Abstract
The 436-amino acid protein enolase 1 from yeast was degraded in vitro by purified wild-type and mutant yeast 20S proteasome particles. Analysis of the cleavage products at different times revealed a processive degradation mechanism and a length distribution of fragments ranging from 3 to 25 amino acids with an average length of 7 to 8 amino acids. Surprisingly, the average fragment length was very similar between wild-type and mutant 20S proteasomes with reduced numbers of active sites. This implies that the fragment length is not influenced by the distance between the active sites, as previously postulated. A detailed analysis of the cleavages also allowed the identification of certain amino acid characteristics in positions flanking the cleavage site that guide the selection of the P1 residues by the three active β subunits. Because yeast and mammalian proteasomes are highly homologous, similar cleavage motifs might be used by mammalian proteasomes. Therefore, our data provide a basis for predicting proteasomal degradation products from which peptides are sampled by major histocompatibility complex class I molecules for presentation to cytotoxic T cells.
The eukaryotic 20S proteasome represents the catalytic core particle of the 26S proteasome, which is an essential component of the ubiquitin-dependent protein degradation pathway. The 20S particle is composed of 14 different but related subunits. Two outer disks, each containing seven α subunits, guard the two inner heptameric β subunit rings, which contain three proteolytically active sites each (1). How substrate molecules gain access to the active sites at the inner surface of the β rings is not known. It has been hypothesized that on association of the 20S particle with regulatory complexes, such as the 19S cap structure or PA28, an opening in the middle of the α subunit rings is induced allowing unfolded proteins to be fed into the 20S particle.
Probably as a consequence of this closed structure, very few proteins have been found to be degraded by 20S proteasomes in vitro, and only very few individual fragments and cleavage sites generated during the degradation of proteins by eukaryotic 20S proteasomes have been analyzed until now. This kind of analysis is of interest for several reasons.
First, it still has to be confirmed that the specificities of the proteasomal β subunits observed in experiments using peptide substrates (3–8), summarized as trypsin-like, chymotrypsin (ChT)-like, and peptidylglutamyl-peptide hydrolyzing activity, correspond to cleavages performed in intact proteins, a situation physiologically more relevant.
Second, the mechanism of protein degradation by the proteasome is not known. For the archebacterial proteasome, which is a less complex structure consisting of 14 identical α and β subunits (9), a processive model has been proposed (2, 10). Because of the high structural homology with the archebacterial particle a similar mechanism can be expected for eukaryotic proteasomes. Furthermore, it has been postulated that the 20S proteasome is equipped with a molecular ruler that determines the length of fragments generated (11). In the archebacterial proteasomes, the ruler was postulated to reflect the distance between each of the seven active β subunits (12). Therefore, a reduction of active sites is predicted to change the average length of fragments generated. In yeast proteasomes carrying only three active sites, another kind of ruler was proposed to reflect the distance of the three active β subunits to putative proteolytically active sites at the C-terminal ends of α-helices at the β annulus (1). Recently, two studies using yeast 20S mutant proteasomes with inactivated β subunits, however, excluded the latter model and revealed instead that no proteolytically active sites exist within the proteasome besides the β subunits containing the active site Thr (13, 14). The factor determining the length of the degradation products is, therefore, imposed either by the distance between the three active sites, analogous to the prediction for the Thermoplasma model, or by as yet unknown mechanisms.
The third reason for an in-depth analysis of fragments resulting from proteasomal protein digests is the lack of knowledge about the selection of cleavage sites during the degradation of substrates. Why the proteasome selects certain cleavage sites and ignores others with identical P1/P′1 composition is not known at all. A compilation of cleavage sites from a large panel of fragments should be very helpful in identifying amino acid motifs that explain the selection and allow the prediction of cleavage sites. Because the subunit topology and structure of mammalian proteasomes is analogous to that of yeast (15), the results will provide crucial information for the general understanding of the proteasomal activity.
We therefore examined in detail peptide fragments generated in digestions of yeast enolase 1 protein by wild-type (wt) and mutant yeast 20S proteasomes containing inactivated β subunits (5). Degradation products of the individual digests were analyzed for processive substrate degradation, fragment length, and the presence of cleavage motifs recognized by the three proteasomal subunits β2/Pup1, β1/Pre3, and β5/Pre2. The data obtained allow us to present proteasomal cleavage maps of a large unmodified protein. We find strong evidence for a processive degradation of the protein substrate and demonstrate that the number of active sites does not affect the average fragment length. From this we can exclude that the rules governing the fragment length in yeast 20S proteasomes are the result of the distance between active sites. The detailed analysis of cleavage sites in digests using wt and mutant yeast proteasomes also allows us to propose distinct cleavage motifs for the three active sites of eukaryotic 20S proteasomes that go far beyond the specificities described by using peptide substrates.
