Abstract
Recombinant mesophilic Escherichia coli (Ec) and thermophilic Bacillus stearothermophilus (Bst) elongation factors EF-Tus, their isolated G-domains, and six chimeric EF-Tus composed of domains of either EF-Tu were prepared, and their GDP/GTP binding activities and thermostability were characterized. BstEF-Tu and BstG-domain bound GDP and GTP with affinities in nanomolar and submicromolar ranges, respectively, fully comparable with those of EcEF-Tu. In contrast, the EcG-domain bound the nucleotides with much lower, micromolar affinities. The exchange of domains 2 and 3 had essentially no effect on the GDP-binding activity; all complexes of chimeric EF-Tus with GDP retained Kd values in the nanomolar range. The final thermostability level of either EF-Tu was the result of a cooperative interaction between the G-domains and domains 2 + 3. The G-domains set up a “basic” level of the thermostability, which was ~20°C higher with the BstG-domain than with the EcG-domain. This correlated with the growth temperature optimum difference of both bacteria and two distinct thermostabilization features of the BstG-domain: an increase of charged residues at the expense of polar uncharged residues (CvP bias), and a decrease in the nonpolar solvent-accessible surface area. Domains 2 + 3 contributed by further stabilization of α-helical regions and, in turn, the functions of the G-domains to the level of the respective growth temperature optima. Their contributions were similar irrespective of their origin but, with Ecdomains 2 + 3, dependent on the guanine nucleotide binding state. It was lower in the GTP conformation, and the mechanism involved the destabilization of the α-helical regions of the G-domain by Ecdomain 2.
Keywords: EF-Tu, thermostability, chimeric protein, EF-Tu domains, G-domain, Escherichia coli, Bacillus stearothermophilus
Elongation factors EF-Tu/EF-1α are abundant, highly homologous cellular GTP-proteins occupying a key position in translation in all organisms as universal carriers of aminoacyl-tRNAs. Their conformation and activity are regulated by GDP and GTP (Jonák and Rychlík 1973; Printz and Miller 1973; Kaziro 1978), and they hydrolyze bound GTP (Krab and Parmeggiani 1998). Their known 3D structures are superimposable, and they share the same catalytic mechanisms (Krab and Parmeggiani 1998). The high structural homology predetermines the elongation factors for the study of evolutionary relationships between organisms (Baldauf et al. 1996) and for elucidation of the structural features of adaptation to various living conditions.
All EF-Tus/EF-1αs are monomeric proteins composed of ~400 amino acid residues (for review, see Krab and Parmeggiani 1998) folded into three clearly distinct domains (Kjeldgaard and Nyborg 1992; Berchtold et al. 1993; Song et al. 1999).
N-terminal domain 1 (~200 residues) of EF-Tu is composed of a predominantly parallel six-stranded β-sheet core surrounded by seven α-helices. It contains the guanine nucleotide-binding site and the GTPase center (Parmeggiani et al. 1987; Kjeldgaard and Nyborg 1992) and is called the catalytic or G-domain (Parmeggiani et al. 1987). Middle domain 2 (~100 residues) and C-terminal domain 3 (~100 residues) are β-barrels of seven and six antiparallel β-strands, respectively, that share an extended interface. The reasons for the arrangement of EF-Tu into three domains are not fully understood except for the fact that formation of the aminoacyl-tRNA-binding site requires participation of all of them. This takes place in the GTP conformation, in which EF-Tus exist in a compact form with tight interfaces between the G-domain and domain 2 as well as the G-domain and domain 3 and which has a high affinity for aminoacyl-tRNAs and ribosomes (Berchtold et al. 1993; Kjeldgaard et al. 1993). Hydrolysis of the bound GTP during the mRNA decoding induces a large conformational change characterized by an opening of the molecule. The interactions between the G-domain and domain 2 are essentially lost, and only interactions between domain 3 and domain 2 are preserved while new interactions are formed between domain 3 and the G-domain. EF-Tu in the open-GDP conformation has a low affinity for aminoacyl-tRNA (Jonák et al. 1980).
Until now, functional evaluation of individual domains of EF-Tu has been carried out mainly by the deletion approach, by examining one- or two-domain proteins (Parmeggiani et al. 1987; Nock et al. 1995; Masullo et al. 1997; Cetin et al. 1998). In this paper, we address the question of how the functions and thermostability of EF-Tu from mesophilic Escherichia coli, growing at ~37°C and EF-Tu from thermophilic Bacillus stearothermophilus growing at temperatures 55°–60°C are built from the contributions of individual domains. Identification of the molecular basis of the increased thermostability of the proteins is expected to help our understanding of protein folding as well as the design of enzymes retaining their activity at high temperature. First, molecules of the EF-Tus were genetically dissected into three corresponding domains, and the domains were combined to form six chimeric EF-Tu proteins. In addition, the G-domains, the functional modules of EF-Tus of both organisms, were prepared and examined. Second, the GDP- and GTP-binding activities and thermostability of the proteins were measured both as the maintenance, at increasing temperatures, of a defined functional state (Jaenicke and Böhm 1998) by the ability to bind GDP and GTP and, independently, using CD spectroscopy, as the maintenance of the α-helix content. There were several reasons for the application of the chimerization approach: (1) The proteins share 75% amino acid sequence identity (Krásný et al. 1998); (2) B. stearothermophilus EF-Tu (Mr = 43,290 D, 394 amino acid residues) is only one amino acid residue longer than E. coli EF-Tu (Mr = 43,200 D, 393 amino acid residues; Jones et al. 1980; Krásný et al. 1998); (3) E. coli and B. stearothermophilus elongation factors and ribosomes are functionally interchangeable (Jonák et al. 1986 and references therein); and (4) the properties of the domains could be evaluated within full-length, three-domain proteins. Finally, we focused our attention on structural features that could lead to different thermal stabilities of both EF-Tus. Our results indicate that their thermostability is the result of cooperative interaction between the G-domains and domains 2 + 3 and provide insight into why the EF-Tu from B. stearothermophilus is more thermostable than the EF-Tu from E. coli.
Results
Recombinant EF-Tu proteins
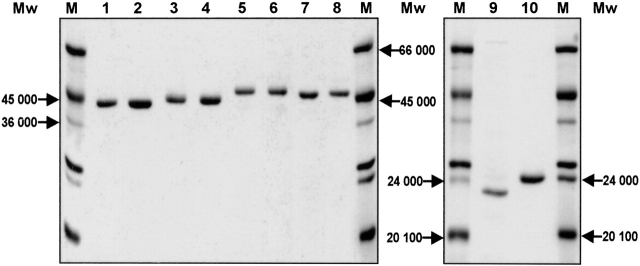
Ten proteins were prepared by the GST-purification technology: recombinant E. coli EF-Tu (EcEF-Tu); recombinant B. stearothermophilus EF-Tu (BstEF-Tu); chimera 1 (CH1, composed of Ecdomains 1 + 3 and Bstdomain 2); chimera 2 (CH2, composed of Ecdomains 1 + 2 and Bstdomain 3); chimera 3 (CH3, composed of Ecdomain 1 and Bstdomains 2 + 3); chimera 4 (CH4, composed of Bstdomains 1 + 3 and Ecdomain 2); chimera 5 (CH5, composed of Bstdomains 1 + 2 and Ecdomain 3); chimera 6 (CH6, composed of Bstdomain 1 and Ecdomains 2 + 3); the EcG-domain; and the BstG-domain. Each protein moved as a single band on SDS-electrophoresis gels (Fig. 1 ▶). Except for the EcG-domain, the proteins were 70%–100% active in binding GDP in the 1 : 1 molar ratio. EcG-domain preparations were only 7%–12% active irrespective of the two different methods of preparation described in Materials and Methods. All the data presented below apply to 100% active proteins.
Figure 1.
12% SDS-PAGE of isolated proteins (stained with Coomassie brilliant blue). EcEF-Tu (lane 1), CH1 (lane 2), CH2 (lane 3), CH3 (lane 4), BstEF-Tu (lane 5), CH4 (lane 6), CH5 (lane 7), CH6 (lane 8), EcG-domain (lane 9), and BstG-domain (lane 10). (M) Molecular weight protein markers.
