kb −/− animals were crossed to Db −/− animals to obtain heterozygotes with the genotype Kb+Db−/Kb−Db+. To detect rare germ-line recombination events (the expected recombination frequency is ≈1 in 500; ref. 42), we crossed these heterozygotes to the BALB/c (H-2d haplotype) strain to obtain animals with the genotype Kb−Db−/H-2d. The screen was done by FACS analysis on blood lymphocytes. Of 155 animals tested, one was a male that had no surface expression of either Kb or Db proteins, as expected for the desired intra-H-2 recombination. The genetic background of these mice is therefore a mixture of the 129 and BALB/c strains.
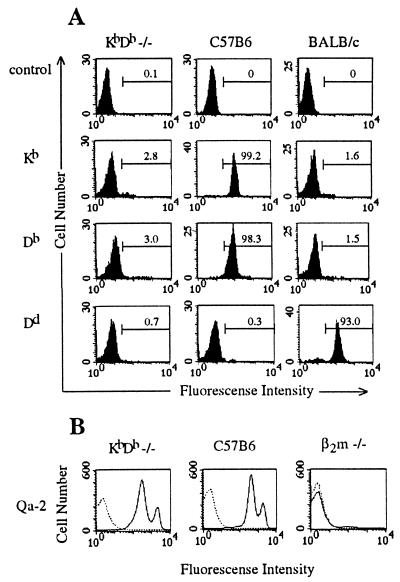
KbDb −/− homozygotes were generated by breeding the Kb−Db−/H-2d male with a Kb −/− female. We screened for lack of expression of H-2d alleles and crossed Kb−Db−/Kb−Db+ offspring to produce the double-knockout animals with a frequency of ≈25% (15 of 58). A line of KbDb −/− double-knockout mice was maintained by brother–sister matings. FACS analysis performed on blood lymphocytes from double-knockout mice showed no cell-surface expression of Kb, Db, or Dd heavy chains (Fig. 1A). Thus, double-knockout animals do not have reduced viability compared with their MHC class Ia-positive littermates, as has also been observed for TAP −/− and β2m −/− animals (6, 7).
Figure 1.
Cell-surface expression of MHC class Ia and class Ib molecules in KbDb −/− mice. (A) Blood lymphocytes from KbDb −/−, C57BL/6, or BALB/c mice were analyzed by direct immunofluorescence with α-Kb-FITC, α-Db-FITC, or α-Dd-FITC antibodies, as indicated. Numbers correspond to percentage of cells in the indicated regions. (B) Splenocytes from KbDb −/−, C57BL/6, or β2m −/− mice were analyzed by indirect immunofluorescence with biotinylated α-Qa-2 antibody and streptavidin-PE (solid line) or, as a control, with streptavidin-PE only (dashed line).
FACS analysis performed on splenocytes and thymocytes from these mice showed no cell-surface expression of Kb, Db, or Dd heavy chains, whereas I-Ab, and Qa-2 were expressed normally (Fig. 1B and data not shown).
Absence of Classical MHC Class I Proteins in KbDb −/− Mice.
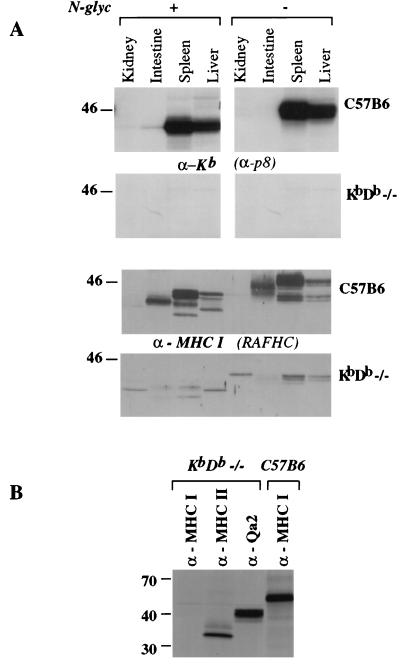
To establish the absence of the classical MHC class I heavy chains in these mice, we analyzed whole-cell lysates from different tissues by immunoblots using RAFHC, also directed against the cytoplasmic tail of Kb (α-p8). As expected from their cell-surface phenotype, KbDb −/− mice do not express the ≈45-kDa glycoprotein in any of the tissues analyzed (Fig. 2A). However, with the antiserum directed against free MHC I heavy chain, we did detect ≈40-kDa immunoreactive glycoproteins in both control and KbDb −/− samples. These glycoproteins likely correspond to the nonclassical class Ib heavy chains which are cross-reactive with the broadly reactive RAFHC.