MATERIALS AND METHODS
Generation of Mutant Proteasomes and Purification of Yeast 20S Proteasomes.
Generation of yeast proteasome mutants and the purification of yeast 20S proteasomes has recently been described (1, 5).
HPLC Separation of Enolase Digests.
For the separation of degradation products, unfractionated enolase digests were subjected to μRP SC 2.1/10 columns (Pharmacia) on a Microbore HPLC-system (SMART system, Pharmacia). Buffer A contained 0.1% trifluoroacetic acid (TFA); buffer B contained 0.081% TFA and 80% acetonitrile. Gradients were 10% B for 5 min, in 35 min to 40% B, in 8 min to 75% B, and up to 85% in another 7 min at a flow rate of 150 μl/min.
Fragmentation of Degradation Products.
Ms/ms experiments were performed on a hybrid quadrupole orthogonal acceleration tandem mass spectrometer (Q- to F; Micromass, Manchester, U.K.). Fragmentation of the parent ions was achieved by collision with argon atoms. Q1 was set to the mass of interest ±0.5 Da, and the collision energy was optimized for each fragment. Integration time for the time of flight analyzer was 1 s with an interscan delay of 0.1 s.
Digestions.
Digests were incubated at 37°C and stopped by freezing at −20°C. All digests were incubated in 0.01% SDS/10 mM EDTA at molar ratios of enolase (10–20 μM) to 20S proteasome (40–60 nM) of 200–400:1. The buffer was 20 mM Hepes⋅KOH (pH 7.6), 2 mM MgAc2, and 1 mM DTT.
Matrix-Associated Laser Desorption Ionization MS Analysis.
One microliter of dihydroxyacetophenon matrix (20 mg of 2,5-dihydroxyacetophenon, 5 mg of ammonium citrate in 1 ml of 80% isopropanol) was mixed with 1 μl of each HPLC peak fraction on a gold target. Measurements were made with a laser-desorption time of flight system (Hewlett–Packard G2025A) at a vacuum of 10−6 torr (1 torr = 133 Pa). For signal generation 50–150 laser shots were added up in the single shot mode.
N-Terminal Sequencing (Edman Degradation).
Routinely, 15 μl of each HPLC peak fraction from digest fractionations were applied to the pulsed-liquid sequencer Procise model 494 (Applied Biosystems).
Statistical Analysis.
Frequencies of amino acids. The probability q(k) for finding a given amino acid exactly k times in a given position by random selection of cleavage sites (considering that each single peptide bond can be randomly selected only once for each enolase molecule) can be calculated according to the hypergeometric distribution.
q(k) values were used to calculate two-sided tail probabilities P to indicate deviation of observed frequencies from a random selection of certain amino acids at a given position. Because we considered positions P6 to P′6 surrounding a cleavage site the total number of potential cleavage sites (N) in enolase 1 was reduced from 435 to 425.
Comparison of amino acid characteristics.
To compare the characteristics of amino acids, a two-sided Student’s t test for two independent data sets was performed. Hydropathicity parameters were taken from Kyte and Doolittle (16), bulkiness parameters were from Zimmerman et al. (17), and normalized frequency parameters for β-turn were from Levitt (18). Results are shown as P values and mean value differences including the 95% confidence limit.
RESULTS
Degradation of Enolase 1.
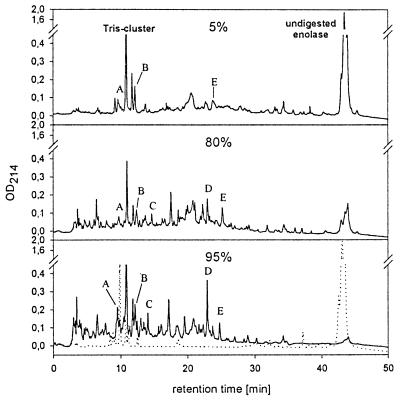
Denaturation of proteins is a prerequisite for their digestion by 20S proteasomes. To perform digestions of proteins without prior covalent modifications, we used protein substrates known for their thermolability, such as yeast enolase 1. Enolase 1 was incubated with yeast 20S proteasomes at a molar ratio of about 300:1. Aliquots were taken at different times and separated by reversed-phase HPLC. Peaks containing degradation products increase over time, while the signal for undigested enolase disappears (Fig. 1). Substrate degradation was linear with a degradation velocity of 6 min (data not shown). The observed stability of the peak pattern over time and the lack of larger enolase 1 fragments, especially at early time points (Fig. 1) argue for a processive substrate degradation mechanism as proposed for the Thermoplasma proteasome (10). Peaks A–E observed at different digestion times indeed contained identical fragments, as determined by matrix-associated laser desorption ionization MS analysis and Edman degradation (data not shown). The fragments in the range of 12 min were coeluted with digestion buffer components, and the peak height, therefore, does not reflect the amount of peptides.
Figure 1.
