Abstract
The HNHc (SMART ID: SM00507) domain (SCOP nomenclature: HNH family) can be subclassified into at least eight subsets by iterative refinement of HMM profiles. An initial clustering of 323 proteins containing the HNHc domain helped identify the subsets. The subsets could be differentiated on the basis of the pattern of occurrence of seven defining features. Domain association is also different between the subsets. The subsets show organism as well as domain-based clustering, suggestive of propagation by both duplication and horizontal transfer events. Structure-based sequence analysis of the subsets led to the identification of common structural and sequence motifs in the HNH family with the other three families under the His-Me endonuclease superfamily.
Keywords: HNHc domain, hidden Markov models, domain classification, duplication, horizontal transfer, temperate bacteriophages, His-Me endonuclease, McrA
The domain HNHc (SMART ID: SM00507, SCOP nomenclature: HNH family) is associated with a range of DNA-binding proteins, performing a variety of binding and cutting functions (Gorbalenya 1994; Shub et al. 1994). Several of the proteins are hypothetical or putative proteins of no well-defined function. The ones with known function are involved in a range of cellular processes including bacterial toxicity, homing functions in groups I and II introns and inteins, recombination, developmentally controlled DNA rearrangement, phage packaging, and restriction endonuclease activity (Dalgaard et al. 1997). These proteins are found in viruses, archaebacteria, eubacteria, and eukaryotes. Interestingly, as with the LAGLI-DADG and the GIY-YIG motifs, the HNHc motif is often associated with endonuclease domains of self-propagating elements like inteins, Group I, and Group II introns (Gorbalenya 1994; Dalgaard et al. 1997).
The HNHc domain is characterized by the presence of a conserved Asp/His residue flanked by conserved His (amino-terminal) and His/Asp/Glu (carboxy-terminal) residues at some distance. A substantial number of these proteins also have a CX2C motif on either side of the central Asp/His residue. Structurally, the HNHc motif appears as a central hairpin of twisted β-strands, which are flanked on each side by an α helix (Kleanthous et al. 1999).
Given the number of HNHc proteins known and the promiscuity of the HNHc domain, we have attempted here to subclassify the HNHc proteins. Such a subclassification aids in functional and structural analysis of this large group of proteins. This work also suggests that other families of the His-Me endonuclease superfamily (SCOP) maintain the HNH motif as in the HNH family and could be mechanistically similar. Although there have been several publications in the past few years suggesting structural similarity within the superfamily (Miller et al. 1999; Raaijmakers et al. 1999; Grishin 2001; Cheng et al. 2002), this is the first report of the sequence motif being identified within the superfamily.
A total of 323 sequences were available at the SMART domain database (http://smart.embl-heidelberg.de) at the time of initiation of this work. HMM profiles were created from the seed alignment in SMART. These helped extract the HNHc domains. Multiple-sequence alignment was done with CLUSTALw (Thompson et al. 1994) and phylogeny analysis with PHYLIP (http://evolution.genetics.washington.edu/phylip.html). The tree files were plotted with Njplot (Perriere and Gouy 1996). Bootstrap values were used to identify the clusters for further analysis. Seed alignments were generated using sequences that showed <80% identity over the full length of the protein sequence in each cluster, except for subsets 7 and 8. Further, sequences requiring addition of gaps in the alignment were excluded from the seed alignment. These alignments were then used to generate individual HMM profiles, which helped identify new family members among the parent set of HNHc domains. The process was repeated iteratively by including each new sequence picked up at E value <0.001 or at a high score (the cut-off score was decided on the basis of a sudden drop in score) until no new sequences could be obtained. A total of 158 sequences are distributed among the eight subsets. Each subset was then analyzed for associated domains (Table 1; Supplemental Material) using SMART and CDART (http://www.ncbi.nlm.nih.gov/Structure/lexington/html/overview.html). The HNHc region of representative proteins from each of the subsets was modeled using the Colicin E7 (PDB ID: 1M08) and the known structurally homologous T4 endonuclease VII (PDB ID: 1en7) as template using the Insight 2000 software (http://www.accelrys.com). DNA was docked from the I-PpoI structure (PDB ID: 1A73).
