Abstract
Copolymer 1 [poly(Y,E,A,K)] is a random synthetic amino acid copolymer of l-tyrosine, l-glutamic acid, l-alanine, and l-lysine that is effective both in suppression of experimental allergic encephalomyelitis and in the treatment of relapsing forms of multiple sclerosis. Copolymer 1 binds promiscuously and very efficiently to purified HLA-DR molecules within the peptide-binding groove. In the present study, YEAK and YEAK-related copolymers and type II collagen (CII) peptide 261–273, a candidate autoantigen in rheumatoid arthritis (RA), competed for binding to RA-associated HLA-DR molecules encoded by DRB1*0101 and DRB1*0401. Moreover, these copolymers (particularly YEAK, YAK, and YEK) inhibited the response of DR1- and DR4-restricted T cell clones to the CII epitope 261–273 by >50%. This direct evidence both for competitive interactions of these copolymers and CII peptide with RA-associated HLA-DR molecules and for inhibition of CII-specific T cell responses suggests that these compounds should be evaluated in animal models for rheumatoid arthritis.
Rheumatoid arthritis (RA) is a common human autoimmune disease with a prevalence of ≈1% among Caucasians (1, 2). It is characterized by a chronic inflammation of the synovial joints and infiltration by activated T cells, macrophages, and plasma cells (3, 4), leading to a progressive destruction of the articular cartilage. Inherited susceptibility to RA is strongly associated with the DRB1 loci encoding HLA-DR1 (DRB1*0101) and -DR4 (DRB1*0401, DRB1*0404, or DRB1*0405) molecules (5–7). Residues 67–71 are polymorphic in HLA-DR proteins, but these RA-related alleles share a common DRβ motif in this region that contributes to the P4 pocket of the peptide-binding groove (8) as well as residues which interact with the T cell receptor of CD4+ T lymphocytes (9–11). It has been proposed that RA-associated HLA-DR molecules confer disease susceptibility by presenting distinct sets of antigenic peptides derived from a synovial joint protein(s) to CD4+ T lymphocytes (12, 13). Although the nature of the autoantigen(s) in RA is unknown, type II collagen (CII) has been proposed as a candidate because it is a major protein of hyaline cartilage and is able to induce arthritis resembling RA in genetically susceptible animals (14–22). Animal models for collagen-induced arthritis, including mice transgenic for HLA-DR1 or -DR4 (21, 22), enabled mapping of T cell determinants implicated in the autoimmune response to CII (23–25). An immunodominant T cell epitope in CII corresponding to residues 261–273 has been identified (24).
Copolymer 1 [Cop 1, poly(Y, E, A, K), called YEAK hereinafter] is a synthetic amino acid copolymer effective both in suppression of experimental allergic encephalomyelitis (26–36) and in the treatment of relapsing forms of multiple sclerosis (37, 38). Recently, the binding of Cop 1 to purified HLA-DR molecules within the peptide-binding groove has been reported (39). Cop 1 inhibited the binding of HA306–318 peptide, a high-affinity epitope of influenza virus, to both HLA-DR1 (DRB1*0101) and -DR4 (DRB1*0401) molecules (39). Copolymers composed of only three amino acids (EAK, YEA, YAK, and YEK) bound to living antigen-presenting cells (APCs) of both mouse and human origin and were cross-reactive with Cop 1 at the T cell level (M.F.-H., R. Aharoni, D. Teitelbaum, R. Arnon, M. Sela, and J.L.S., unpublished observations). In view of the possible therapeutic applications of Cop 1 or related copolymers in RA, it was important to determine whether these compounds compete with CII for binding to HLA-DR1 and -DR4 molecules. In the present report, the competition of Cop 1 and other copolymers with CII261–273 peptide for binding to RA-associated HLA-DR1 and -DR4 molecules was established. Moreover, these copolymers (particularly YEAK, YAK, and YEK) inhibited the response of DR1- and DR4-restricted T cell clones to the CII261–273 epitope. These findings provide direct evidence for competitive interactions between these copolymers and CII peptide for binding to RA-associated HLA-DR molecules and for inhibition of the CII-specific T cell response, suggesting the possible utility of these compounds in the treatment of RA.