Enolase 1 degradation by yeast wt 20S proteasomes. Percent values represent the degree of enolase degradation as determined by linear regression. (Bottom) Dotted line shows incubation of enolase without proteasome.
Digestion Map of Enolase 1 by wt Yeast 20S Proteasomes.
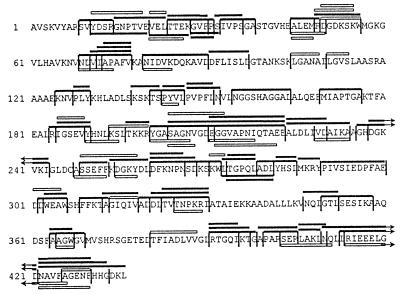
All peak fractions obtained during reversed-phase HPLC separation of digests were analyzed by matrix-associated laser desorption ionization MS, Edman sequencing, and hybrid quadrupole orthogonal acceleration tandem mass spectrometer MS/MS analysis and compiled into a digestion map (Fig. 2). The digests were reproduced two times, yielding comparable HPLC profiles. As summarized earlier (19), proteasomes are able to perform cleavages after almost every amino acid, but some residues are disfavored (such as Pro, Gln, Asn, Gly, Thr, Ser, and Lys) and other residues are favored (Ile, Leu, Trp, Tyr, Asp, and Arg) (Table 1). The selection of amino acids at positions flanking the cleavage sites will be discussed below.
Figure 2.
Digestion map generated from degradation of enolase 1 by yeast wt 20S proteasomes. Vertical lines, cleavage sites determined by Edman degradation and MS; solid bars, degradation products identified by Edman degradation and mass spectrometry; open bars, degradation products identified by Edman degradation. The C terminus is not yet confirmed by MS.
Table 1.
Frequencies of amino acids in the P1 position
| P1 residue | Total no. in enolase | No. of observed cleavages
|
||||
|---|---|---|---|---|---|---|
| wt | β2/Pup1− | β1/Pre3− | β2/Pup1− β1/Pre3− | β5/Pre2− | ||
| (n = 105) | (n = 80) | (n = 88) | (n = 73) | (n = 96) | ||
| Ala | 55 | 11 | 10 | 13 | 10 | 10 |
| Arg | 14 | 8 | 0 | 5 | 0 | 9 |
| Asn | 19 | 3 | 1 | 3 | 1 | 3 |
| Asp | 31 | 12 | 9 | 1 | 0 | 14 |
| Cys | 1 | 1 | 0 | 0 | 0 | 0 |
| Gln | 9 | 0 | 1 | 2 | 1 | 1 |
| Glu | 25 | 6 | 8 | 1 | 0 | 9 |
| Gly | 37 | 2 | 0 | 2 | 1 | 2 |
| His | 11 | 2 | 0 | 1 | 1 | 2 |
| Ile | 22 | 7 | 7 | 6 | 8 | 3 |
| Leu | 40 | 19 | 16 | 18 | 20 | 10 |
| Lys | 37 | 4 | 0 | 3 | 0 | 4 |
| Met | 5 | 1 | 0 | 1 | 1 | 1 |
| Phe | 16 | 5 | 8 | 9 | 8 | 2 |
| Pro | 15 | 0 | 0 | 0 | 0 | 0 |
| Ser | 31 | 5 | 2 | 3 | 4 | 8 |
| Thr | 20 | 2 | 1 | 2 | 1 | 0 |
| Trp | 5 | 4 | 4 | 4 | 4 | 2 |
| Tyr | 9 | 3 | 7 | 4 | 7 | 5 |
| Val | 34 | 10 | 6 | 10 | 6 | 11 |
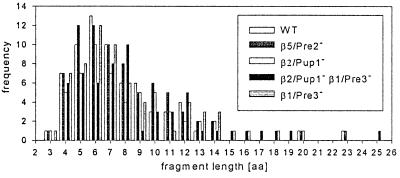
Three conclusions can be drawn from the observed cleavages: (i) the yeast 20S wt proteasome clearly prefers certain peptide bonds over others as cleavage sites; (ii) not all possible cleavage sites are used in one individual substrate molecule, resulting in the production of overlapping peptides; and (iii) some regions of the enolase sequence are characterized by a high number but others are characterized by a low number of cleavage sites. This clustering cannot be correlated to charge clusters and repetitive elements, which were not detected in the enolase 1 sequence by using the saps program (20). The fragments identified (n = 66) in a single digest of enolase by wt 20S proteasomes cover a range of 3–23 amino acids (see Fig. 4). Fragments with a size of 3–9 amino acids clearly dominate with a mean fragment length of 7.5 amino acids (SD = 3.5 residues). The digestion of enolase 1 using human 20S proteasomes resulted in a similar set of fragments, indicating a highly conserved selection of cleavage sites (A.K.N., T.P.D., M.S., W.K., S.S., H.-G.R., and H.S., unpublished results).