Table 1.
Table lists the number of proteins in each subset, the maximum and minimum identity between full-length proteins within each subset, the taxonomic distribution, and the associated domains
| Subset | No. of Proteins | Max. Identity (%) | Min. Identity (%) | Taxonomic Distribution | Associated Domains |
| 1 | 15 | 99.4 | 37.4 | Bacteria | Cloacin (PF03515) |
| 2 | 31 | 99.6 | 18.8 | 23 phage, 2 bacteria, 3 eukaryote, 3 virus | AP2 (PF00847), IENR1 (SM00497) |
| 3 | 22 | 99.8 | 18.8 | Bacteria | Duf222 (PF02720) |
| 4 | 19 | 99.9 | 23.3 | 17 eukaryote (predominantly mitochondrion), 2 bacteria | — |
| 5 | 20 | 99.5 | 22.4 | 17 bacteria, 3 eukaryote | Rvt (PF00078) |
| 6 | 17 | 89.2 | 34.8 | Bacteria | Rvt (PF00078) |
| 7 | 11 | 99.9 | 50.9 | Bacteria | — |
| 8 | 18 | 98.3 | 78.3 | Bacteria | — |
Results and Discussion
Proteins with the HNHc domain could be divided into at least eight subsets on the basis of this analysis (Fig. 1 ▶; Table 1). These are not identified in any of the protein motifs/ domains/family databases (InterPro:IPR002711, IPR003615, Pfam:PF01844). Each subset shows a unique signature sequence in the HNHc region apart from the common HNH/HNHc domain signature (Fig. 1 ▶). Modeling and structural analysis of representative proteins from each of the eight subsets was done (Supplemental Material).
Figure 1.
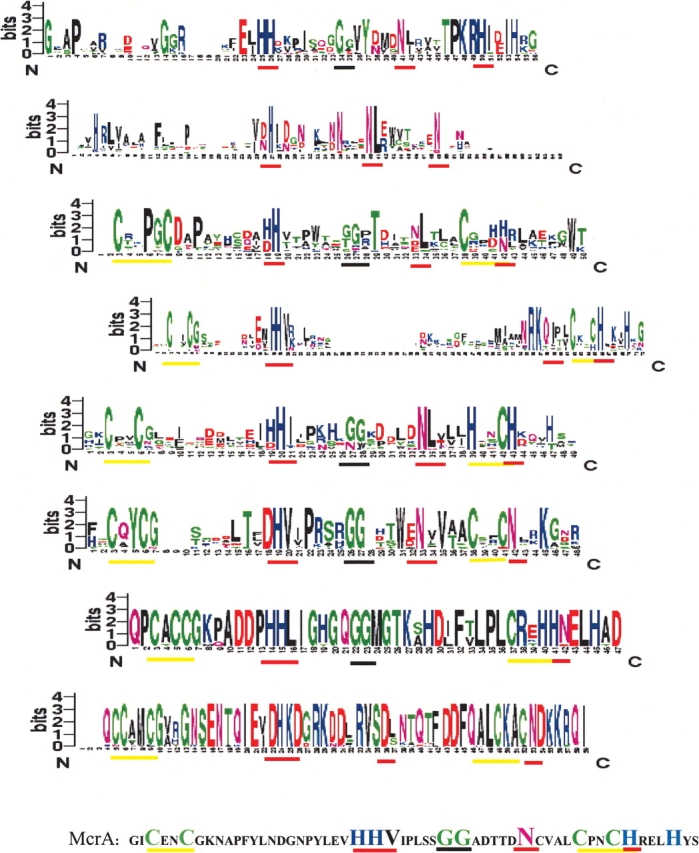
Sequence logos (Schneider and Stephens 1990) for all eight subsets. The subsets are arranged in numerical order. The logos are displayed such that the central Histidine residue of the HNHc motif in the logos is aligned. The McrA sequence is given below with the important residues highlighted. The position of the key features in each of the subset logos are highlighted with a bar below them. The red bars depict the H-N-H, which forms the core HNH element. The cysteine dyads and their equivalents are highlighted with a yellow bar. The black bar highlights the GG motif.