Figure 2.
Expression of MHC-encoded molecules in KbDb −/− mice. (A) Whole-cell lysates from the indicated tissues were homogenized and adjusted for protein concentration (100 μg per lane). As a control, half of the samples were treated with N-glycosidase (Left). Lysates were analyzed by immunoblots with α-Kb (α-p8, Upper) or α-MHC class I free heavy chain (RAFHC, Lower) antibodies. (B) Spleen-cell Con A blasts were labeled biosynthetically for 45 min and chased for 2 h. Cell lysates were made, and H-2-encoded proteins were immunoprecipitated with α-Kb (α-p8), α-MHC class I free heavy chain (RAFHC), α-Qa-2, or α-MHC class II (N22) antibodies.
Immunoblots with the RAFHC antiserum showed an intestine-specific glycoprotein of ≈43 kDa in C57BL/6 but not in KbDb −/− animals. Because this species was present only in wild-type animals and was not detected with the α-p8 antiserum, it might correspond to an H-2 Kb splice variant (46). The use of alternative-splice acceptor sites during the removal of intron 7 in pre-mRNA splicing produces two forms of H-2Kb protein: the predominant form, derived from a transcript that was spliced at the upstream splice acceptor site for long exon 8, and a Kb molecule, derived from a transcript that was spliced at the downstream acceptor site for short exon 8 (46).
Immunoprecipitation experiments from 35S-labeled KbDb −/− splenocytes, Con A blasts (Fig. 2B), or T cell lines obtained from these animals confirmed the data obtained by immunoblots. Whereas MHC class I specific antiserum did not recover any immunoreactive species from cell lysates prepared from KbDb −/− mice, antibodies against MHC class II proteins or Qa-2 molecules recovered proteins of expected molecular masses (data not shown).
Thus, KbDb −/− animals lack expression of the classical MHC class Ia heavy chain, whereas MHC class II and nonclassical MHC class Ib expression seems normal.
Severe Deficiency in CD8+ T Cell Maturation in KbDb −/− Mice.
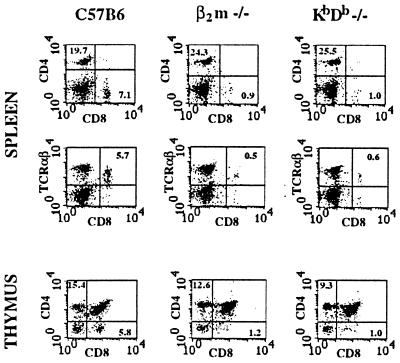
We analyzed the numbers of CD8+TCRαβ+ T cells in splenocytes and thymocytes from double KbDb −/− animals. FACS analysis showed ≈5- to 10-fold reduction in the number of peripheral CD8+ T cells, with some compensatory increase in the CD4+ T cell compartment (Fig. 3). This reduction was comparable to that seen in β2m −/− animals (Fig. 3; refs. 6–8). The most obvious explanation for the small peripheral CD8+ pool is a defect in thymic development caused by the lack of classical MHC class I molecules. Indeed, the thymic CD4-CD8+ population was reduced ≈5-fold (to 1% of total), whereas the numbers of CD4+CD8- cells were comparable to those in control animals (Fig. 3).
Figure 3.
Reduction in CD8+TCRαβ T cell numbers in KbDb −/− mice. FACS analysis was performed on C57BL/6, β2m −/−, or KbDb −/− splenocytes (Top and Middle) or thymocytes (Bottom) with α-CD8-FITC, α-CD4-PE, and α-TCRαβ-PE antibodies, as indicated. Numbers correspond to percentage of cells in the indicated regions.