Figure 4.
Distribution of fragment lengths generated by wt and mutant proteasomes from enolase 1. For each proteasome only internal fragments with known C terminus derived from one single digestion were included.
Digestion of Enolase 1 by Mutant Yeast 20S Proteasomes.
To analyze the contribution of the three active β subunits to the observed cleavages, we digested enolase 1 with mutant yeast 20S proteasomes carrying inactivated β subunits, which were purified from the following mutant strains: pre2-K33A, pup1-T1A, pup1-T1A pre3-T1A, and pre3-T1A (5). Cleavage activity was analyzed as described for wt proteasomes. The β2/Pup1− proteasome did not cleave peptide bonds after basic amino acids (Fig. 3A and Table 1), the β1/Pre3− particle performed only one of the 17 cleavages found in wt proteasomes after acidic residues (Fig. 3B and Table 1) and the β2/Pup1−β1/Pre3− proteasome completely lacked proteolytic activity after basic and acidic residues (Fig. 3C and Table 1). The β5/Pre2− proteasome, completely lacking ChT-like activity as tested with fluorogenic substrates (5, 13), surprisingly performed a high number of cleavages after hydrophobic amino acids (Fig. 3D and Table 1). Because yeast 20S proteasomes contain no proteolytically active sites besides the three active β subunits, the β1/Pre3 and β2/Pup1 subunits, previously thought to cleave only after charged residues, also harbor ChT-like activity. These features were already observed during the analysis of synthetic peptide substrates digested with the same mutant proteasomes (13) and during experiments using inhibitors of the trypsin-like and ChT-like activities (21, 22). In addition, the number of cleavages after basic and acidic residues performed by the β5/Pre2− particle was increased compared with wt proteasomes, most likely due to lack of competing cuts normally performed by the β5/Pre2 subunit. Even more surprising, all mutant proteasomes generated fragments with an average length of 7–9 amino acids (7.9, SD = 3.5 for β5/Pre2−; 7.9, SD = 3.3 for β2/Pup1−; 8.8, SD = 4.3 for β2/Pup1−β1/Pre3−; 7.8, SD = 3.5 for β1/Pre3−), which is in the range of fragments generated by wild-type proteasomes (Fig. 4). Therefore, the size of fragments generated by yeast proteasomes is not related to the distance between the active β subunits, as postulated for the Thermoplasma proteasome (11).
Figure 3.
Digestion maps generated from degradation of enolase 1 by yeast mutant 20S proteasomes. The maps include all the degradation products of enolase 1 identified after digestion by β5/Pre2− (A) , β2/Pup1− (B), β2/Pup1−β1/Pre3− (C), β1/Pre3− (D) 20S proteasomes. Degradation products were identified by Edman degradation and MS. Symbols are as in Fig. 2.
P1 Amino Acids and Statistical Analysis of Flanking Residues.
The observed frequencies of amino acids in the P1 position of fragments generated by wt and mutant proteasomes indicate dominant cleavages after Leu or Arg, which are used approximately every second time these residues appear in enolase 1. On the other hand, only 2 of 37 Gly residues are used for cleavage (Table 1). Despite exerting a significant influence, the P1 amino acid is obviously not the only factor determining the location of a cleavage site. By considering only the P1 position, for example, it remains unclear why cleavages were detected after certain Leu residues but not after other Leu residues.
To statistically evaluate the frequency of all amino acids in positions ±6 surrounding cleavable peptide bonds, we compared the observed frequencies with those frequencies expected from a random cleavage distribution. By using the hypergeometric distribution, a value is assigned to each amino acid in every position, representing the probability (P) for the observed frequency to be the result of randomly distributed cleavages. A statistically significant accumulation or scarcity of particular amino acids in certain positions is taken as an indication for a positive or negative influence on the selection of peptide bonds hydrolyzed by the proteasomal subunits. To justify this approach, a set of cleavages of the same size as the data set was generated by a random number generator and statistically evaluated as a control, resulting in no significant accumulation of amino acids (data not shown).
Specificity of the wt Proteasome.
Examination of cleavages performed by the wt proteasome (n = 105) revealed the following deviations from randomness in the cleavage pattern (P values are two-sided tail probabilities). Leu as a P1 residue has the highest significance (P < 0.00082) and reflects pronounced ChT-like activity within wt proteasomes. Weaker preferences for Arg in P1 (P < 0.0084), Pro in P4 (P < 0.0033), and Gln in P5 (P < 0.0011) are also evident. On the other hand, certain residues appear to exert negative influence in certain positions. In the P1 position, Gly is less abundant than expected (P < 0.0011). When observed cleavage sites (n = 105) are compared with the remaining sites (not observed as cleavage sites, n = 320) using a two-sided Student’s t test, a preference for small residues becomes apparent in P′1 (P < 0.0072; 1.57 ± 1.15).