MATERIALS AND METHODS
Protein Expression and Purification.
Recombinant HLA-DR1 and -DR4 molecules were expressed in Drosophila S2 cells as described (11, 40). Cells were grown in roller bottles in ExCell 401 medium (Sigma) supplemented with 0–5% fetal bovine serum (Sigma) at 26°C. Cells were harvested 4–5 days after induction by 1 mM CuSO4. Immunoaffinity purification of recombinant HLA-DR1 and -DR4 molecules was performed as reported (11). Briefly, supernatant from harvested cells was sequentially passed through protein A, protein G, and protein A-LB3.1 columns, followed by elution of the bound HLA-DR with 50 mM 3-[cyclohexylamino]-1-propanesulfonic acid (CAPS) (pH 11.5); next, the supernatant was neutralized with 200 mM phosphate (pH 6.0). Proteins were concentrated on a Centriprep 10 membrane (Amicon).
Peptides and Proteins.
Cop 1 (YEAK) is a synthetic random copolymer prepared by polymerization of the N-carboxyanhydrides of l-tyrosine, γ-benzyl-l-glutamate, l-alanine, and ɛ,N-trifluoroacetyl-l-lysine (26); the end product is a mixture of acetate salts of random polypeptides. EAK, YEA, YAK, and YEK were synthesized similarly. The following copolymers were synthesized by Teva Pharmaceutical Industries (Petach Tikva, Israel): Cop 1, YEAK, batch 55495 in the molar ratio of 1 Y: 1.5 E: 4.8 A: 3.7 K, with an average molecular weight (MW) of 5,800 or batch 52596 in the molar ratio of 1 Y: 1.5 E: 4.3 A: 3.3 K, MW 8,150; EAK, batch SD-1689 in the molar ratio of 1.2 E: 4.3 A: 3 K, MW 8,850; YEA, batch SD-1690 in the molar ratio of 1 Y: 0.6 E: 3 A, MW 7,600; YAK, batch SD-1691 in the molar ratio of 1 Y: 4.3 A: 3.1 K, MW 20,000; and YEK, batch SD-1697 in the molar ratio of 1 Y: 1 E: 3 K, MW 11,050.
Peptides were synthesized by using solid phase techniques (41) on an Applied Biosystems Peptide Synthesizer and were purified by RP-HPLC. Peptide sequences were HA306–318, PKYVKQNTLKLAT (MW 1,718) and CII261–273, AGFKGEQGPKGEP (MW 1,516).
Peptide Labeling.
Biotinylation of YEAK, EAK, YEA, YAK, and YEK was performed with excess N-hydroxysuccinimide biotin (Sigma) in dimethyl sulfoxide as described (34). Unreacted biotin was removed by dialysis (Spectra/Por membrane MWCO 500, Spectrum Medical Industries).
Assays for Peptide Binding to Class II Major Histocompatibility Complex Proteins.
Solutions. The solutions used in this assay are the following: binding buffer (20 mM Mes/140 mM NaCl/0.05% NaN3, pH 5.0) unless otherwise specified, PBS (150 mM sodium chloride/7.5 mM sodium phosphate, dibasic/2.5 mM sodium phosphate, monobasic, pH 7.2), Tris-buffered saline (TBS) (137 mM sodium chloride/25 mM Tris, pH 8.0/2.7 mM potassium chloride); and TBS plus 0.05% Tween 20.
Microtiter assay plate preparation.
The 96-well microtiter immunoassay plates (Pro-Bind, Falcon) were coated with 1 μg/well of affinity-purified LB3.1 mAb in PBS (100 μl total) for 18 h at 4°C. The wells were then blocked with TBS/3% BSA for 1 h at 37°C and washed three times with TBS plus 0.05% Tween 20. Before sample addition, 50 μl of TBS/1% BSA was added to each well.
Inhibition reactions.
Biotinylated YEAK, YEA, YAK, EAK, or YEK at a final concentration of 1.5 μM in 50 μl of the binding buffer was coincubated with unlabeled inhibitors (YEAK, YEA, YAK, EAK, YEK, CII261–273, or HA306–318) and HLA-DR molecules for 40 h at 37°C.