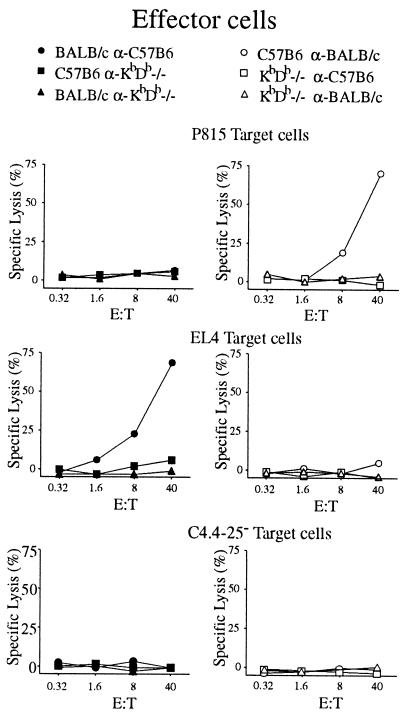
To test the function of KbDb −/− lymphocytes, we performed a series of primary MLCs with splenocytes from the KbDb −/−, BALB/c (allogeneic, H-2d), and C57BL/6 (KbDb +/+ control) animals. Cytotoxic responses were then examined on targets with the relevant haplotypes. Spleen cells from KbDb −/− mice showed neither cytotoxic nor stimulatory activity (Fig. 4), consistent with the low numbers of peripheral CD8+ T cells observed by flow cytometry. We conclude that KbDb −/− animals are severely deficient in CD8+ T cell maturation; this deficiency is caused by a block in thymic development.
Figure 4.
Absence of primary cytotoxic response in MLC by spleen cells from unprimed KbDb −/− mice. Effector cells were obtained by the standard 7-day MLC with spleen cells from C57BL/6, BALB/c, or KbDb −/− used as stimulators and responders (as indicated). CTL-mediated cytotoxicity was measured in a standard 4-h 51Cr release assay with H-2d P815 (Top), H-2b EL4 (Middle), or β2m −/− C4.4–25− (Bottom) targets. E:T, effector:target ratio.
Cytotoxic Activity of CD8+ T Cells from KbDb −/− Mice After in Vivo Priming.
Although functional CD8+ T cells are seemingly absent from KbDb −/− mice, it might be possible to achieve activity from a small residual CD8+ repertoire by in vivo immunization.
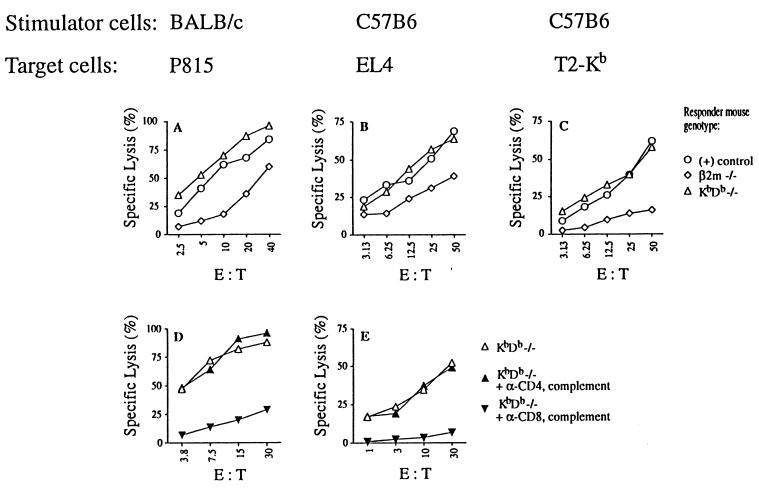
The KbDb −/− mice were injected (primed) twice with splenocytes from either BALB/c or C57BL/6 mice (fully allogeneic or KbDb +/+ control, respectively). The splenocytes from these mice were then used as responder cells for in vitro MLC. After 5 days of MLC, we found strong cytolytic activity mediated by KbDb −/− CD8+ T cells against both H-2d (Fig. 5 A and D) and H-2b (Fig. 5 B and E) mouse target cells. This response was even stronger than that from β2m −/− animals (Fig. 5). Similar levels of lytic activity were observed when human T2 targets transfected with mouse MHC class I molecules (H-2Kb) were used, whereas untransfected T2 cells were not killed (Fig. 5C and data not shown). These results suggest that KbDb −/− CD8+ T cells directly recognize classical MHC class I antigens. Further, because of the absence of MHC class Ia-mediated selection in KbDb −/− mice, such MHC class Ia-specificity may be germ-line-encoded (47). Similar results were obtained with CTL lines specific for H-2b and H-2d derived from KbDb −/− mice (data not shown).
Figure 5.