Specificity of the β5/Pre2 Active Site.
To evaluate a “cleavage motif” for a single active β subunit, in this case β5/Pre2, cleavages performed by the β2/Pup1−β1/Pre3− 20S particle were analyzed (n = 73). The statistical analysis clarifies the order of preference for the different hydrophobic P1 residues relative to their abundance in the substrate: Leu (P < 0.0000003) > Tyr (P < 0.0001) > Phe (P < 0.002) > Trp (P < 0.0035) > Ile (P < 0.02), but no accumulation is apparent for Val. This is, however, in agreement with recent studies on the cleavage of peptide substrates, indicating that cleavage after Val can be efficiently catalyzed by β2/Pup1 (13). Similar to the wt proteasome, Gly is disfavored in the P1 position. When amino acids are grouped (hydrophobic, basic, acidic, and others) the preference for hydrophobic residues in P1 is even more impressive (P < 3 × 10−17) with a weak preference for basic residues at P′1 (P < 0.0012). When comparing the observed cleavages (n = 73) to the remaining sites (n = 352), the following preferences are observed: in positions P3 and P′5 polar residues are clearly favored over nonpolar ones (P < 0.000072; 1.54 ± 0.75 and P < 0.0032; 1.15 ± 0.76, respectively), and a preference for β-turn-promoting residues is seen in position P3 (P < 0.0093; 0.13 ± 0.10). To determine more precisely why β5/Pre2 selects certain Leu as P1 residues, but not others, observed cleavage sites with Leu in P1 (n = 20) were compared with the corresponding unobserved sites with Leu in P1 (n = 19): Again there is a positive correlation between cleavage and the presence of polar (P < 0.00076; 3.2 ± 1.8) and turn-promoting residues (P < 0.0022; 0.38 ± 0.24) in the P3 position.
Certain Cleavages Are Not Restricted to a Single Active Site.
β5/Pre2-catalyzed cleavages as defined by the β2/Pup1−β1/Pre3− double mutant (n = 73) should belong to a set complementary to that provided by the β5/Pre2− mutant (n = 96). However, there is considerable overlap between these two sets of cleavages. Cleavages common to both β5/Pre2− and β2/Pup1−β1/Pre3− proteasome digests (n = 31), therefore, represent features not dependent on a single subunit. We find Tyr preferred at the P1 position (P < 0.0002) and Pro at P4 (P < 0.0003). When comparing cleavage sites of this intersecting subset (n = 31) against the remaining sites (n = 394), P1 residues tend to be more hydrophobic (P < 0.0013; 1.8 ± 1.1) and P3 residues more hydrophilic (P < 0.00023; 2.1 ± 1.1).
Specificity of the β2/Pup1 Active Site.
Because the respective double mutant proteasomes were not available, the motifs for β2/Pup1 and β1/Pre3 subunits must be approximated. One possibility for approximating β2/Pup1 specificity is to consider those cleavages catalyzed exclusively by β1/Pre3− proteasomes (n = 88) but not by β1/Pre3β2/Pup1− proteasomes (n = 73). For these cleavages (n = 41), analysis points out weak preferences for Gly in position P′1 (P < 0.001) and Pro in position P′3 (P < 0.001). Another possibility of approaching the β2/Pup1 motif is to look at those cleavages common to both β1/Pre3− (n = 88) and β5/Pre2− (n = 96) proteasomes, which should mostly originate from the β2/Pup1 subunit (n = 46). Analysis of amino acid frequency reveals a strong preference for Pro in P4 (P < 0.00004) and P′3 and Lys in P3 (P < 0.0031). However, this set of 46 cleavages includes 31 cleavages that can be found in either β1/Pre3−β2/Pup1−, or β2/Pup1− proteasomes and, therefore, belong to the category of cleavages that are not subunit-specific. Analysis of the remaining 15 strictly β2/Pup1-specific cleavages reveals Arg in P1 (P < 0.000044) Pro in P′3, Gly in P′1, but not Pro in P4, again showing that the preference for Pro in P4 is strongly associated with those cleavages that can be catalyzed by more than a single subunit.
Specificity of the β1/Pre3 Active Site.
Again, there are several possibilities of approximating a β1/pre3 cleavage motif. Looking at those cleavages common to both digests with proteasomes lacking active β5/Pre2 (n = 96) and active β2/Pup1 subunits (n = 80), the intersecting set includes 44 cleavages: Pro in P4 is the most prominent feature (P < 0.000002), followed by Tyr in P1 (P < 0.0009). If those cleavages not specific to a single subunit are eliminated, only 14 cleavages remain as truly β1/Pre3-specific. Asp now is significant in P1 (P < 0.0000007) and Glu in P1 also becomes evident (P < 0.0063). Comparing the cleavages common to proteasomes with active β1/pre3 subunits (n = 44) against the remaining sites of enolase (n = 381), the P5 position turns out to prefer hydrophilic residues (P < 0.0001; 3.2 ± 1.6). The P3 position favors hydrophobic residues (P < 0.00061; 2.82 ± 1.6) and disfavors turn-promoting residues (P < 0.0076; 0.29 ± 0.21). If cleavages with Asp or Glu in P1 (n = 31) are compared against the remaining Asp and Glu sites (n = 24), amino acids at the P’4 position tend to be more hydrophobic (P < 0.0056; 2.07 ± 1.43). The proposed cleavage motifs are summarized in Table 2.