Detection of class II major histocompatibility complex protein/peptide complexes.
Bound peptide–biotin was detected by using streptavidin-conjugated alkaline phosphatase as follows: Plates were washed three times with TBS plus 0.05% Tween 20 and incubated with 100 μl of streptavidin-conjugated alkaline phosphatase (1:3,000, Bio-Rad) for 1 h at 37°C, followed by addition of p-nitrophenyl phosphate in triethanolamine buffer (Bio-Rad). The absorbance at 410 nm caused by released p-nitrophenyl phosphate was monitored by a microplate reader (Dynatech MR4000).
T Cell Hybridomas and Antigen Presentation Assays.
The following mouse T cell hybridomas specific for CII were used: HLA-DR1-restricted 3.19 and 19.3 clones (22) and HLA-DR4-restricted 3838 and D3 clones (25). APCs were L57.23 [L cells transfected with HLA-DR1 (22)], L cells transfected with HLA-DR4 (42), and Priess cells (DRB1*0401/DRB4*0101). T cell stimulation experiments were performed in 96-well microtiter plates in a total volume of 0.2 ml. Irradiated (3,000 rad) APCs (2.5 × 104/well) were coincubated with CII261–273 (40 μg/ml) and varying concentrations of copolymers for 2 h at 37°C; next T cells (5 × 104/well) were added and incubated for 24 h at 37°C. Supernatants (30 μl) were taken and incubated with interleukin 2 (IL-2)-dependent CTLL cells (5 × 104/well) for 12 h, followed by labeling with [3H]thymidine (1 μCi/well) for 12 h. Plates were harvested, and the radioactivity was monitored by using a 1450 Microbeta Plus liquid scintillation counter (Wallac, Gaithersburg, MD).
RESULTS
Inhibition of Binding of Random Synthetic Copolymers to Recombinant HLA-DR1 and -DR4 Molecules by CII261–273 Epitope.
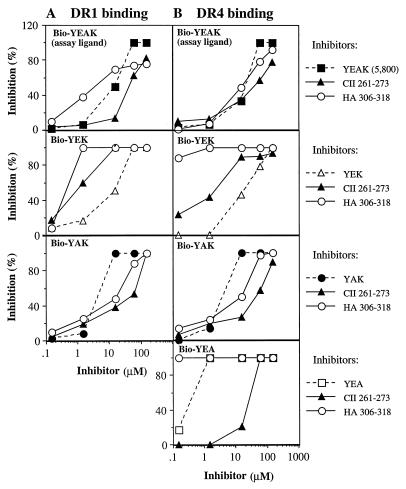
Three different preparations of Cop 1 (YEAK) bound to purified HLA-DR1, -DR2, and -DR4 molecules with high affinity and in a peptide-specific manner (39). Recently, copolymers composed of only three amino acids YEA, YEK, YAK, and EAK were also shown to bind these HLA-DR molecules with high affinity and to compete with Cop 1 for binding (M.F.-H., R. Aharoni, D. Teitelbaum, R. Arnon, M. Sela, and J.L.S., unpublished observations). To determine whether YAK, YEK, YEA, or YEAK competed with the RA-associated epitope CII261–273 for binding to HLA-DR1 or -DR4 molecules, recombinant water-soluble HLA-DR1 and -DR4 proteins (encoded by DRA/DRB1*0101 and *0401, respectively) were employed (EAK was excluded from the study because its binding to and competitive efficiency for HLA-DR1 and -DR4 was low). Competitive binding assays were carried out with biotinylated YEAK, YAK, YEA, or YEK and unlabeled inhibitors (YEAK, YAK, YEA, YEK, CII261–273, or HA306–318 peptide) (Fig. 1).The binding of biotinylated YEAK, YEA, or YAK to HLA-DR1 or -DR4 molecules was less well inhibited by the CII261–273 epitope than was the binding of YEK, as expressed by higher IC50 values for the CII peptide. HA306–318 inhibited the binding of each copolymer more efficiently than did CII261–273 (Fig. 1 A and B and Table 1), but the inhibition of binding of the various biotinylated copolymers was in the same order with YAK and YEAK as the best competitors (i.e., higher IC50 values for the HA peptide). These experiments indicate that random copolymers compete with and prevent binding of the autoantigenic epitope CII261–273 to RA-associated HLA-DR1 or -DR4 molecules.