Cytotoxic activity of CD8+ T cells from KbDb −/− mice after in vivo priming. C57BL/6, BALB/c, β2m −/−, or KbDb −/− mice were injected twice in vivo with either BALB/c (A and D) or C57BL/6 (B, C, and E) splenocytes. Effector cells were obtained by a standard 6-day MLC with the indicated splenocytes used as stimulators. KbDb −/− effector cells (anti-BALB/c in D and anti-C57BL/6 in E) were depleted of CD8+ or CD4+ T cells with α-CD8 or α-CD4 antibodies and complement (D and E). CTL-mediated cytotoxicity was measured in a standard 4-h 51Cr release assay using H-2d P815 (A and D), H-2b EL4 (B and E), or T2-Kb (C) target cells. The controls for A were C57BL/6 anti-BALB/c effectors, and the controls for B and C were BALB/c anti-C57BL/6 effectors (○).
A minor yet fully functional peripheral T cell population can thus develop and be activated in vivo in the complete absence of classical MHC class Ia molecules.
NK Cells in KbDb −/− Mice.
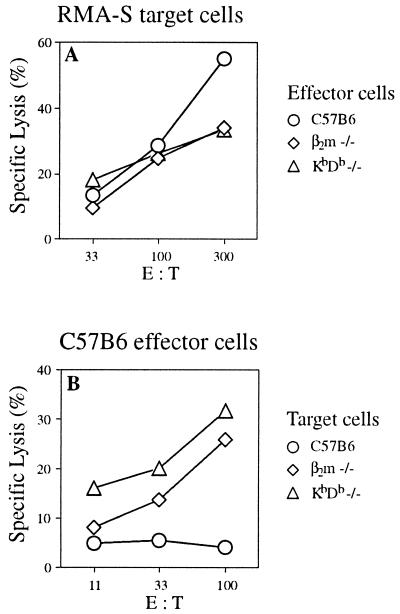
MHC class I proteins are important for protection against NK-cell lysis (3, 34–36). Therefore, we tested whether KbDb −/− animals possess functional NK cells and whether lack of heavy chain expression would render KbDb −/− cells susceptible to lysis by wild-type NK cells. NK-cell activity from KbDb −/− mice stimulated with the interferon inducer tilorone was tested on NK-sensitive TAP-deficient RMA-S target cells. NK cells from KbDb −/− mice were able to lyse RMA-S cells (Fig. 6A), indicating that NK cells can develop in the complete absence of classical MHC class I molecules and retain at least some of their cytotoxic function. A similar result was obtained with YAC-1 target cells (data not shown). KbDb −/− Con A blasts and β2m −/− Con A blasts show similar levels of sensitivity, indicating that neither MHC class I free heavy chains nor β2m-associated MHC class Ib molecules strongly influence sensitivity of Con A blasts to NK-cell lysis (Fig. 6B; ref. 49).
Figure 6.
KbDb −/− mice as donors of effector or target cells in NK-mediated cytotoxicity in vitro. (A) C57BL/6, β2m −/−, and KbDb −/− mice were injected with the interferon inducer tilorone, and splenocytes from these mice were used as effector cells 1 day after injection. NK-mediated cytotoxicity was measured with 4-h 51Cr release assay on RMA-S target cells. (B) C57BL/6 lymphokine-activated killer cells were obtained by 5-day culture of erythrocyte-depleted splenocytes. NK-mediated cytotoxicity was measured with 4-h 51Cr release assay on C57BL/6, β2m −/−, and KbDb −/− Con A blast target cells.
DISCUSSION
We have generated mice deficient for the classical MHC class Ia molecules (Kb and Db) by intercrossing mice defective in either Kb (H-2Kb −/−) or Db (H-2Db −/−) and screening for rare intra-H-2 recombinants. These doubly deficient KbDb −/− mice have a profound reduction in the numbers of peripheral CD8+ T cells, but can nonetheless generate strong MHC-specific CD8+ T cell responses after priming in vivo.
Several conclusions may be drawn from these observations. Classical MHC class I molecules are required for selection of at least 90% of the CD8+ T cells. The contribution by β2m-associated, TAP-loaded class I molecules of other provenances (e.g., class Ib molecules) can thus account for no more than 10% of CD8+ T cells. Although KbDb −/− mice have few peripheral CD8+ T cells, these cells are fully functional, and they must have arisen in a developmental pathway that is independent of classical MHC class Ia molecules. The apparent class I specificity of these CD8+ T cells may be germ-line-encoded, as shown by Zerrahn et al. (47), and does not depend on the presence of host class Ia molecules. The low numbers of CD8+ T cells can be ascribed to a block in thymic T cell development from the CD4+CD8+ to the CD4-CD8+ stage.