Table 2.
Summary of cleavage motif
| Position
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P5 | P4 | P3 | P2 | P1 | P′1 | P′2 | P′3 | P′4 | P′5 | ||
| wt | (Pro)* | Leu | β-turn | ||||||||
| (Arg) | |||||||||||
| −Gly | small | ||||||||||
| β5/Pre2 | polar, | Leu | basic | β-turn | |||||||
| β-turn | Tyr | polar | |||||||||
| Phe | |||||||||||
| Trp | |||||||||||
| −Gly | |||||||||||
| β2/Pup1 | (Lys) | Arg | (Gly) | (Pro) | |||||||
| (Lys) | |||||||||||
| β1/Pre3 | polar | hydrophobic | Asp | hydrophobic | |||||||
| (Glu) | |||||||||||
| Subunit | Pro | hydrophobic | Tyr | ||||||||
| independent | hydrophobic | ||||||||||
*Amino acids in parentheses indicate P values below the significance level after the adjustment for multiple testing according to the Bonferroni method (23). Significances are, however, supported by amino acid characteristic analysis or crystal structure data. A minus indicates a disfavoured amino acid.
DISCUSSION
The specificity of 20S proteasomes has been investigated so far either by the use of peptide substrates and inhibitors of the proteasomal activities. To analyze the proteasomal specificity under more physiological conditions, we established a protocol that allowed the digestion of an entire protein, yeast enolase 1, without denaturation by covalent modification of amino acids. The identification of more than 400 enolase 1 fragments generated in digests using wt and mutated yeast 20S proteasomes carrying different combinations of inactivated β subunits allowed a detailed analysis of the length of fragments generated, proteolytic specificity, β subunit contribution to the observed cleavages, and a description of motifs that explain the selection of cleavage sites.
Protein Degradation and Fragment Length.
We determined a duration of 6 min for the degradation of one molecule of enolase by one yeast wt 20S proteasome. This value is in the range of activities found so far for degradation of whole proteins by eukaryotic and archaeal 20S proteasomes (2, 12, 24, 25) but is likely to be far from physiological levels. In vivo it takes less than 60 min from viral infection to major histocompatibility complex class I-restricted presentation of viral peptides to cytotoxic T cells (26). Numerous possibilities can explain this discrepancy, ranging from the lack of crucial components in the digestion buffer to missing in vivo “helpers of proteasomal proteolysis,” such as the 19S cap complex, hsc70 (27), or activators such as PA28 in mammalian cells (8, 28–30).
Our experiments using yeast 20S proteasomes and enolase 1 as a substrate show a very stable pattern of degradation products in HPLC-separated digests over time (Fig. 1), as reported for the Thermoplasma 20S proteasome (2, 10). In addition, the same degradation products could be identified at early time points when only 5% of the substrate is degraded and also at late time points when 95% of the substrate molecules are digested (data not shown). This demonstrates that final degradation products are generated without the presence of large degradation intermediates.
The finding that all proteasomes analyzed so far generate peptides with an average length of 7–9 amino acids led to the proposal of an intrinsic molecular ruler determining the fragment size (1, 9, 11). Our finding that proteasomes carrying six, four, or two active β subunits generate fragments of similar length strongly argues against the distance between the active sites influencing the length of proteasomal digestion products (Fig. 4). The small fragment length increase from the wild-type proteasomes to the β1/Pre3−β2/Pup1− proteasomes is most likely due to lack of cleavage activity after charged amino acids, which represent 25% of the amino acids in enolase 1. Our observed fragment length distribution is also in agreement with recent results using partially modified protein substrates and archaeal proteasomes (2). An explanation might be that proteasomes either have an exit size filter preventing the dissociation of products above a certain size or that active β subunits preferentially bind polypeptide stretches of 7 or 8 amino acids. The nonsymmetrical shape of the fragment length distributions (Fig. 4) might indicate a log-normal distribution of the fragment size, as postulated (2, 10). The fact that the main part of cleavage products does not fall into the range of major histocompatibility complex ligands (8–10 amino acids) probably mirrors the predominant contribution of the proteasome to general intracellular proteolysis, which preceded its participation in antigen processing in evolution.
Specificity and Selectivity of the Active β Subunits.