Figure 1.
Inhibition of binding of biotinylated copolymers to recombinant HLA-DR1 (A) and -DR4 (B) molecules by different competitors. Recombinant water-soluble HLA-DR1 or -DR4 molecules (0.15 μM) were incubated with biotinylated YEAK (MW 5,800), YEK (MW 11,050), YAK (MW 20,000), or YEA (MW 7,600) (1.5 μM) in the presence of unlabeled copolymers, HA306–318, or CII261–273 at a range of concentrations. All incubations were carried out in duplicate at pH 5.0 for 40 h at 37°C. The amount of bound biotinylated copolymer (assay ligand) was measured as described. Specific binding is expressed as the percentage of inhibition by using the following formula: percentage of inhibition = 100% − [(absorbance at 410 nm with competitor − background)/(absorbance without competitor − background) × 100]. The signals at 410 nm without competitor were 0.91–0.95, and the background was 0.17.
Table 1.
IC50 for inhibition of binding of copolymers to recombinant HLA-DR1 and -DR4 molecules by antigenic peptides
| Biotinylated copolymer* | HLA–DR1†
|
HLA–DR4
|
||
|---|---|---|---|---|
| CII261–273 | HA306–318 | CII261–273 | HA306–318 | |
| YEAK | 40.0‡ | 3.5 | 40.0 | 10.5 |
| YEK | 0.8 | 0.4 | 2.0 | ≪0.1 |
| YAK | 40.0 | 10.5 | 40.0 | 10.5 |
| YEA | ND§ | ND | 23.0 | ≪0.1 |
Recombinant HLA-DR1 and -DR4 molecules were purified as described.
YEAK with an average MW of 5,800; YAK, MW 20,000; EAK, MW 8,850; YEA, MW 7,600; and YEK, MW 11,050 (1.5 μM) were incubated with CII261–273 or HA306–318 peptides at a range of concentrations and with purified HLA-DR1 and -DR4 molecules at pH 5.0 followed by capture with class II-specific mAb and peptide detection with alkaline phosphatase-streptavidin.
Data are presented as IC50 (μM, inhibitory concentration of CII or HA peptide giving 50% inhibition of the binding of biotinylated copolymers). These values were calculated based on the competitive binding assays (Fig. 1) (i.e. the higher the IC50 of the peptide, the more effective the copolymer in competing for binding of the peptide).
ND, not determined.
Inhibition of HLA-DR1- and -DR4-Restricted CII-Specific T Cell Response by Random Copolymers.
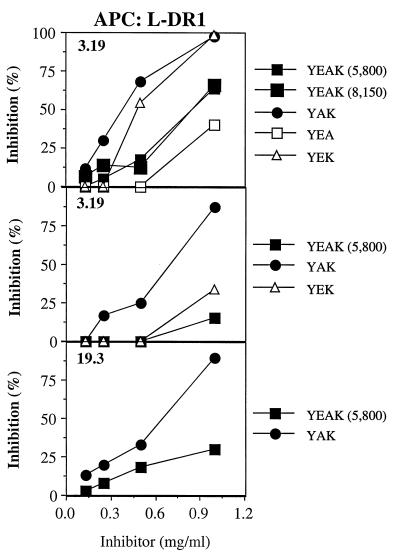
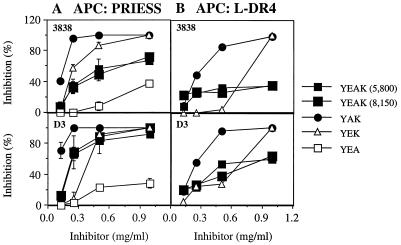
To determine whether YAK, YEK, YEA, or YEAK could also inhibit presentation of the CII261–273 peptide to autoreactive T cells, CII-specific T cell hybridomas restricted to HLA-DR1 (3.19 and 19.3) (22) and HLA-DR4 (3838 and D3) (25) were examined. Irradiated APCs were incubated with CII261–273 and relevant copolymer for 2 h before T cells were added for 24 h, and supernatants were tested for IL-2 secretion by these hybridomas. YAK was the most potent inhibitor of HLA-DR1-restricted T cells, by using L fibroblasts transfected with HLA-DR1 as APCs for the CII peptide, whereas the other copolymers inhibited the response less efficiently (Fig. 2). A similar pattern of activity was obtained for HLA-DR4-restricted T cells by using either Priess or L fibroblasts transfected with HLA-DR4 as APCs (Fig. 3 A and B). Thus, based on these data, the ability of random copolymers to compete with the potential autoantigenic CII261–273 peptide presented to the T cell hybridomas is expressed in the following order: YAK > YEK > YEAK ≫ YEA.