KbDb −/− mice would be expected to lack almost all peripheral CD8+ T cells, based on the comparison with β2m −/− and TAP −/− animals (6–8). Nevertheless, we find the low numbers of CD8+ T cells striking, because class Ib molecules are structurally very similar to the classical MHC class Ia products (22, 23) and are expressed in hematopoietic cells, which have some capacity for positive selection (49). There are no arguments that would prohibit the involvement of class Ib molecules in positive selection of CD8+ T cells. However, class Ib molecules require both β2m and peptide for their expression and are not expressed properly at the cell surface of β2m −/− or TAP −/− animals. For the study of T cell MHC class Ia- and Ib-mediated T cell selection, KbDb −/− mice are clearly the preferred model, because their deficiency is complete and limited to only MHC class Ia molecules. Our results show that the contribution of nonclassical class Ib molecules to the development of the CD8+ T cell repertoire is minor.
The remaining pool of functional CD8+ T cells in β2m −/− or TAP −/− mice has been explained by the low levels of cell-surface MHC class I still expressed in these mice. This explanation is supported by a number of studies showing a biased specificity in these T cells, which show increased avidity for self-MHC class I molecules (14, 19). In KbDb −/− mice, no MHC class Ia is present to influence the development of T cells, but we still observe the presence of a minor CD8+ T cell pool. However, β2m −/− or TAP −/− mice are deficient in expression of MHC class Ib molecules, whereas levels of class Ib molecules seem to be normal in KbDb −/− mice. Thus, the remaining population of CD8+ T cells in KbDb −/− mice may be explained by their selection on MHC class Ib molecules, but this hypothesis will have to be addressed experimentally. The importance of MHC class I products in positive selection of CD8+ T cells is well established from β2m −/− and TAP −/− models (2, 6–8). Our results argue for the existence of a developmental pathway of CD8+ T cells that is entirely independent of the H-2K and D locus products. It remains to be established whether this pathway requires expression of nonclassical class Ib products or is a consequence of stochastic down-regulation of CD4 coreceptor after the CD4+CD8+ stage (2). At least one class Ib gene product, called CD1, has been implicated in the development of mouse T cells and plays a crucial role in positive selection and function of NK1.1 CD4+ T cells (26, 29).
The NK cells from KbDb −/− animals are of considerable interest, because these develop in the selective absence of MHC class Ia molecules. KbDb −/− spleen NK cells are able to lyse the standard NK-cell tumor target cells, such as YAC-1 and RMA-S (Fig. 6A), efficiently and have thus retained at least some cytolytic function. NK cells in KbDb −/− and β2m −/− animals probably have similar mechanisms of self-tolerance, because NK cells from β2m −/− animals do not kill KbDb −/− targets and vice versa (49). It is possible that nonclassical class Ib molecules, acting through inhibitory NK receptors, are involved in self-tolerance of KbDb −/− as well as in β2m −/− NK cells. Of interest in this respect is the Qa-1 class Ib molecule, which presents the signal peptide derived from H-2D to CTL in most H-2 haplotypes, except H-2k (50, 51). Furthermore, some functions of the Qa-1 molecule are dependent on its association with β2m (52). In human cells, the NK-protective function of HLA-G can now be understood as the presentation of HLA-G’s signal sequence by HLA-E. This may present a situation analogous to that proposed for Qa-1, because HLA-E and Qa-1 have similarities in their antigen binding clefts and present similar types of peptides (52, 53). Qa-1 cannot present the H-2D-derived signal sequence in KbDb −/− mice, because the H-2Db gene product is absent (the genomic deletion in the H-2Db gene includes exons 1–3, coding for the signal peptide, α1 and α2), but perhaps other signal sequences could substitute. Thus, it is unlikely that signal peptide presentation by Qa-1 plays a role in self-tolerance in β2m −/− or KbDb −/− mice. What is the mechanism of this self-tolerance? Recent reports have suggested down-regulation of positive signaling pathways as an explanation for tolerance (40, 54, 55). Killing of class I-deficient tumor targets may be explained by unregulated expression of molecules (distinct from class I) by these targets. In any case, KbDb −/− animals provide a tool to study activation, inhibition, and development of NK cells.