EcEF-Tu, BstEF-Tu, and their chimeric variants differed in their electrophoretic mobility in 12% SDS-polyacrylamide gel (Fig. 1 ▶) even though, according to the amino acid composition, their Mrs are essentially identical. BstEF-Tu moved considerably more slowly than EcEF-Tu, indicating the Mr of BstEF-Tu to be ~7 kD higher than that of EcEF-Tu (see also Wittinghofer and Leberman 1976; Wormer et al. 1983; Jonák et al. 1986). Our experiments show that the mobility difference between EcEF-Tu and BstEF-Tu can already be traced down to their G-domains: (1) The isolated BstG-domain was markedly slower than the isolated EcG-domain even though the G-domains did not essentially differ in Mr or pI (data not shown); (2) chimeric EF-Tus comprising the BstG-domain were slower than those with the EcG-domain. Exchange of domains 3 but not domains 2 further modulated the mobility.
Interaction of EF-Tus, chimeric forms of elongation factors Tu, and isolated G-domains with GDP and GTP
The parameters governing the interaction between the proteins and GDP or GTP are summarized in Table 1. The Kd of the BstEF-Tu • GDP complex lay in the nanomolar range (4.17 nM) and the Kd of the BstEF-Tu • GTP complex in the submicromolar range (295 nM). They closely correlated with the Kds of the analogous complexes of EF-Tu from E. coli and so did the association and dissociation rates of the complexes of both organisms.
Table 1.
Kinetic parameters of interaction of E. coli and B. stearothermophilus EF-Tus, G-domains, and CH1–CH6 chimeric EF-Tus with GDP and GTP
| GDP (0°C) | GTP (0°C) | |||||
| Protein | k−1 (×104 sec−1) | k+1 (×104 M−1 sec−1) | Kd (nM) | k−1 (×104 sec−1) | k+1 (×10−4 M−1 sec−1) | Kd (nM) |
| Ec G-domain | 23.73 | 0.13 | 1825.38 | 58–115 | 0.09–0.18 | 5100–8000 |
| EC EF-Tu | 6.58 | 14.20 | 4.63 | 141.11 | 5.06 | 309.15 |
| CH1 | 7.37 | 43.12 | 1.71 | n.d. | n.d. | n.d. |
| CH2 | 2.37 | 29.94 | 0.79 | n.d. | n.d. | n.d. |
| CH3 | 6.97 | 37.43 | 1.86 | n.d. | n.d. | n.d. |
| Bst G-domain | 16.44 | 40.16 | 6.25 | 121.38 | 5.55 | 218.58 |
| Bst EF-Tu | 6.90 | 21.56 | 4.17 | 94.21 | 3.20 | 294.73 |
| CH4 | 3.42 | 17.98 | 1.90 | n.d. | n.d. | n.d. |
| CH5 | 6.25 | 17.94 | 3.48 | n.d. | n.d. | n.d. |
| CH6 | 2.14 | 26.22 | 0.82 | n.d. | n.d. | n.d. |
Reaction conditions are described in Materials and Methods. All results were obtained in several independent assays, and the values are the averages. Values in italics were calculated using k+1 = k−1/Kd. Values of k+1, k−1, and Kd for the E. coli G-domain • GTP complex were obtained from the references of Parmeggiani et al. (1987), Cetin et al. (1998), and Jensen et al. (1989). (n.d.) Not determined.
The kinetic parameters of the GDP complexes of chimeric EF-Tus indicate that the exchange of domains did not compromise the GDP-binding ability of the six new artificial proteins, chimeras CH1–CH6, as compared with EF-Tu of E. coli or B. stearothermophilus (Table 1). All chimeric EF-Tus were found to bind GDP with high affinity, with Kds in the nanomolar range (0.8–3.5 nM).
On the other hand, the kinetic parameters of the GDP and GTP complexes of the isolated EcG-domain, in particular, k+1 and Kd, strongly differed from those of the BstG-domain (Table 1). The BstG-domain bound GDP and GTP with the affinity almost as strong as that of BstEF-Tu. In contrast, the affinity of the EcG-domain for GDP and GTP was about three orders of magnitude and more than one order of magnitude lower, respectively, than those of EcEF-Tu (Table 1; Fasano et al. 1978; Jensen et al. 1989; Cetin et al. 1998). The difference in the affinity for GDP or GTP between the BstG-domain and the EcG-domain was mainly caused by the fact that the k+1s of the BstG-domain • GDP/GTP complexes were more than one to two orders of magnitude greater than those of the EcG-domain • GDP/GTP complexes.
Heat stability of GDP forms of the G-domains, EF-Tus, and their variants
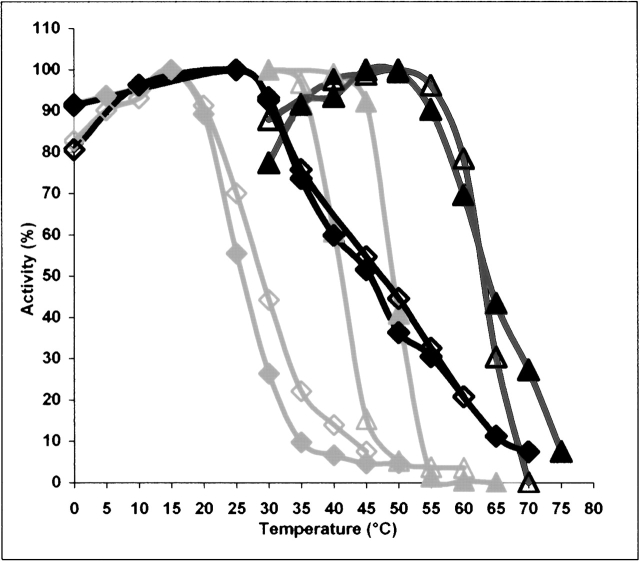
The heat inactivation profiles of all individual proteins were determined and their θ½s, the temperatures at which half of their maximal GDP binding activity was lost, are summarized in Table 2. According to the θ½ values, the BstG-domain (θ½ = 45.5°C) was ~20°C more thermostable than the EcG-domain (θ½ = 25.8°C) and ~18°C less thermostable than BstEF-Tu (θ½ = 63.8°C). The difference in θ½ between EcG-domain • GDP and EF-Tu • GDP (θ½ = 49.1°C) was ~23°C. With respect to θ½, the mesophilic EcEF-Tu in the GDP conformation was ~15°C less thermostable than the thermophilic BstEF-Tu • GDP (Fig. 2 ▶). The exchange of one noncatalytic domain of BstEF-Tu for the homologous Ecdomain, as in CH4 and CH5, was already sufficient to decrease the thermostability of its GDP form. In contrast, the substitution of one noncatalytic domain of EcEF-Tu • GDP for the homologous Bstdomain, as in CH1 and CH2, was not sufficient, contrary to expectation, to increase it, but actually decreased it. Only the exchange of both noncatalytic domains 2 + 3 for the homologous Bstdomains, as in CH3, resulted in a slight enhancement (2.7°C) of the thermostability as compared with EcEF-Tu • GDP.
Table 2.
Parameters characterizing the thermal stability of E. coli and B. stearothermophilus EF-Tus, G-domains, and CH1–CH6 chimeric EF-Tus
| Protein | θ1/2 (GDP) (°C) | θ1/2 (GTP) (°C) | Tm CD (GDP) (°C) | Tm CD (GTP) (°C) |
| Ec G-domain | 25.8 | 29 | n.d. | n.d. |
| Ec EF-Tu | 49.1 | 41.2 | 46.5 | 42.2 |
| CH1 (GEc − 2Bst-3Ec) | 47.8 | 42.6 | 46.2 | n.d. |
| CH2 (G +2Ec − 3Bst) | 47.8 | 39 | 45.5 | n.d. |
| CH3 (GEc-2 +3bst) | 51.8 | 51.6 | 46.9 | 47 |
| Bst G-domain | 45.5 | 47.3 | 42 | 44 |
| Bst EF-Tu | 63.8 | 63 | 57.5 | 58 |
| CH4 (GBst − 2Ec − 3Bst) | 62 | 58.2 | 57.1 | n.d. |
| CH5 (G +2Bst − 3Ec) | 55.2 | 55.2 | 54 | n.d. |
| CH6 (GBst-2 +3Ec) | 59 | 53.8 | 55.5 | 50.8 |
Reaction conditions are described in Materials and Methods. θ1/2 is the temperature of half-inactivation of the protein in GDP or GTP binding. Tm is the temperature of 50% change in the α-helix content of the protein in the GDP or GTP conformation. (n.d.) Not determined. All values are the means from at least three independent experiments.