Analysis of the specificity of the different β subunits revealed that the β5/Pre2 subunit is responsible for the ChT-like activity, as expected. Proteasomes containing inactivated β2/Pup1 or β1/Pre3 subunits lacked only cleavage activity after basic or acidic residues, respectively. By analyzing the cleavage sites performed by the double mutant containing β5/Pre2 as the only active subunit, we found the strongest selection for Leu in P1 followed by Tyr, Phe, and Trp. By comparing the flanking residues around the β5/Pre2 cleavage sites with the rest of the observed cleavages, we found a strong preference for polar and β-turn-promoting amino acids at P3. This observation possibly correlates to characteristics of amino acids at the inner surface of the proteasome surrounding the active site Thr-1 of β5/Pre2. A polar pocket formed by His-98, Asp-114, Glu-120, and Arg-125 of the β6/C5 subunit might accommodate P3 residues of substrates. A similar situation might exist at the P′1 position, where basic residues are enriched and might interact with the negatively charged side chains of Asp-114 and Asp-116 of β5/Pre2.
The analysis of cleavages performed by β2/Pup1 revealed a preference for Arg over Lys in P1. The reasons for this selectivity are not known but have already been observed (31). Flanking residue analysis revealed also an enrichment of Lys at P3 of the fragments generated by β2/Pup1. The basic side chain might interact with an acidic pocket formed by Asp-28 (β2/Pup1) and Asp-114, Glu-120, and Glu-121 of β3/Pup3. On the P′ side, flexible and β-turn-promoting amino acids such as Gly at P′1 and Pro at P′3 are enriched. This feature of β2/Pup1 substrates might be imposed by amino acids, including His-35 of β2/Pup1, which block the end of a putative substrate binding cleft and thus require substrate bending.
The β1/Pre3 subunit favors Asp over Glu in P1 and at position P5 polar amino acids and at position P3 hydrophobic residues are enriched. Whether these features are reflected by the amino acids surrounding the active site Thr-1 remains to be determined.
Surprisingly, the β2/Pup1 and β1/Pre3 subunits also contribute substantially to the ChT-like activity as evident from the analysis of the β5/Pre2− proteasome, where only a slight reduction in the number of cleavages after hydrophobic amino acids was found. After analyzing the cleavages performed by the β5/Pre2− and the β2/Pup1−β1/Pre3− proteasomes, expected to contain complementary sets of cleavage sites, 31 identical cleavages were found in both groups. By examining the residues flanking these cleavage sites, we found Pro at P4 significantly enriched possibly favoring a turn at this position. When we extended the analysis of amino acid frequencies and characteristics to 10 amino acids flanking observed cleavage sites in both directions, we did not observe any additional significant amino acid preferences. This finding might indicate that a stretch of about 10 amino acids (P5 to P′5) is critical for the selection of a cleavage site by the β subunits.
The cleavage motifs for proteasomal cleavages described herein represent tools for developing an algorithm for the prediction of proteasomal cleavage sites. These motifs will be relevant for the prediction of peptides generated by mammalian proteasomes because the cleavages in enolase 1 generated by yeast or human 20S proteasomes overlap to a large extent (A.K.N., T.P.D., M.S., W.K., S.S., H.-G.R., and H.S., unpublished results). Nevertheless, detailed studies using human 20S proteasomes, lacking or containing interferon-inducible β subunits, should be performed to assess their contribution to the generation of epitopes for cytotoxic T cells and to refine the cleavage motifs.
Acknowledgments
This work was supported by grants of the Deutsche Forschungsgemeinschaft [Leibnizprogram to H.G.R. (Ra369/4–1); Sonderforschungsbereich 510, C1 and SCHI 301/2–1 to H.S.], the European Union (Biotech 95–1627 and Biomed 95–0263), Merck KGaA (Darmstadt, Germany), and the Fonds der chemischen Industrie, Frankfurt, Germany.