Figure 2.
Inhibition of IL-2 production by HLA-DR1-restricted, CII-specific T cell hybridomas in the presence of different copolymers. Irradiated L57.23 cells were coincubated in duplicate with CII261–273 (40 μg/ml) and varying concentrations of copolymers for 2 h at 37°C; next, T cells (clones 3.19 or 19.3) were added and incubated for 24 h at 37°C. Supernatants (30 μl) were incubated with IL-2-dependent CTL-L as described. Data are presented as the percentage of inhibition of CTL-L proliferation by using the following formula: percentage of inhibition = 1 − [(cpm in the presence of inhibitor − background)/(cpm in the absence of inhibitor − background) × 100].
Figure 3.
Inhibition of IL-2 production by HLA-DR4-restricted, CII-specific T cell hybridomas in the presence of different copolymers. Irradiated Priess cells (A) or L cells transfected with HLA-DR4 (B) were coincubated in duplicate with CII261–273 (40 μg/ml) and varying concentrations of copolymers for 2 h at 37°C; next, T cells (clones 3838 or D3) were added and incubated for 24 h at 37°C. Results represent the mean ± SD of two independent experiments. Other details are described in the legend to Fig. 2.
DISCUSSION
In this report, direct evidence for inhibition of the CII-specific T cell response by 4- and 3-aa random synthetic copolymers is provided, based on competition for binding to RA-associated HLA-DR1 and -DR4 molecules and on decrease in IL-2 production by HLA-DR1- and -DR4-restricted T cell hybridomas in the presence of these copolymers.
Previous findings suggested that the activity of Cop 1 in experimental allergic encephalomyelitis and multiple sclerosis involves binding to class II major histocompatibility complex molecules within the peptide-binding groove, where it may act either as a blocking peptide or as an antagonist or partial agonist, resulting in suppression of autoimmune T cell responses (39). Here, the binding of Cop 1 to RA-associated HLA-DR1 (DRB1*0101) and -DR4 (DRB1*0401) molecules was competed by the peptide determinant CII261–273, a candidate autoantigen in RA. In addition to Cop 1 (YEAK), copolymers of three amino acids, in particular YAK and YEK, which were shown to bind purified HLA-DR1 and -DR4 molecules with high affinity (M.F.-H., R. Aharoni, D. Teitelbaum, R. Arnon, M. Sela, and J.L.S., unpublished observations), competed for binding with CII peptide very efficiently. In these binding experiments, recombinant “empty” HLA-DR1 and -DR4 molecules were employed, with no interference from the endogenous peptides, as opposed to previously analyzed HLA-DR1 and -DR4 molecules, of which only 10–20% are available for exogenous peptide binding (43); this alteration resulted in different binding affinities for the copolymers in these two reports. In addition, the use of empty molecules may account for differences in the affinity of CII261–273 binding in the competitive assays with various copolymers. Previous reports have determined the requirements for CII261–273 epitope binding to DRB1*0401 as well as T cell receptor contact residues by using human DR4 purified from Priess cells (24, 25). In these studies, Phe 263 of the CII261–273 peptide was aligned to the P1 pocket and Gln 266 was aligned to the P4 pocket of the HLA-DR4-binding groove, in agreement with peptide-binding requirements for these pockets (44). The core of the T cell determinant was found to be similar for HLA-DR1 and -DR4 alleles (22, 24). Because the P1 pocket of both DR1 and DR4 molecules can accommodate Y, any of the random copolymers (YEAK, YAK, or YEK) would have the potential to block the CII261–273 binding.