Figure 2.
Heat inactivation profiles of GDP (closed symbols) and GTP (open symbols) forms of E. coli and B. stearothermophilus EF-Tus and G-domains. EcEF-Tu (gray ▴), EcG-domain (gray ♦), BstEF-Tu (black ▴), and BstG-domain (black ♦).
The thermostability profiles of the wild-type EF-Tus isolated from E. coli or B. stearothermophilus were essentially identical to the profiles of the recombinant EF-Tus (data not shown).
Heat stability of GTP forms of the G-domains, EF-Tus, and their variants
Conversion of the proteins from GDP to GTP conformation differently affected their heat stability. The θ½ of EcEF-Tu decreased by 8°C from 49.1°C to 41.2°C, whereas the θ½ of BstEF-Tu remained essentially unchanged. In contrast, both the EcG-domain and BstG-domain displayed a higher thermostability in the GTP conformation than in the GDP conformation (Fig. 2 ▶; Table 2). The replacement in EcEF-Tu of domains 2 + 3 by Bstdomains 2 + 3, as in CH3, protected the protein from the decrease in thermostability of its GTP form. Values of θ½ equal to 51.8°C for CH3 • GDP and 51.6°C for CH3 • GTP were obtained (Table 2). The replacement in BstEF-Tu of domains 2 + 3 by Ecdomains 2 + 3, as in CH6, had an opposite effect, rendering its thermostability sensitive to the type of the bound guanine nucleotide, similarly as in EcEF-Tu. The θ½ of CH6 • GTP was more than 5°C lower than the θ½ of CH6 • GDP (Table 2). The swapping of only one domain provided a pattern of results similar to that obtained with the proteins in the GDP conformation.
Unfolding of the G-domains, EF-Tus, and their variants by heat
To compare the functional thermostability profiles of individual proteins and their structural stability, preservation of the α-helix content of the proteins at increasing temperature was measured by CD spectroscopy.
As the first result, each product showed a CD spectrum of a protein with a defined structure. The CD spectra of chimeric EF-Tus matched those of BstEF-Tu or EcEF-Tu (data not shown), implying that the organization of the secondary structural elements in the chimeras was not dramatically changed. The negative CD at 220 nm, indicative of the presence of α-helices, was larger in G-domains than in the three-domain EF-Tu variants (Fig. 3 ▶). This difference is to be expected because the deletion of the middle and C-terminal domain removes parts of EF-Tu that contain only β-strands. Such secondary structure composition of domains 2 and 3 revealed by X-ray diffraction analysis for EcEF-Tu (Kjeldgaard and Nyborg 1992; Berchtold et al. 1993; Song et al. 1999) was also proposed to hold for domains 2 and 3 of BstEF-Tu (Krásný et al. 1998).
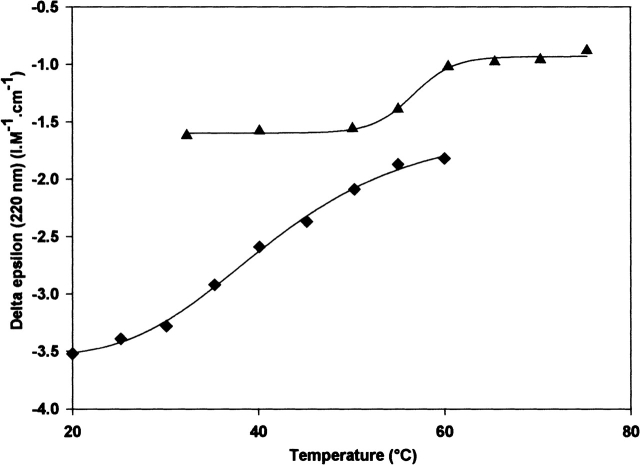
Figure 3.
The change in CD of BstEF-Tu (▴) and BstG-domain (♦) measured at 220 nm as a function of increasing temperature.
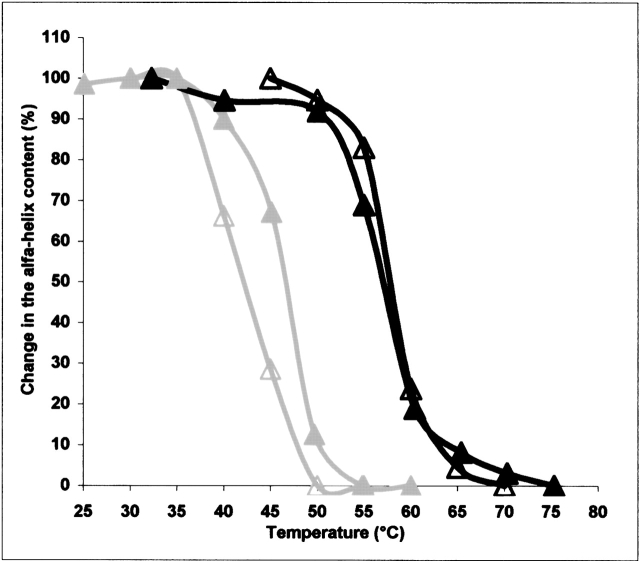
The CD spectrum of each protein displayed a characteristic temperature-induced transition change. The change (in percent) in the helicity of the proteins in GDP or GTP conformation detected at 220 nm was plotted against the temperature. The mid-temperatures (Tm) of the transition from the α-helical conformation to disordered conformation (unfolding) determined for every protein are summarized in Table 2. The Tm of the isolated BstG-domain • GDP was 42°C. Its conversion into the GTP conformation increased the Tm by 2°C. The CD spectrum of the EcG-domain could not be considered as representative because the concentration of the protein in an active conformation was low (see above). The Tms of GDP forms of the three-domain proteins comprising the BstG-domain (BstEF-Tu, chimeras CH4–CH6) were 12°–15.5°C higher than the Tm of the BstG-domain alone. They lay in the range of 54°–57.5°C. The Tms of the GDP forms of EcEF-Tu and chimeras CH1–CH3, comprising the EcG-domain, lay in the range of 45.5°–46.9°C (Table 2). The mid-temperature of α-helix unfolding of EcEF-Tu decreased by 4.3°C, to 42.2°C, when the protein in the GTP conformation was assayed. On the other hand, upon conversion of BstEF-Tu from the GDP to GTP conformation, an ~0.5°C increase in the Tm was observed (Fig. 4 ▶; Table 2). Swapping of domains 2 + 3 between both EF-Tus influenced sensitivity of the unfolding to the type of bound guanine nucleotide analogously as in the case of the functional thermostability. The Tm displayed by CH6 • GTP, composed of the BstG-domain and Ecdomains 2 + 3, was ~5°C lower than the Tm of CH6 • GDP. On the other hand, the Tm of CH3, consisting of the EcG-domain and Bst domains 2 + 3, was not influenced by bound guanine nucleotide. The thermal stability of α-helices in CH3 • GDP and CH3 • GTP was essentially identical (Table 2). The temperature-induced change in the helicity of the three-domain proteins was typically completed within a temperature range of ~15°C. However, an ~30°C temperature range was required to complete the denaturation of α-helices of the separated BstG-domain (Fig. 3 ▶). The figure also shows that the α-helix denaturation started in the isolated BstG-domain at a lower temperature than in the three-domain proteins.
Figure 4.
Temperature-dependent change in the α-helix content of GDP (closed symbols) and GTP (open symbols) forms of E. coli EF-Tu (gray lines) and B. stearothermophilus EF-Tu (black lines).