ABBREVIATIONS
- ChT
chymotrypsin
- wt
wild type
References
- 1. Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik H D, Huber R. Nature (London) 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 2.Kisselev A F, Akopian T N, Goldberg A L. J Biol Chem. 1998;273:1982–1989. doi: 10.1074/jbc.273.4.1982. [DOI] [PubMed] [Google Scholar]
- 3.Orlowski M, Michaud C. Biochemistry. 1989;28:9270–9278. doi: 10.1021/bi00450a006. [DOI] [PubMed] [Google Scholar]
- 4.Cardozo C, Vinitsky A, Hidalgo M C, Michaud C, Orlowski M. Biochemistry. 1992;31:7373–7380. doi: 10.1021/bi00147a023. [DOI] [PubMed] [Google Scholar]
- 5.Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf D H. J Biol Chem. 1997;272:25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- 6.Niedermann G, King G, Butz S, Birsner U, Grimm R, Shabanowitz J, Hunt D F, Eichmann K. Proc Natl Acad Sci USA. 1996;93:8572–8577. doi: 10.1073/pnas.93.16.8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niedermann G, Grimm R, Geier E, Maurer M, Realini C, Gartmann C, Soll J, Omura S, Rechsteiner M C, Baumeister W, Eichmann K. J Exp Med. 1997;186:209–220. doi: 10.1084/jem.186.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dick T P, Ruppert T, Groettrup M, Kloetzel P M, Kuehn L, Koszinowski U H, Stevanovic S, Schild H, Rammensee H G. Cell. 1996;86:253–262. doi: 10.1016/s0092-8674(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 9.Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 10.Akopian T N, Kisselev A F, Goldberg A L. J Biol Chem. 1997;272:1791–1798. doi: 10.1074/jbc.272.3.1791. [DOI] [PubMed] [Google Scholar]
- 11.Wenzel T, Eckerskorn C, Lottspeich F, Baumeister W. FEBS Lett. 1994;349:205–209. doi: 10.1016/0014-5793(94)00665-2. [DOI] [PubMed] [Google Scholar]
- 12.Dick L R, Aldrich C, Jameson S C, Moomaw C R, Pramanik B C, Doyle C K, DeMartino G N, Bevan M J, Forman J M, Slaughter C A. J Immunol. 1994;152:3884–3894. [PMC free article] [PubMed] [Google Scholar]
- 13.Dick, T. P., Nussbaum, S. K., Deeg, M., Heinemeyer, W., Groll, M., Schirle, M., Keilholz, W., Stevanovic, S., Wolf., D. H., Huber, R. et al. (1998) J. Biol. Chem., in press. [DOI] [PubMed]
- 14.Ditzel L, Huber R, Mann K, Heinemeyer W, Wolf D H, Groll M. J Mol Biol. 1998;279:1187–1191. doi: 10.1006/jmbi.1998.1818. [DOI] [PubMed] [Google Scholar]
- 15.Kopp F, Hendil K B, Dahlmann B, Kristensen P, Sobek A, Uerkvitz W. Proc Natl Acad Sci USA. 1997;94:2939–2944. doi: 10.1073/pnas.94.7.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 17.Zimmerman J M, Eliezer N, Simha R. J Theor Biol. 1968;21:170–201. doi: 10.1016/0022-5193(68)90069-6. [DOI] [PubMed] [Google Scholar]
- 18.Levitt M. Biochemistry. 1978;17:4277–4285. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- 19.Coux O, Tanaka K, Goldberg A L. Annu Rev Immunol. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 20.Brendel V, Bucher P, Nourbakhsh I R, Blaisdell B E, Karlin S. Proc Natl Acad Sci USA. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savory P J, Djaballah H, Angliker H, Shaw E, Rivett A J. Biochem J. 1993;296:601–605. doi: 10.1042/bj2960601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reidlinger J, Pike A M, Savory P J, Murray R Z, Rivett A J. J Biol Chem. 1997;272:24899–24905. doi: 10.1074/jbc.272.40.24899. [DOI] [PubMed] [Google Scholar]
- 23.Bland J M, Altman D G. Br Med J. 1995;310:170–175. [Google Scholar]
- 24.McGuire M J, McCullough M L, Croall D E, DeMartino G N. Biochim Biophys Acta. 1989;995:181–186. doi: 10.1016/0167-4838(89)90078-2. [DOI] [PubMed] [Google Scholar]
- 25.Dick L R, Moomaw C R, DeMartino G N, Slaughter C A. Biochemistry. 1991;30:2725–2734. doi: 10.1021/bi00224a022. [DOI] [PubMed] [Google Scholar]
- 26.Hosken N A, Bevan M J, Carbone F R. J Immunol. 1989;142:1079–1083. [PubMed] [Google Scholar]
- 27.Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz A L, Ciechanover A. J Biol Chem. 1997;272:9002–9010. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- 28.Dubiel W, Pratt G, Ferrell K, Rechsteiner M. J Biol Chem. 1992;267:22369–22377. [PubMed] [Google Scholar]
- 29.Ma C P, Slaughter C A, DeMartino G N. J Biol Chem. 1992;267:10515–10523. [PubMed] [Google Scholar]
- 30.Groettrup M, Soza A, Eggers M, Kuehn L, Dick T P, Schild H, Rammensee H G, Koszinowski U H, Kloetzel P M. Nature (London) 1996;381:166–168. doi: 10.1038/381166a0. [DOI] [PubMed] [Google Scholar]
- 31.Ossendorp F, Eggers M, Neisig A, Ruppert T, Groettrup M, Sijts A, Mengede E, Kloetzel P M, Neefjes J, Koszinowski U, Melief C. Immunity. 1996;5:115–124. doi: 10.1016/s1074-7613(00)80488-4. [DOI] [PubMed] [Google Scholar]