KbDb −/− mice, in which the responses dominated by classical MHC class I are absent, provide a unique system for studying the functions and immunological activities of class Ib molecules. This type of study cannot be accomplished cleanly at present in β2m or TAP deficient mice, in which surface-expressed free MHC class I heavy chains can serve as restriction elements for immune responses (15, 16, 19, 21). Although the expression of properly folded MHC class I molecules on β2m −/− or TAP −/− cells may be low, T cells are often able to recognize target cells with very low MHC ligand densities (56). Furthermore, β2m- and TAP-deficient animals mount protective immune responses against certain intracellular pathogens as well (8). Some of these responses arise via the CD8+ MHC class Ia-restricted mechanism (21), whereas others employ compensatory responses by CD4+ CTL or γδ T cells (57). KbDb −/− animals will be useful in understanding the mechanisms underlying such compensatory immune responses.
Acknowledgments
We thank Kirsten Fischer Lindahl and David Raulet for helpful comments on the manuscript. This work was supported by National Institutes of Health Grants 5P01AI-AG37833 and R01 AI33456.
ABBREVIATIONS
- β2m
β2-microglobulin
- CTL
cytotoxic T lymphocyte
- FACS
fluorescence-activated cell sorter
- FITC
fluorescein isothiocyanate
- Ia
I-region associated
- MHC
major histocompatibility complex
- MLC
mixed-lymphocyte culture
- NK
natural killer
- PE
phycoerythrin
- RAFHC
rabbit anti-MHC I free heavy chain
- TAP
peptide transporter
- TCR
T cell receptor
References
- 1. Bijlmakers M J, Neefjes J J, Wojcik-Jacobs E H, Ploegh H L. Eur J Immunol. 1993;23:1305–1313. doi: 10.1002/eji.1830230618. [DOI] [PubMed] [Google Scholar]
- 2.Jameson S C, Hogquist K A, Bevan M J. Annu Rev Immunol. 1995;13:93–126. doi: 10.1146/annurev.iy.13.040195.000521. [DOI] [PubMed] [Google Scholar]
- 3.Ljunggren H G, Karre K. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 4.Tanchot C, Lemonnier F A, Perarnau B, Freitas A A, Rocha B. Science. 1997;276:2057–2062. doi: 10.1126/science.276.5321.2057. [DOI] [PubMed] [Google Scholar]
- 5.Markiewicz M A, Girao C, Opferman J T, Sun J, Hu Q, Agulnik A A, Bishop C E, Thompson C B, Ashton-Rickardt P G. Proc Natl Acad Sci USA. 1998;95:3065–3070. doi: 10.1073/pnas.95.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zijlstra M, Bix M, Simister N E, Loring J M, Raulet D H, Jaenisch R. Nature (London) 1990;344:742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- 7.Van Kaer L, Ashton-Rickardt P G, Ploegh H L, Tonegawa S. Cell. 1992;71:1205–1214. doi: 10.1016/s0092-8674(05)80068-6. [DOI] [PubMed] [Google Scholar]
- 8.Raulet D H. Adv Immunol. 1994;55:381–421. doi: 10.1016/s0065-2776(08)60514-3. [DOI] [PubMed] [Google Scholar]
- 9.Allen H, Fraser J, Flyer D, Calvin S, Flavell R. Proc Natl Acad Sci USA. 1986;83:7447–7451. doi: 10.1073/pnas.83.19.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bix M, Raulet D. J Exp Med. 1992;176:829–834. doi: 10.1084/jem.176.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glas R, Franksson L, Ohlen C, Hoglund P, Koller B, Ljunggren H G, Karre K. Proc Natl Acad Sci USA. 1992;89:11381–11385. doi: 10.1073/pnas.89.23.11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ljunggren H G, Stam N S, Ohlen C, Neefjes J J, Hoglund P, Heemels M T, Bastin J, Schumacher T N M, Townsend A, Karre K, et al. Nature (London) 1990;346:476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- 13.Machold R P, Andree S, Van Kaer L, Ljunggren H G, Ploegh H L. J Exp Med. 1995;181:1111–1122. doi: 10.1084/jem.181.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ljunggren H G, Van Kaer L, Ashton-Rickardt P G, Tonegawa S, Ploegh H L. Eur J Immunol. 1995;25:174–178. doi: 10.1002/eji.1830250129. [DOI] [PubMed] [Google Scholar]