Characterization of subsets
The first and second subsets lack the characteristic cysteine double dyad. The first subset is comprised of the toxin group of HNHc proteins, including the colicins, pyocins, klebsiellins, and the uropathogenicity-specific proteins. All of the proteins in Subset 1 are bacterial proteins (Table 1). Structure of the HNHc region of two of the colicins is known (Kleanthous et al. 1999), and both show the typical hairpin of twisted β strands flanked by α helices on either side. The second subset has mostly phage proteins that associate with at least two other DNA-binding domains commonly found in eukaryotic proteins. This includes the AP2 domains associated with DNA repair enzymes and IENR1 domain. This subset, in contrast to the other subsets, has the HNHc domain closer to the amino-terminal end of the protein (Supplemental Material). Most of the annotated proteins in this subset are intron-encoded site-specific endonucleases (SwissProt ID: Q8WRA0, TEV3_BPR03, etc.), suggesting that the unannotated proteins in this subset could also perform a similar function.
Subset3 includes ~22 proteins, most of which reside in Mycobacterium tuberculosis, and could have arisen by serial duplication as pointed out earlier by Aravind et al. (2000). Horizontal transfer might be responsible for the presence of similar proteins in Caulobacter crestentus and Rhodococcus erythropolis. These nucleases contain a four-residue insertion between the first pair of cysteines, and in 16 of the 22 cases, contain only one cysteine of the second dyad. The domain is usually associated with another domain Duf222 of unknown function (Table 1; Supplemental Material). One of the proteins in the subsets is annotated to be a transposase, which corroborates well with their multiple presence in the same organism.
The next two subsets both associate with the reverse transcriptase domains, but differ in the relative position of the Rvt domain with respect to the HNHc domain. The fourth subset consists of Group II intron-encoded Zn domains of the mitochondrial lineage (Zimmerly et al. 2001), whereas the fifth subset includes proteins of bacterial, phage, and algal origin. The two-cysteine dyads are intact in most of the proteins belonging to the fourth subset, as in the case of T4 endonuclease VII. Therefore, the proteins of this subset should have at least two metal-binding sites formed by the cysteine dyads and the conserved HNH. The fifth subset maintains the first cysteine dyad, but has only one cysteine of the second dyad. The other cysteine is replaced by a histidine residue, which would probably allow Zinc binding, even in the absence of the first cysteine residue. Zimmerly et al. (2001) suggested that the Zn domains of mitochondrial introns have a separate lineage compared with the chloroplast and bacterial lineage. This observation is corroborated in the present study.
Most proteins belonging to subset 6 are hypothetical proteins with no assigned function and no known associated domains. This set of proteins lacks two of the otherwise critical histidines (H102 of colicins, which is a ‘D’ in this case, and H127 of colicins is usually a H/P in this subset). The four cysteines are, however, preserved as in the case of T4 endonuclease VII along with N118 equivalent of colicin that is involved in structural stabilization of colicins. Subset 7 is comprised of the Type II endonucleases belonging to the IceA group of conserved proteins in Helicobacter pylori and the Nla II restriction endonuclease in Neisseria. Subset 8 consists of a conserved group of Salmonella proteins. The sequence has CxCC at the first dyad, whereas the second dyad has only a single ‘C’; the other ‘C’ is replaced by an H/K, which might still allow Zn binding by the pseudo zinc finger usually formed by the four cysteines. The asparagine residue of the HNHc motif is replaced by histidine in this case. Subsets 7 and 8 have a high conservation of residues in the HNH region in each of the subsets. However, within the sets for the full protein sequences, the percentage identity varies from 99% to 50% in the case of Subset 7, and from 98% to 78% in the case of Subset 8. The HNHc region seems to be more conserved than the rest of the protein post-duplication/horizontal transfer.