The inhibitory concentrations of YAK (0.05–0.7 mg/ml), YEAK (0.2–0.85 mg/ml), and YEK (0.23–0.73 mg/ml) that reduced the secretion of IL-2 by HLA-DR1- and -DR4-restricted T cell hybridomas (IC50), were higher than those of different defined antigenic peptides usually used for inhibition of binding to major histocompatibility complex proteins (45–47). However, these concentrations were within the range reported to inhibit both myelin basic protein-specific (32, 33) or proteolipid protein-specific (35) T cell lines and clones by Cop 1 (YEAK) and antigen-specific T cell activation by different random copolymers (48) (0.1–0.5 mg/ml). This may be caused by the fact that these copolymers are mixtures of random polypeptides; consequently, each component is underrepresented in the mixture, requiring higher molar amounts to obtain the inhibitory effect. Further studies, particularly in vivo studies in murine arthritis models, will indicate whether any of these compounds are potentially useful in the treatment of RA in humans.
Acknowledgments
We thank Dr. Michael Sela for continued stimulation and support. We thank Mrs. Anastasia Haykov and Mrs. Michal Mandelboim for expert technical assistance. This work was supported by a grant from the National Institutes of Health (CA-47554). M.F.-H. is the recipient of a National Multiple Sclerosis Society Advanced Postdoctoral Fellowship.
ABBREVIATIONS
- RA
rheumatoid arthritis
- CII
type II collagen
- Cop 1
copolymer 1, YEAK
- HA
influenza virus hemagglutinin
- MW
molecular weight
- IL
interleukin
- APC
antigen-presenting cell
References
- 1. Feldmann M, Brennan F M, Maini R N. Cell. 1996;85:307–310. doi: 10.1016/s0092-8674(00)81109-5. [DOI] [PubMed] [Google Scholar]
- 2.Harris E J. In: Textbook of Rheumatology. Kelley W, Harris E, Ruddy S, Sledge C, editors. Philadelphia: Saunders; 1997. pp. 898–932. [Google Scholar]
- 3.Janossy G, Panayai G, Duke O, Bofill M, Poulter L W, Goldstein G. Lancet. 1981;ii:839–841. doi: 10.1016/s0140-6736(81)91107-7. [DOI] [PubMed] [Google Scholar]
- 4.Cush J J, Lipsky P E. Arthritis Rheum. 1988;31:1230–1238. doi: 10.1002/art.1780311003. [DOI] [PubMed] [Google Scholar]
- 5.Gregersen P K, Silver J, Winchester R J. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 6.Todd J A, Acha-Orbea H, Bell J I, Chao N, Fronek Z, Jacob C O, McDermott M, Sinha A A, Timmerman L, McDevitt H O. Science. 1988;240:1003–1009. doi: 10.1126/science.3368786. [DOI] [PubMed] [Google Scholar]
- 7.Wordsworth B P, Lanchbury J S, Sakkas L I, Welsh K I, Panayi G S, Bell J I. Proc Natl Acad Sci USA. 1989;86:10049–10053. doi: 10.1073/pnas.86.24.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stern L J, Brown J H, Jardetzky T S, Gorga J C, Urban R G, Strominger J L, Wiley D C. Nature (London) 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 9.Hammer J, Gallazzi F, Bono E, Karr R W, Guenot J, Valsasnini P, Nagy Z A, Sinigaglia F. J Exp Med. 1995;181:1847–1855. doi: 10.1084/jem.181.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penzotti J E, Doherty D, Lybrand T P, Nepom G T. J Autoimmun. 1996;9:287–293. doi: 10.1006/jaut.1996.0037. [DOI] [PubMed] [Google Scholar]