Discussion
Domains of EcEF-Tu and BstEF-Tu and guanine nucleotide-binding activity
All 10 recombinant proteins were active in the binding of GDP and GTP and GTP hydrolysis (data not shown). The affinity characteristics of the thermophilic BstEF-Tu for GDP and GTP, determined here for the first time, closely matched those of the mesophilic EcEF-Tu (Table 1; Arai et al. 1974; Miller and Weissbach 1977; Fasano et al. 1978; Sanangelantoni et al. 1996; Cetin et al. 1998). GDP was bound by either factor about 100 times more strongly than GTP, mainly as the consequence of about 10–20 times greater k−1 of EF-Tu • GTP complexes. This behavior has been considered typical of bacterial EF-Tus. As it is shown here, it holds for BstEF-Tu as well, although GTP, owing to the extra phosphate residue, has additional interactions with the proteins as compared with GDP (Berchtold et al. 1993; Kjeldgaard et al. 1993). No explanation in terms of the 3D structure for the different affinity of EF-Tus for GDP and GTP has been found as yet.
Comparison of the GDP- and GTP-binding properties of isolated G-domains provided different results. The BstG-domain possessed affinities for GDP and for GTP quite comparable with those of the intact BstEF-Tu. In contrast, the removal of domains 2 + 3 from EcEF-Tu resulted in a profound loss in the binding affinity for guanine nucleotides, particularly for GDP. The Kd increased almost 1000 times from the nanomolar range to the micromolar range (Table 1; Parmeggiani et al. 1987; Cetin et al. 1998). As reported from other laboratories, the affinity of the EcG-domain for GTP also decreased as compared with that of EcEF-Tu • GTP, but only about 20 times (Parmeggiani et al. 1987; Jensen et al. 1989; Cetin et al. 1998), so that the binding of GDP and GTP by the isolated EcG-domain took place with almost the same (low) affinity (Table 1).
These data imply that whereas in BstEF-Tu, the high and different affinity for GDP and GTP appears to be intrinsic to the G-domain itself, in E. coli, the cooperation involving all three domains of EF-Tu is required to establish this phenotype (cf. also Cetin et al. 1998). Nock et al. (1995) reported that in Thermus thermophilus EF-Tu, the deletion of domains 2/3 or only of domain 3 provoked a lower affinity for guanine nucleotides similarly as in the E. coli system. In contrast, the same truncation of Sulfolobus solfataricus EF-1α increased the affinity for GDP and GTP by about one order of magnitude compared with the intact protein (Masullo et al. 1997). This implies that the same level of affinity to guanine nucleotides can be attained by different strategies in different EF-Tus.
We did not check whether the separated G-domains mixed with domains 2/3 could form any functional complexes. It was demonstrated that a mixture of the G-domain and domains 2/3 of T. thermophilus EF-Tu could neither form any detectable complexes nor function in poly(U)-dependent poly(Phe) synthesis (Nock et al. 1995). This indicated that proper covalent linking of the three domains of EF-Tu is required for the formation of specific interdomain interactions found in the crystal structure of the protein and, in turn, for its functioning.
Thermostability of EF-Tu: Domain cooperation
Our studies with EcEF-Tu and BstEF-Tu and their domains indicate that the final level of thermostability of the three-domain EF-Tus is attained by a cooperative interaction between G-domains and domains 2 + 3. The G-domains set up a “basic level” of the thermal stability of the EF-Tus (cf. also Sanangelantoni et al. 1996; Masullo et al. 1997). The level set up by the thermophilic BstG-domain was ~20°C higher than that of the EcG-domain. In turn, this difference in θ½ between the G-domains fully correlated with the difference in θ½ between intact EF-Tus. In absolute terms, the θ½ of the G-domains were still ~20°C below those of the respective EF-Tus.
Both the EcG-domain and the BstG-domain were a little more thermostable in the GTP conformation than in the GDP conformation, which may be because of the extra interaction with the γ-phosphate residue of GTP (Kjeldgaard et al. 1993).
The domains 2 + 3 were found to enhance the thermal stability of the α-helical structure of the G-domain to the level consistent with the bacterial growth temperature optimum. The unfolding of the α-helical regions of the G-domain within the three-domain EF-Tu started at higher temperatures than in the G-domain alone (Fig. 3 ▶). In contrast to the G-domains, the gain in thermostability due to domains 2 + 3 indicated by θ½ was (1) similar, irrespective of the domain origin, and (2) smaller in the GTP complexes than in the GDP complexes. The conversion to the GTP conformation decreased particularly the θ½ and Tm of those proteins comprising Ecdomains 2 + 3 as was the case for EcEF-Tu and chimera CH6, composed of the BstG-domain and Ecdomains 2 + 3. The physiological consequences of the different stability of the GDP and GTP forms of E. coli EF-Tu have not been understood. Comparison of θ½ of GDP and GTP forms of the tested proteins revealed that the assumed tight interaction of either G-domain with Ecdomain 2 that takes place in the GTP conformation always resulted in a decrease of thermostability as compared with the GDP forms. The results provide evidence for the capacity of domain 2 to modulate the state of the G-domain. The mechanism of the modulation was indicated by the CD measurements. The changes in Tms of the proteins correlated with the changes of their θ½ even though the former were absolutely smaller (Table 2). (Values of θ½ and Tm cannot be compared directly because they were determined under different experimental conditions, e.g., incubation time; see Materials and Methods.) Our results are consistent with the view that (1) the integrity of the α-helical regions of the G-domain and the function of the protein are closely related; and (2) in the GTP conformation, Ecdomain 2 decreases the thermostability of the G-domain by affecting the stability of its α-helical regions and/or by affecting the stabilizing interactions of the α-helical regions with nonhelical regions of the protein. The substitution of Cys 81 by Gly in EcEF-Tu was shown to abolish this GTP-sensitive phenotype (Anborgh et al. 1992), similarly as in EF-1α, which naturally carries Ala instead of Cys at this position (Nagata et al. 1976). Thus, Cys 81 might be involved in the transmission pathway of a signal from domain 2.
In contrast to EcEF-Tu, the thermostability of BstEF-Tu and CH3, composed of the EcG-domain and Bst domains 2 + 3, was not a function of the guanine nucleotide state, and the stabilizing effect of Bst domains 2 + 3 was not sensitive to the GDP/GTP exchange. All these results indicate that differences exist between Bst and Ec domains 2 in the way they contact and, in turn, modulate (the function of) the G-domain.
The opposing effects of Ec and Bstdomains 2 + 3 on the thermostability in response to GTP also became clearly apparent in experiments aimed at the determination of the GTPase temperature optimum of each protein. The optimum of CH3 was ~10°C higher than that of EcEF-Tu, and vice versa, the GTPase of CH6 had an ~9°C lower temperature optimum than BstEF-Tu (data not shown). Such intermediate thermostability was already reported for a construct composed of the G-domain of archeal S. solfataricus and domains 2 + 3 of E. coli (Arcari et al. 1999).
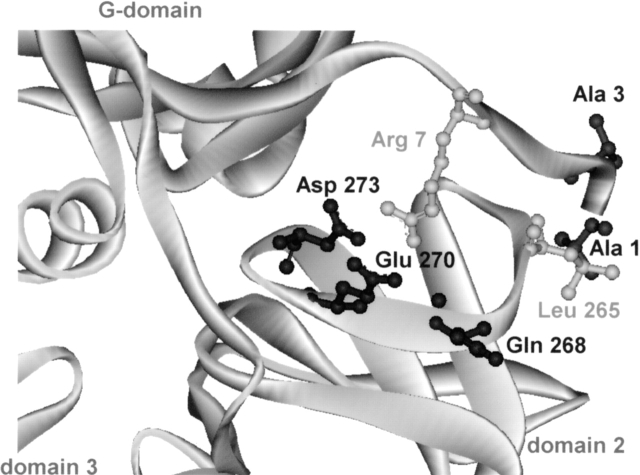
The crystal structures of BstEF-Tu • GTP and EcEF-Tu • GTP could help in the identification of interactions involved in the modulation of stability of the G-domain by domain 2. However, in the absence of these crystal structures, a mere correlation between amino acid residues of the G-domain–domain-2 interface, distant up to 4.0 Å and thus likely to be involved in domain interaction in T. thermophilus (Tt) EF-Tu • GPPNP (Berchtold et al. 1993), and amino acid residues occurring at homologous positions in the G-domain and domain 2 of Ec and Bst EF-Tus was established. The correlation indicates that 21 amino acid residues of the Bst- or EcG-domains might contact, usually more than once, 16 residues in the respective domain 2, and they are, in most cases, identical with amino acid residues of TtEF-Tu • GPPNP. Only in five cases (Fig. 5 ▶) did the amino acid residues of these pairs differ between Ec and Bst. These pairs were in Ec/Bst: (1) Ser1/Ala1–Leu264/Leu265; (2) Glu3/Ala3–Leu264/Leu265; (3) Arg7/Arg7–Glu272/Asp273; (4) Arg7/Arg7–Glu267/Gln268; and (5) Arg7/Arg7–Arg269/Glu270. Three of the five pairs listed above involve amino acid residues different (underlined) from those detected in homologous positions of TtEF-Tu • GPPNP. Therefore, their pairing in Bst- or EcEF-Tus is not certain. The remaining two pairs, Arg7–Glu267 (Ec) and Arg7–Asp273 (Bst), also exist in the T. thermophilus protein. In Bst, Glu 267 is replaced by Gln 268, and in Ec, Asp 273 is replaced by Glu 272. Thus, at present, consideration of only these structural differences between Ec and Bst domain 2 interfaces, as relevant to different contact properties of Ec and Bstdomain 2, might be justified. Altogether, Ecdomain 2 (residues 200–295) and Bstdomain 2 exhibit ~69% identity in the amino acid sequence, which corresponds to 30 different substitutions. Only three of them are situated at the interface with domain 1 (see above).