- 15.Cook J R, Solheim J C, Connolly J M, Hansen T H. J Immunol. 1995;154:47–57. [PubMed] [Google Scholar]
- 16.Sandberg J K, Chambers B J, Van Kaer L, Karre K, Ljunggren H G. Eur J Immunol. 1996;26:288–293. doi: 10.1002/eji.1830260203. [DOI] [PubMed] [Google Scholar]
- 17.Apasov S, Sitkovsky M. Proc Natl Acad Sci USA. 1993;90:2837–2841. doi: 10.1073/pnas.90.7.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamouse-Smith E, Clements V K, Ostrand-Rosenberg S. J Immunol. 1993;151:6283–6290. [PubMed] [Google Scholar]
- 19.Glas R, Ohlen C, Hoglund P, Karre K. J Exp Med. 1994;179:661–672. doi: 10.1084/jem.179.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aldrich C, Ljunggren H G, Van Kaer L, Ashton-Rickardt P G, Tonegawa S, Forman J. Proc Natl Acad Sci USA. 1994;91:6525–6528. doi: 10.1073/pnas.91.14.6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinn D G, Zajac A J, Hioe C E, Frelinger J A. J Virol. 1997;71:8392–8396. doi: 10.1128/jvi.71.11.8392-8396.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shawar S M, Vyas J M, Rodgers J R, Rich R R. Annu Rev Immunol. 1994;12:839–880. doi: 10.1146/annurev.iy.12.040194.004203. [DOI] [PubMed] [Google Scholar]
- 23.Fischer Lindahl K, Hermel E, Loveland B E, Wang C R. Annu Rev Immunol. 1991;9:351–372. doi: 10.1146/annurev.iy.09.040191.002031. [DOI] [PubMed] [Google Scholar]
- 24.Simister N E, Jacobowitz Israel E, Ahouse J C, Story C M. Biochem Soc Trans. 1997;25:481–486. doi: 10.1042/bst0250481. [DOI] [PubMed] [Google Scholar]
- 25.Kurlander R J, Shawar S M, Brown M L, Rich R R. Science. 1992;257:678–679. doi: 10.1126/science.1496381. [DOI] [PubMed] [Google Scholar]
- 26.Mendiratta S K, Martin W D, Hong S, Boesteanu A, Joyce S, Van Kaer L. Immunity. 1997;6:469–477. doi: 10.1016/s1074-7613(00)80290-3. [DOI] [PubMed] [Google Scholar]
- 27.Zhou X Y, Tomatsu S, Fleming R E, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt E M, Ruddy D A, Prass C E, et al. Proc Natl Acad Sci USA. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strominger J L. Cell. 1989;57:895–898. doi: 10.1016/0092-8674(89)90326-7. [DOI] [PubMed] [Google Scholar]
- 29.Chen H, Paul W E. J Immunol. 1997;159:2240–2249. [PubMed] [Google Scholar]
- 30.Rohren E M, Pease L R, Ploegh H L, Schumacher T N. J Exp Med. 1993;177:1713–1721. doi: 10.1084/jem.177.6.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pamer E G, Wang C R, Flaherty L, Lindahl K F, Bevan M J. Cell. 1992;70:215–223. doi: 10.1016/0092-8674(92)90097-v. [DOI] [PubMed] [Google Scholar]
- 32.Tian Z, Xu Y, Warner C M. Biol Reprod. 1992;47:271–276. doi: 10.1095/biolreprod47.2.271. [DOI] [PubMed] [Google Scholar]
- 33.Tabaczewski P, Stroynowski I. J Immunol. 1994;152:5268–5274. [PubMed] [Google Scholar]
- 34.Raulet D H, Held W. Cell. 1995;82:697–700. doi: 10.1016/0092-8674(95)90466-2. [DOI] [PubMed] [Google Scholar]
- 35.Lanier L L, Phillips J H. Semin Immunol. 1995;7:75–82. doi: 10.1006/smim.1995.0011. [DOI] [PubMed] [Google Scholar]