- 11.Dessen A, Lawrence C M, Cupo S, Zaller D M, Wiley D C. Immunity. 1997;7:473–481. doi: 10.1016/s1074-7613(00)80369-6. [DOI] [PubMed] [Google Scholar]
- 12.Woulfe S L, Bono C P, Zacheis D A, Kirschmann T A, Baudino C S, Karr R W, Schwartz B D. Arthritis Rheum. 1995;38:1744–1753. doi: 10.1002/art.1780381207. [DOI] [PubMed] [Google Scholar]
- 13.Kirschmann D A, Duffin K L, Smith K E, Welply J K, Howard S C, Schwartz B D, Woulfe S L. J Immunol. 1995;155:5655–5662. [PubMed] [Google Scholar]
- 14.Courtenay J S, Dallman M J, Dayan A D, Martin A, Mosedale B. Nature (London) 1980;283:666–668. doi: 10.1038/283666a0. [DOI] [PubMed] [Google Scholar]
- 15.Trentham D E, Townes A S, Kang A H. J Exp Med. 1977;146:857–868. doi: 10.1084/jem.146.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cathcart E S, Hayes K C, Gonnerman W A, Lazzari A A, Franzblau C. Lab Invest. 1986;54:26–31. [PubMed] [Google Scholar]
- 17.Iwakura Y, Tosu M, Yoshida E, Takiguchi M, Sato K, Kitajima I, Nishioka K, Yamamoto K, Takeda T, Hatanaka M, Yamamoto H, Sekiguchi T. Science. 1991;253:1026–1028. doi: 10.1126/science.1887217. [DOI] [PubMed] [Google Scholar]
- 18.Keffer J, Probert L, Cazlaris H, Georgopoulos S, Kaslaris E, Kioussis D, Kollias G. EMBO J. 1991;13:4025–4031. doi: 10.1002/j.1460-2075.1991.tb04978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kouskoff V, Korganow A-S, Duchatelle V, Dedott C, Benoist C, Mathis D. Cell. 1996;87:811–822. doi: 10.1016/s0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- 20.Nabozny G H, Baisch J, Cheng S, Cosgrove D, Griffiths M M, Luthra H S, David C S. J Exp Med. 1996;183:27–37. doi: 10.1084/jem.183.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fugger L, Michie S A, Rulifson I, Lock C B, Sonderstrup-McDevitt G. Proc Natl Acad Sci USA. 1994;91:6151–6155. doi: 10.1073/pnas.91.13.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosloniec E F, Brand D D, Myers L K, Whittington K B, Gumanovskaya M, Zaller D M, Woods A, Altmann D M, Stuart J M, Kang A H. J Exp Med. 1997;185:1113–1122. doi: 10.1084/jem.185.6.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brand D D, Myers L K, Terato K, Whittington K B, Stuart J M, Kang A H, Rosloniec E F. J Immunol. 1994;152:3088–3097. [PubMed] [Google Scholar]
- 24.Fugger L, Rothbard J B, Sonderstrup-McDevitt G. Eur J Immunol. 1996;26:928–933. doi: 10.1002/eji.1830260431. [DOI] [PubMed] [Google Scholar]
- 25.Andersson E C, Hansen B E, Jacobsen H, Madsen L S, Andersen C B, Engberg J, Rothbard J B, Sonderstrup-McDevitt G, Malmstrom V, Holmdahl R, Svejgaard A, Fugger L. Proc Natl Acad Sci USA. 1998;95:7574–7579. doi: 10.1073/pnas.95.13.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teitelbaum D, Meshorer A, Hirshfeld T, Arnon R, Sela M. Eur J Immunol. 1971;1:242–248. doi: 10.1002/eji.1830010406. [DOI] [PubMed] [Google Scholar]
- 27.Teitelbaum D, Webb C, Meshorer A, Arnon R, Sela M. Eur J Immunol. 1973;3:273–279. doi: 10.1002/eji.1830030505. [DOI] [PubMed] [Google Scholar]