Figure 5.
Differences in G-domain–domain 2 interactions between B. stearothermophilus and E. coli EF-Tus. A homology model of the G-domain–domain 2 interface of B. stearothermophilus EF-Tu in GTP conformation. Amino acid residues different from E. coli EF-Tu are shown in dark gray; identical residues, in light gray. This figure was made using DS ViewerPro program.
Basis for the difference in E. coli and B. stearothermophilus G-domain stability
Differences in ΔGstab as small as 3–6.5 kcal/mole were reported to account for thermostability increases between 10°C and 20°C. This indicates that, in principle, only few changes in the noncovalent forces responsible for ΔGstab can distinguish mesophilic from thermophilic proteins (for review, see Vieille and Zeikus 2001).
In the absence of the 3D structure of BstEF-Tu, its amino acid sequence was aligned with that of EcEF-Tu to look for the features known to stabilize proteins (Cambillau and Claverie 2000; Vieille and Zeikus 2001).
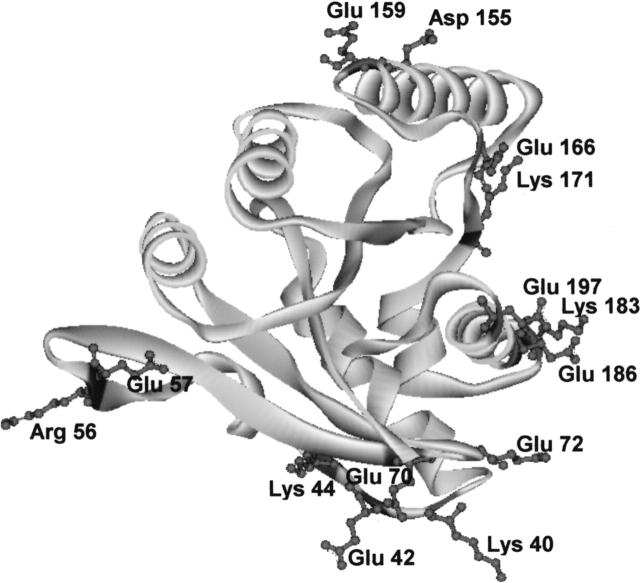
There are 97 differences in the amino acid structure between both proteins, or the sequence identity is ~75%. As a whole, BstEF-Tu contains three negatively charged residues more and four positively charged residues less than EcEF-Tu. This corresponds with a lower isoelectric point of 4.91 for BstEF-Tu as compared with 5.3 for EcEF-Tu. As shown in our experiments, the G-domains were mainly responsible for the difference in the thermostability between both EF-Tus. However, the G-domains differ from each other at only 38 amino acid positions and are the most similar (81% identity) of all three domains. Nevertheless, the structural features that could lead to different thermal stabilization of the G-domains are prominent. Among 38 amino acid differences, the ratio between charged versus polar, uncharged amino acid residues is 8/11 in the EcG-domain and 14/6 in the BstG-domain. The difference (in percent) between charged and polar uncharged amino acids (CvP bias), which has been reported to be the best indicator of the organism’s lifestyle (Cambillau and Claverie 2000; Suhre and Claverie 2003), clearly classifies the BstG-domain (CvP value = 15.1%) as much more thermostable than the EcG-domain (CvP value = 9.5%). According to Cambillau and Claverie plots, the BstG-domain and the EcG-domain “would belong” to the class of hyperthermophiles and moderate thermophiles, respectively. Although this classification provided exaggerated results in absolute terms (see below), the difference in CvP values between the G-domains is fully proportional to the difference (~20°C) in thermostability between mesophilic and thermophilic classes of microorganisms. Among amino acid substitutions that occurred between the EcG-domain and the BstG-domain, there were seven replacements by new charged amino acid residues (Gly40Lys, Ala42Glu, Ala57Glu, Pro72Glu, Gln159Glu, Ala186Glu, and Ser197Glu) and seven substitutions of one charged amino acid for another one (Arg44Lys, Lys56Arg, Asp70Glu, Glu155Asp, Asp166Glu, Arg171Lys, and Glu183Lys). The distribution of the charged residues of the BstG-domain different from those present in the EcG-domain is shown on a computer-generated homology model of the BstG-domain (Fig. 6 ▶). Most of the replacements by charged residues occurred at the surface of one side of the G-domain, which does not get into contact with domains 2 and 3. The presence of solvent-accessible charged residues at the surface is considered to be a characteristic feature of (hyper)thermophilic proteins because it may allow protein stabilization through ion bonds (Cambillau and Claverie 2000; Suhre and Claverie 2003). The Glu183Lys and Ala186Glu changes are the most likely example of an introduction of a new ion pair as i, i + 3 spacing along an α-helix is consistent with ion pair formation. The same number of charged amino acid residues as in the BstG-domain was found in the G-domain of thermophilic EF-Tu from T. thermophilus, and the G-domain of another thermophilic EF-Tu from Thermus aquaticus had one additional charged residue.
Figure 6.
Charged amino acid residues in the G-domain of EF-Tu of B. stearothermophilus newly introduced or different from those in the G-domain of EF-Tu of E. coli. This figure was made using DS ViewerPro program.
When the entire molecules of the EF-Tus were subjected to the CvP-bias analysis, the values 13.0% for EcEF-Tu and 11.7% for BstEF-Tu were obtained. This was surprising because the reported CvP-bias values obtained by the whole-genome analysis of E. coli and B. stearothermophilus were 2.63% and 9.05%, respectively (Suhre and Claverie 2003), thus fully consistent with the mesophilic character of E. coli and the moderately thermophilic character of B. stearothermophilus. The discrepancy between the EF-Tu and the whole-genome CvP values indicates that both EF-Tu proteins, and EcEF-Tu in particular, significantly differ in the CvP-bias features from the majority of cellular proteins and indicates that other strategies, besides the increased ion-pair formation in the G-domain of BstEF-Tu, come into play in the thermostabilization process of the EF-Tu proteins. The CvP value of the mesophilic EcEF-Tu being higher than that of the thermophilic BstEF-Tu adds to the recent observation of Suhre and Claviere (2003), demonstrating that the strict correspondence between the highest CvP bias and the highest optimal growth temperature breaks down below 80°C. Analysis of CvP features of five more members of the EF-Tu family (B. subtilis, T. aquaticus, T. thermophilus, S. solfataricus, and Thermotoga maritima) provided no simple results either. They will be described elsewhere (H. Šanderová and J. Jonák, in prep.).