- 36.Hoglund P, Sundback J, Olsson-Alheim M Y, Johansson M, Salcedo M, Ohlen C, Ljunggren H G, Sentman C L, Karre K. Immunol Rev. 1997;155:11–28. doi: 10.1111/j.1600-065x.1997.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 37.Liao N S, Bix M, Zijlstra M, Jaenisch R, Raulet D. Science. 1991;253:199–202. doi: 10.1126/science.1853205. [DOI] [PubMed] [Google Scholar]
- 38.Hoglund P, Ohlen C, Carbone E, Franksson L, Ljunggren H G, Latour A, Koller B, Karre K. Proc Natl Acad Sci USA. 1991;88:10332–10336. doi: 10.1073/pnas.88.22.10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braud V M, Allan D S J, O’Callaghan C A, Soderstrom K, D’Andrea A, Ogg G S, Lazetic S, Young N T, Bell J I, Phillips J H, et al. Nature (London) 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 40.O’Callaghan C A, Bell J I. Immunol Rev. 1998;163:129–138. doi: 10.1111/j.1600-065x.1998.tb01192.x. [DOI] [PubMed] [Google Scholar]
- 41.Pascolo S, Bervas N, Ure J M, Smith A G, Lemonnier F A, Perarnau B. J Exp Med. 1997;185:2043–2051. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein J, Figueroa F, David C S. Immunogenetics. 1983;17:553–596. doi: 10.1007/BF00366126. [DOI] [PubMed] [Google Scholar]
- 43.Alexander J, Payne J A, Murray R, Frelinger J A, Cresswell P. Immunogenetics. 1989;29:380–388. doi: 10.1007/BF00375866. [DOI] [PubMed] [Google Scholar]
- 44.Glas R, Sturmhofel K, Hammerling G, Karre K, Ljunggren H G. J Exp Med. 1992;175:843–846. doi: 10.1084/jem.175.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machold R P, Ploegh H L. J Exp Med. 1996;184:2251–2259. doi: 10.1084/jem.184.6.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Handy D E, McCluskey J, Lew A M, Coligan J E, Margulies D H. Immunogenetics. 1988;28:81–90. doi: 10.1007/BF00346155. [DOI] [PubMed] [Google Scholar]
- 47.Zerrahn J, Held W, Raulet D H. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- 48.Bix M, Raulet D. Nature (London) 1992;359:330–333. doi: 10.1038/359330a0. [DOI] [PubMed] [Google Scholar]
- 49.Hoglund P, Glas R, Menard C, Kase A, Johansson M H, Franksson L, Lemonnier F, Karre K. Eur J Immunol. 1998;28:370–378. doi: 10.1002/(SICI)1521-4141(199801)28:01<370::AID-IMMU370>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 50.Aldrich C J, DeCloux A, Woods A S, Cotter R J, Soloski M J, Forman J. Cell. 1994;79:649–658. doi: 10.1016/0092-8674(94)90550-9. [DOI] [PubMed] [Google Scholar]
- 51.Soloski M J, DeCloux A, Aldrich C J, Forman J. Immunol Rev. 1995;147:67–89. doi: 10.1111/j.1600-065x.1995.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 52.Jiang H, Ware R, Stall A, Flaherty L, Chess L, Pernis B. Immunity. 1995;2:185–194. doi: 10.1016/s1074-7613(95)80079-4. [DOI] [PubMed] [Google Scholar]
- 53.Lee N, Goodlett D R, Ishitani A, Marquardt H, Geraghty D E. J Immunol. 1998;160:4951–4960. [PubMed] [Google Scholar]
- 54.Dorfman J R, Zerrahn J, Coles M C, Raulet D H. J Immunol. 1997;159:5219–5225. [PubMed] [Google Scholar]
- 55.Salcedo M, Andersson M, Lemieux S, Van Kaer L, Chambers B J, Ljunggren H G. Eur J Immunol. 1998;28:1315–1321. doi: 10.1002/(SICI)1521-4141(199804)28:04<1315::AID-IMMU1315>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 56.Sykulev Y, Joo M, Vturina I, Tsomides T J, Eisen H N. Immunity. 1996;4:565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 57.Muller D, Koller B H, Whitton J L, LaPan K E, Brigman K K, Frelinger J A. Science. 1992;255:1576–1578. doi: 10.1126/science.1347959. [DOI] [PubMed] [Google Scholar]