- 28.Teitelbaum D, Webb C, Bree M, Meshorer A, Arnon R, Sela M. Clin Immunol Immunopathol. 1974;3:256–262. doi: 10.1016/0090-1229(74)90012-9. [DOI] [PubMed] [Google Scholar]
- 29.Lando Z, Teitelbaum D, Arnon R. J Immunol. 1979;123:2156–2160. [PubMed] [Google Scholar]
- 30.Sela M, Arnon R, Teitelbaum D. Bull Inst Pasteur (Paris) 1990;88:303–314. [Google Scholar]
- 31.Aharoni R, Teitelbaum D, Arnon R. Eur J Immunol. 1993;23:17–25. doi: 10.1002/eji.1830230105. [DOI] [PubMed] [Google Scholar]
- 32.Teitelbaum D, Aharoni R, Arnon R, Sela M. Proc Natl Acad Sci USA. 1988;85:9724–9728. doi: 10.1073/pnas.85.24.9724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teitelbaum D, Milo R, Arnon R, Sela M. Proc Natl Acad Sci USA. 1992;89:137–141. doi: 10.1073/pnas.89.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fridkis-Hareli M, Teitelbaum D, Gurevich E, Pecht I, Brautbar C, Kwon O J, Brenner T, Arnon R, Sela M. Proc Natl Acad Sci USA. 1994;91:4872–4876. doi: 10.1073/pnas.91.11.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teitelbaum D, Fridkis-Hareli M, Arnon R, Sela M. J Neuroimmunol. 1996;64:209–217. doi: 10.1016/0165-5728(95)00180-8. [DOI] [PubMed] [Google Scholar]
- 36.Fridkis-Hareli M, Teitelbaum D, Arnon R, Sela M. Cell Immunol. 1995;163:229–236. doi: 10.1006/cimm.1995.1121. [DOI] [PubMed] [Google Scholar]
- 37.Bornstein M B, Miller A, Slagle S, Weitzman M, Crystal H, Drexler E, Keilson M, Merriam A, Wassertheil-Smoller S, Spada V, et al. N Engl J Med. 1987;317:408–414. doi: 10.1056/NEJM198708133170703. [DOI] [PubMed] [Google Scholar]
- 38.Johnson K P, Brooks B R, Cohen J A, Ford C C, Goldstein J, Lisak R P, Myers L W, Panitch H S, Rose J W, Schiffer R B, et al. Neurology. 1995;45:1268–1276. doi: 10.1212/wnl.45.7.1268. [DOI] [PubMed] [Google Scholar]
- 39.Fridkis-Hareli M, Strominger J L. J Immunol. 1998;160:4386–4397. [PubMed] [Google Scholar]
- 40.Stern L J, Wiley D C. Cell. 1992;68:465–477. doi: 10.1016/0092-8674(92)90184-e. [DOI] [PubMed] [Google Scholar]
- 41.Barany G, Merrifield R. In: The Peptides. Gross E, Meinhofer J, editors. New York: Academic; 1979. pp. 1–284. [Google Scholar]
- 42.Woods A, Chen H Y, Trumbauer M E, Sirotina A, Cummings R, Zaller D M. J Exp Med. 1994;180:173–181. doi: 10.1084/jem.180.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammer J, Valsasnini P, Tolba K, Bolin D, Higelin J, Takacs B, Sinigaglia F. Cell. 1993;74:197–203. doi: 10.1016/0092-8674(93)90306-b. [DOI] [PubMed] [Google Scholar]
- 44.Rammensee H-G, Friede T, Stevanovic S. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 45.Wraith D C, Smilek D E, Mitchell D J, Steinman L, McDevitt H O. Cell. 1989;59:247–255. doi: 10.1016/0092-8674(89)90287-0. [DOI] [PubMed] [Google Scholar]
- 46.Gautam A M, Glynn P. J Immunol. 1990;144:1177–1180. [PubMed] [Google Scholar]
- 47.Lamont A G, Sette A, Fujinami R, Colon S M, Miles C, Grey H M. J Immunol. 1990;145:1687–1693. [PubMed] [Google Scholar]
- 48.Rock K L, Benacerraf B. J Exp Med. 1983;157:1618–1634. doi: 10.1084/jem.157.5.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]