B. stearothermophilus is a moderate thermophile, and it has been proposed that in these organisms, the hydrophobic interactions may be greatly involved in the adaptation to high temperatures (Szilagyi and Zavodszky 2000). Indeed, the calculated nonpolar solvent-accessible surface area (Hubbard and Thornton 1993) of the BstG-domain molecule, upon folding, was found to be ~4% smaller than that of EcG-domain • GDP. A similar difference was observed between domains 2 and between domains 3 of both EF-Tus, and the nonpolar solvent-accessible surface area of BstEF-Tu • GDP was calculated to be ~5% smaller than that of EcEF-Tu • GDP. Thus, in contrast to the CvP results, a decrease in nonpolar solvent-accessible surface area was detected to occur throughout the entire BstEF-Tu, in all three domains. This implies that an increase in hydrophobic interactions could be an essential reason for the higher thermostability of BstEF-Tu in comparison to EcEF-Tu. Comparative studies between Ec- and BstG-domains revealed that hydrophobic substitutions Val14Ile and Ser65Ala occurred in the core of the BstG-domain. To directly address a possible role of these substitutions in the stabilization of the G-domains, the residues were mutated and the thermostability of the mutated proteins was determined. Four mutants were prepared and examined: BstG-domain Ile14Val, BstG-domain Ala65Ser, EcG-domain Val14Ile, and EcG-domain Ser65Ala. The Ec mutants in the GDP conformation displayed about the same θ½, and in the GTP conformation, the Val14Ile mutant had θ½ ~2°C higher than the wild-type (wt) EcG-domain. The Bst mutants displayed ~4°–6°C lower θ½ in both conformations than the wild-type form of the domain (data not shown). These preliminary experiments at least partially support the involvement of hydrophobic interactions in the stabilization of the G-domains.
The decrease in nonpolar solvent-accessible surface area and a 4% increase in polar solvent-accessible surface area (data not shown) might also be consistent with the observed better solubility of the BstG-domain as compared with the EcG-domain. Whether the change in hydrophobic burial also relates to the slower electrophoretic mobility of BstG-domain/BstEF-Tu in comparison with EcG-domain/EcEF-Tu (Fig. 1 ▶) remains to be elucidated.
EF-Tu proteins are unique from the point of view of the division into two moieties, the G-domain and domains 2 + 3, fully unrelated both functionally and structurally. The thermostability of either moiety is considerably lower than the optimum growth temperature of the respective organisms, and the level of thermostability, consistent with it, can only be achieved by the interaction of both moieties in a cooperative way. To elucidate this mechanism, structural–functional studies on more EF-Tus are required to reveal dynamic aspects of the interdomain contacts that have not yet been explained by the static 3D models to date.
Materials and methods
Materials and reagents
The Expand High Fidelity PCR System was purchased from Roche Molecular Biochemicals. The pGEX-5X-3 expression vector, Glutathione Sepharose 4B, reduced glutathione, factor Xa, and [3H]GDP (10 Ci/mmole) were purchased from Amersham Pharmacia Biotech. Phosphoenolpyruvate was obtained from Sigma. 2-Mercaptoethanol, phenylmethyl sulphonylfluoride (PMSF), GDP (Na-salt), and GTP (Na-salt) were from Serva. [γ-32P]GTP (5000 Ci/mmole) was provided by ICN or Lacomed, and pyruvate kinase was purchased from Calbiochem (Merck). Nitrocellulose membrane filters (HAWP, 0.45 μm; BA85, 0.45 μm) were obtained from Millipore and Schleicher & Schuell Biosciences (Marketing Consulting), respectively. ChromaSpin TE-10 columns were purchased from Clontech (I.T.A.-Intertact).
Bacterial strains and plasmids
Plasmid pEMBL/tufA containing the E. coli tufA gene (Parmeggiani et al. 1987; Jensen et al. 1989) and chromosomal DNA of B. stearothermophilus strain CCM 2184 were used as templates for preparation of recombinant and chimeric genes. E. coli strain DH5α was used for cloning procedures, and E. coli strain BL21 and expression plasmid pGEX-5X-3 were used for overproduction of recombinant proteins.
Construction of recombinant and chimeric genes
The primers 5′-CGGGATCCCCTCTAAAGAAAAATTTGAAC GTAC-3′ (EcG, forward primer for the E. coli G-domain), 5′-CGAATTCTTAGCCCAGAACTTTAGCAACA-3′ (EcD3, reverse primer for E. coli domain 3), and 5′-CGGGATCCCCGC TAAAGCGAAATTTGAGCG-3′ (BstG), and 5′-CGAATTCT TACTCGATGATTTCCGATACG-3′ (BstD3) were designed for the amplification of E. coli and B. stearothermophilus tuf genes for overexpression of recombinant EF-Tu proteins (the flanking regions in italics contain recognition sites of BamHI and EcoRI restriction enzymes). The tufA gene of E. coli carried by the plasmid pEMBL/tufA (Parmeggiani et al. 1987; Jensen et al. 1989) was used for the preparation, by PCR reaction, of gene fragments coding for the E. coli G-domain (EcG-domain, amino acids residues 1–199), Ecdomain 2 (amino acids residues 200–295), Ecdomain 3 (amino acids residues 296–393), as well as the fragments coding for the EcG-domain and Ecdomain 2 (amino acids residues 1–295) and Ecdomains 2 + 3 (amino acids residues 200–393). The chromosomal tuf gene of B. stearothermophilus was used for the preparation, by PCR reaction, of the gene fragments coding for the B. stearothermophilus G-domain (BstG-domain, amino acids residues 1–199), Bstdomain 2 (amino acids residues 200–296), Bstdomain 3 (amino acids residues 297–394), as well as the gene fragments coding for the BstG-domain and Bstdomain 2 (amino acids residues 1–296) and Bstdomains 2 + 3 (amino acids residues 200–394). A further eight synthetic oligonucleotides were designed to synthesize the above fragments of the tufA gene of E. coli (Ec) and the tuf gene of B. stearothermophilus (Bst) for construction of chimeric genes: BstG/EcD2, 5′-GCGGTTGATGAG TACATCCCGGAACCAGAGCGTGCGAT-3′ (the flanking region in italics represents the 3′ part of the BstG-domain, and the second part codes for the 5′ part of Ecdomain 2); EcD2/BstG, 5′-CGCACGCTCTGGTTCCGGGATGTACTCATCAACCGCG T-3′; BstD2/EcG, 5′-TTCACGTTGCGGAGTCGGAATATAA GAATCCAGGAAGCC-3′; EcG/BstD2, 5′-TTCCTGGATTCT TATATTCCGACTCCGCAACGTGAAGT-3′; BstD2/EcD3, 5′-CAAGTATTGGCAAAACCGGGCACCATCAAGCCGCACAC-3′; EcD3/BstD2, 5′-GTGCGGCTTGATGGTGCCCGGTTTTGCCAA TACTTGGC-3′; EcD2/BstD3, 5′-CAGGTACTGGCTAAGCCGG GCTCAATCACGCCGCATAC-3′; and BstD3/EcD2, 5-′ATGCGGCGTGATTGAGCCCGGCTTAGCCAGTACCTGAC-3′.
The 5′ flanking region (18 nt) of these primers coding for the 3′-end or 5′-beginning of the preceding or following domain, respectively, from the “opposite” gene allowed junction of individual domains to form a template for amplification of chimeric genes. By this procedure, six chimeric forms of EF-Tu, representing all possible combinations of protein domains of EF-Tu from both organisms, were prepared. In addition, free recombinant G-domains of EF-Tu of both organisms were also prepared by overexpression, from the PCR product synthesized using primers 5′-CGGGATCCCCTCTAAAGAAAAATTTGAACGTAC-3′ (EcG), 5′-CGAATTCTTAAATATAAGAATCCAGGAAGCC-3′ (for EcG-domain), 5′-CGGGATCCCCGCTAAAGCGAAATTT GAGCG-3′ (BstG), and 5′-CGAATTCTTAGATGTACTCATCA ACCGCGT-3′ (for the BstG-domain). Using the BamHI and EcoRI restriction sites introduced by primers, the PCR products were cloned in the expression vector pGEX-5X-3 downstream of the recognition site of the serine protease factor Xa. The nucleotide structures of cloned genes were verified by sequencing double-stranded DNA in the ABI PRISM 310 Genetic Analyser. All proteins were produced without N-terminal methionine and with natural stop codons. Because of the cloning in the pGEX vector polylinker and the fusion protein cleavage by factor Xa, the isolated proteins contained three additional N-terminal residues (Gly, Ile, Pro).
Expression and purification of proteins
All recombinant proteins fused with Glutathione S-transferase (GST) were expressed in E. coli strain BL21 and separated from the native E. coli EF-Tu by affinity chromatography on Glutathione Sepharose 4B using the GST (glutathione-S-transferase) technology. Untagged, GST-free products were obtained after cleavage of the fused proteins with factor Xa, essentially as described in Tomincová et al. (2002).
The GST-G-domain of E. coli was isolated by two methods. The soluble fusion protein was purified according to Tomincová et al. (2002) with the following modifications: All buffers contained 10% glycerol; a 100,000g supernatant of sonicated cell crude extract was used, and binding of the fusion protein to Glutathione Sepharose 4B was performed on ice for 40 min.
The insoluble fusion protein fraction in inclusion bodies was purified under urea denaturation conditions, essentially as described in Sambrook and Russell (1989).
The concentration of proteins was determined by the procedure of Bradford (1976) using bovine serum albumin as a standard. The concentration of biologically active protein (specific activity) was determined by measuring the maximum binding of [3H]GDP at optimal temperature (Miller and Weissbach 1977). The purity was examined by SDS-PAGE (Laemmli 1970).
Formation of GTP complexes
To obtain proteins in the GTP form, the protein • GDP complex was incubated at 30°C for 20 min (G-domains at 15°C for 8 min) with 4.5 mM phosphoenolpyruvate and 34 mg/L pyruvate kinase. The manufactured GTP and [γ-32P]GTP were incubated under the same conditions to eliminate any GDP contamination (Anborgh et al. 1992).
Preparation of nucleotide-free protein
Nucleotide-free proteins were prepared according to Knudsen et al. (1995) using ChromaSpin TE-10 spin columns. Concentration of the active protein in the resulting eluate was determined using the nucleotide binding assay.
Determination of the association and dissociation rate constant and equilibrium dissociation constant of the protein • GDP/GTP complexes
The association rate constant k+1 of the [3H]GDP • protein or [3H]GTP • protein complexes was determined by incubating the nucleotide-free protein (5 pmole) and [3H]GDP (5 pmole, specific activity 6000 cpm/pmole) or [3H]GTP (100 pmole, specific activity 8000 cpm/pmole) in 0.5 mL of buffer A (50 mM Tris-Cl at pH 7.6, 10 mM MgCl2, 60 mM NH4Cl, 1 mM DTT) on ice, as described (Fasano et al. 1978; Knudsen et al. 1995). Aliquots (100 μL) were withdrawn at 15, 30, 45, 60, and 75 sec and filtered through a nitrocellulose filter, and the filters were washed with 3 mL of ice-cold buffer A. The results were analyzed according to the second-order rate equation [1/(b − a)] • ln[a(b − x)/b(a − x)] = k+1t, where a is the initial concentration of [3H]GDP/GTP, b the initial concentration of protein, and x the concentration of [3H]GDP • protein or [3H]GTP • protein complexes at time t.
The determination of the dissociation rate constant, k−1, was carried out by incubating 50 pmole of preformed binary complexes of proteins prepared by preincubation with [3H]GDP (500 pmole, specific activity 800 cpm/pmole) or [3H]GTP (5000 pmole, specific activity 4800 cpm/pmole) in 0.5 mL of buffer A on ice for 90 min, essentially as described (Knudsen et al. 1995; Laurberg et al. 1998); the dissociation reaction was started by adding a 1000-fold or 100-fold molar excess of unlabeled GDP or GTP, respectively, in 0.5 mL of buffer A and followed kinetically. Aliquots (100 μL) were withdrawn at regular time intervals and filtered through a nitrocellulose filter; the filters were washed with 3 mL of ice-cold buffer A. The k−1 was calculated according to the first-order rate equation ln(ct/c0) = −k−1t, where c0 is the initial concentration of preformed binary complexes and ct the concentration at different times t.
The apparent equilibrium dissociation constant Kd of the protein • GDP/GTP complexes was determined (Anborgh et al. 1992) by incubating 2 pmole of nucleotide-free protein with 0.5–15 pmole of [3H]GDP (specific activity 2500 cpm/pmole) in 1 mL of buffer A or incubating 50 pmole of nucleotide-free protein with 5–500 pmole of [3H]GTP (specific activity 300–8500 cpm/pmole) in 100 μL of buffer A on ice for 60 min. Then 1 mL or 100 μL from the [3H]GDP or [3H]GTP mixture, respectively, was filtered through a nitrocellulose filter, and the filters were washed as described above. The dissociation constants were calculated using Scatchard plot according to the equation r/[nucleotide]free = 1/Kd(n − r), where r is the number of moles of nucleotide bound per mole of protein and n is the number of binding sites (Créchet and Parmeggiani 1986).
Heat stability test
For the heat stability test (Anborgh et al. 1992), protein • GDP or protein • GTP complexes (23 pmole) were incubated in the presence of 230 pmole of [3H]GDP (specific activity 750 cpm/pmole) or 230 pmole of [γ-32P]GTP (920 pmole in the experiments with EcG-domain; specific activity 2000 cpm/pmole), respectively, in 40 μL of buffer B (50 mM Tris-Cl at pH 7.6; 60 mM NH4Cl, 10 mM MgCl2, 10 mM 2-mercaptoethanol) in the range of 0°–75°C for 8 min, then cooled on ice. Aliquots of 30 μL were spotted on nitrocellulose filters, which were washed three times with 2 mL of cold buffer B. Filters were dried, and the amount of bound nucleotide was measured in a scintillation counter. The temperature-dependent profile of the residual nucleotide binding activity of each protein was determined at least three times.
The circular dichroism (CD) measurement
Measurements were carried out using a JOBIN-YVON CD6 spectrometer in the range of temperatures 10°–75°C. The protein concentration in buffer B was 2.9 μM. Sample cells with 1 mm pathlengths were used. The spectra were accumulated three times at a given temperature. The scanning rate was ~0.3°C/min. The values of molar ellipticity obtained at wavelength 220 nm were plotted against temperature.
Calculations
Solvent-accessible surface areas were calculated using the program NACCESS (Hubbard and Thornton 1993). The default values were used (probe radius, 1.4 Å, z-slices, 0.05 Å, van der Waals radii) in the calculations.
Acknowledgments
This work was supported by grants 204/98/0863 and 303/02/0689 from the Grant Agency of the Czech Republic (to J.J.). The plasmid pEMBL/tufA was kindly supplied by A. Parmeggiani. We thank R. Hilgenfeld for providing us with data on amino acid residues situated at the G-domain–domain 2 interface of T. thermophilus EF-Tu • GPPNP; P. Řezáčová for computer facility to calculate solvent-accessible surface areas; and L. Výborná, K. Zemanová, and H. Dlouhá for skillful technical assistance. The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
EF-Tu, elongation factor Tu
EF-1α, eukaryotic elongation factor 1α
GST, glutathione S-transferase
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03272504.
References
- Anborgh, P.H., Parmeggiani, A., and Jonák, J. 1992. Site-directed mutagenesis of elongation factor Tu. The functional and structural role of residue Cys 81. Eur. J. Biochem. 208 251–257. [DOI] [PubMed] [Google Scholar]
- Arai, K., Kawakita, M., and Kaziro, Y. 1974. Studies on the polypeptide elongation factors from E. coli. V. Properties of various complexes containing EF-Tu and EF-Ts. J. Biochem. (Tokyo) 76 293–306. [DOI] [PubMed] [Google Scholar]
- Arcari, P., Masullo, M., Arcucci, A., Ianniciello, G., de Paola, B., and Bocchini, V. 1999. A chimeric elongation factor containing the putative guanine nucleotide binding domain of archeal EF-1α and the M and C domains of eubacterial EF-Tu. Biochemistry 38 12288–12295. [DOI] [PubMed] [Google Scholar]
- Baldauf, S.L., Palmer, J.D., and Doolitle, W.F. 1996. The root of the universal three and the origin of eukaryotes based on elongation factor phylogeny. Proc. Natl. Acad. Sci. 93 7749–7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold, H., Reshetnikova, L., Reiser, C.O.A., Schirmer, N.K., Sprinzl, M., and Hilgenfeld, R. 1993. Crystal structure of active elongation factor Tu reveals major domain rearrangement. Nature 365 126–132. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Cambillau, C. and Claverie, J.M. 2000. Structural and genomic correlates of hyperthermostability. J. Biol. Chem. 275 32383–32386. [DOI] [PubMed] [Google Scholar]
- Cetin, R., Anborgh, P.H., Cool, R.H., and Parmeggiani, A. 1998. Functional role of the noncatalytic domains of elongation factor Tu in the interactions with ligands. Biochemistry 37 486–495. [DOI] [PubMed] [Google Scholar]
- Créchet, J.B. and Parmeggiani, A. 1986. Characterization of the elongation factors from calf brain. 3. Properties of the GTPase activity of EF-1 α and mode of action of kirromycin. Eur. J. Biochem. 161 647–653. [DOI] [PubMed] [Google Scholar]
- Fasano, O., Bruns, W., Crechet, J.B., Sander, G., and Parmeggiani, A. 1978. Modification of elongation-factor-Tu • guanine-nucleotide interaction by kirromycin. A comparison with the effect of aminoacyl-tRNA and elongation factor Ts. Eur. J. Biochem. 89 557–565. [DOI] [PubMed] [Google Scholar]
- Hubbard, S.J. and Thornton, J.M. 1993. NACCESS computer program. Department of Biochemistry and Molecular Biology, University College, London.
- Jaenicke, R. and Böhm, G. 1998. The stability of proteins in extreme environments. Curr. Opin. Struct. Biol. 8 738–748. [DOI] [PubMed] [Google Scholar]
- Jensen, M., Cool, R.H., Mortensen, K.K., Clark, B.F.C., and Parmeggiani, A. 1989. Structure–function relationships of elongation factor Tu. Isolation and activity of the guanine-nucleotide-binding domain. Eur. J. Biochem. 182 247–255. [DOI] [PubMed] [Google Scholar]
- Jonák, J. and Rychlík, I. 1973. Study of conditions for the inhibitory effect of N-tosyl-L-phenylalanyl chloromethane on protein synthesis and the possibility of existence of different forms of the elongation factor S3 in Bacillus stearothermophilus. Biochim. Biophys. Acta 324 554–562. [DOI] [PubMed] [Google Scholar]
- Jonák, J., Smrt, J., Holý, A., and Rychlík, I. 1980. Interaction of Escherichia coli EF-Tu • GTP and EF-Tu • GDP with analogues of the 3′ terminus of aminoacyl-tRNA. Eur. J. Biochem. 105 315–320. [DOI] [PubMed] [Google Scholar]
- Jonák, J., Pokorná, K., Meloun, B., and Karas, K. 1986. Structural homology between elongation factors EF-Tu from Bacillus stearothermophilus and Escherichia coli in the binding site for aminoacyl-tRNA. Eur. J. Biochem. 154 355–362. [DOI] [PubMed] [Google Scholar]
- Jones, M.D., Petersen, T.E., Nielsen, K.M., Magnusson, S., Sottrup-Jensen, L., Gausing, K., and Clark, B.F.C. 1980. The complete amino-acid sequence of elongation factor Tu from Escherichia coli. Eur. J. Biochem. 108 507–526. [DOI] [PubMed] [Google Scholar]
- Kaziro, Y. 1978. The role of guanosine 5-triphosphate in polypeptide chain elongation. Biochim. Biophys. Acta 505 95–127. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard, M. and Nyborg, J. 1992. Refined structure of elongation factor EF-Tu from Escherichia coli. J. Mol. Biol. 223 721–742. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard, M., Nissen, P., Thirup, S., and Nyborg, J. 1993. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure 1 35–50. [DOI] [PubMed] [Google Scholar]
- Knudsen, C.R., Kjaersgard, I.V., Wiborg, O., and Clark, B.F. 1995. Mutation of the conserved Gly94 and Gly126 in elongation factor Tu from Escherichia coli. Elucidation of their structural and functional roles. Eur. J. Biochem. 228 176–183. [PubMed] [Google Scholar]
- Krab, J.M. and Parmeggiani, A. 1998. EF-Tu, a GTPase odyssey. Biochim. Biophys. Acta 1443 1–22. [DOI] [PubMed] [Google Scholar]
- Krásný, L., Mesters, J.R., Tieleman, L.N., Kraal, B., Fučík, V., Hilgenfeld, R., and Jonák, J. 1998. Structure and expression of elongation factor Tu from Bacillus stearothermophilus. J. Mol. Biol. 283 371–381. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Laurberg, M., Mansilla, F., Clark, B.F., and Knudsen, C.R. 1998. Investigation of functional aspects of the N-terminal region of elongation factor Tu from Escherichia coli using a protein engineering approach. J. Biol. Chem. 273 4387–4391. [DOI] [PubMed] [Google Scholar]
- Masullo, M., Ianniciello, G., Arcari, P., and Bocchini, V. 1997. Properties of truncated forms of the elongation factor 1α from the archaeon Sulfolobus solfataricus. Eur. J. Biochem. 243 468–473. [DOI] [PubMed] [Google Scholar]
- Miller, D.L. and Weissbach, H. 1977. Factors involved in the transfer of aminoacyl-tRNA to the ribosome. In Molecular mechanisms in protein biosynthesis (eds. H. Weissbach and S. Pestka), pp. 323–373. Academic Press, New York.
- Nagata, S., Iwasaki, K., and Kaziro, Y. 1976. Interaction of the low molecular weight form of elongation factor 1 with guanine nucleotides and aminoacyl-tRNA. Arch. Biochem. Biophys. 172 168–177. [DOI] [PubMed] [Google Scholar]
- Nock, S., Grillenbeck, N., Ahmadian, M.R., Ribeiro, S., Kreutzer, R., and Sprinzl, M. 1995. Properties of isolated domains of the elongation factor Tu from Thermus thermophilus HB8. Eur. J. Biochem. 234 132–139. [DOI] [PubMed] [Google Scholar]
- Parmeggiani, A., Swart, G.W.M., Mortensen, K.K., Jensen, M., Clark, B.F.C., Dente, L., and Cortese, R. 1987. Properties of genetically engineered G domain of elongation factor Tu. Proc. Natl. Acad. Sci. 84 3141–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Printz, M.P. and Miller, D.L. 1973. Evidence for conformational changes in elongation factor Tu induced by GTP and GDP. Biochem. Biophys. Res. Commun. 53 149–156. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 1989. Molecular cloning: A laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sanangelantoni, A.M., Cammarano, P., and Tiboni, O. 1996. Manipulation of the tuf gene provides clues to the localization of sequence element(s) involved in the thermal stability of Thermotoga maritima elongation factor Tu. Microbiology 142 2525–2532. [DOI] [PubMed] [Google Scholar]
- Song, H., Parsons, M.R., Rowsell, S., Leonard, G., and Phillips, S.E.V. 1999. Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 Å resolution. J. Mol. Biol. 285 1245–1256. [DOI] [PubMed] [Google Scholar]
- Suhre, K. and Claverie, J.M. 2003. Genomic correlates of hyperthermostability, an update. J. Biol. Chem. 278 17198–17202. [DOI] [PubMed] [Google Scholar]
- Szilagyi, A. and Zavodszky, P. 2000. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: Results of a comprehensive survey. Structure Fold. Des. 8 493–504. [DOI] [PubMed] [Google Scholar]
- Tomincová, H., Krásný, L., and Jonák, J. 2002. Isolation of chimaeric forms of elongation factor EF-Tu by affinity chromatography. J. Chromat. B 770 129–135. [DOI] [PubMed] [Google Scholar]
- Vieille, C. and Zeikus, G.J. 2001. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65 1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittinghofer, A. and Leberman, R. 1976. Elongation factor T from Bacillus stearothermophilus and Escherichia coli. Purification and some properties of EF-Tu and EF-Ts from Bacillus stearothermophilus. Eur. J. Biochem. 62 373–382. [DOI] [PubMed] [Google Scholar]
- Wormer, W., Glöckner, C., Mierzowski, M., and Wolf, H. 1983. On heterogenity of elongation factor Tu among eubacteria. FEMS Microbiol. Lett. 18 69–73. [Google Scholar]