Structural analysis and the His-Me endonuclease superfamily
A representative protein from each of the subsets was chosen, and the HNH region was modeled on the basis of the Colicin E7 and T4 endonuclease VII structure. At this time, it was noticed that a good structural superposition in the HNH region could be achieved for Colicin E7 (1M08), I-PpoI (1A73), T4 endonuclase VII (1EN7), and Sm endonuclease (1SMN). This has also been pointed out earlier by Grishin (2001). However, a detailed structure-based sequence alignment suggesting the conservation of the H-N-H motif in the four structures had not been made earlier (Fig. 2 ▶; Supplemental Material). The structural equivalents identified here showed that the residues implicated (1SMN: D86, H89, E114, N119) in the Sm endonuclease mechanism (Miller et al. 1999) had the appropriate spatial equivalence (1CE7: E542, H545, V564, H569; 1A73: T95, H98, E114, N119; 1EN7: D40, H43, L57, N62) in the other three structures, and could be superimposed with an rmsd of 0.8, 1.06, and 0.98 Å, respectively, on the equivalent residues of Serratia endonuclease. The spatial equivalence of the active residues suggests a mechanistic equivalence, implying a possible common reaction mechanism for the entire superfamily. However, it is not clear how some of these proteins manage to be sequence specific, whereas the rest are sequence independent. The patterns of variation seen in the subsets are similar to the variations seen in the alignment of the His-Me structures.
Figure 2.
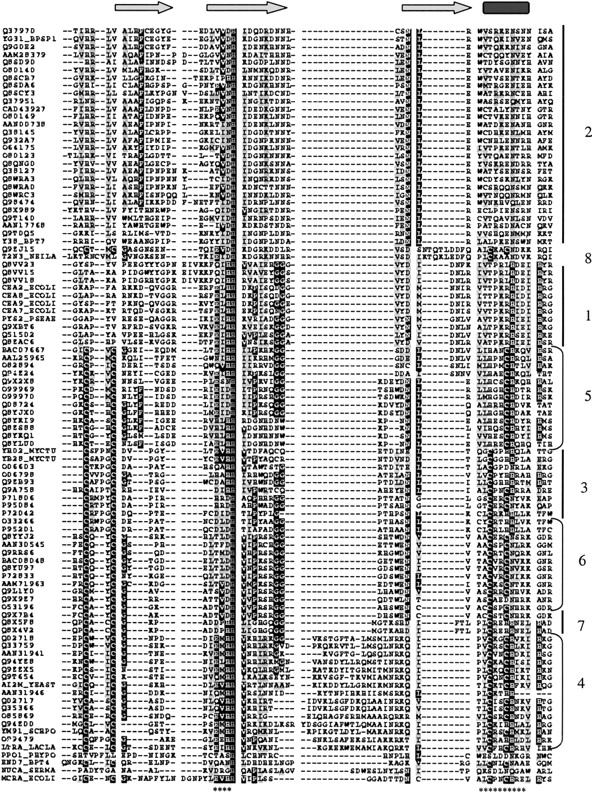
Alignment of the eight subsets on the basis of structure-based sequence alignment of the four His-Me endonuclease structures. The complete alignment is available as Supplemental Material. The subsets are marked in the following order: subset2, subset8, subset1, subset5, subset3, subset6, subset7, and subset4. (*) The 13 residues used for the final superposition. The alignment was generated using a structure-based sequence alignment obtained from the Biosym software and was curated manually to include the subset sequences.
Following the analyses of Colicin E7 using the superposition with the I-PpoI DNA structure by Cheng et al. (2002), we analyzed the subset structure models for structure-function relations using the PpoI–DNA structure (see Supplemental Material). As is evident from Figure 1 ▶, each of these subsets maintain certain residues constant within the group apart from the H-N-H and other conserved residues in the family. From the structural models, it is seen that these subset-specific conserved residues point toward the DNA backbone in the modeled structure. The possible models suggest that all of the subsets maintain a charged pocket around the DNA. These residues form good candidates for site-directed mutagenesis and biochemical studies.
Conclusions
We have subclassified proteins containing the HNHc domain into eight subsets on the basis of clustering on a tree, followed by refinement of clusters using HMMER profiles. This study should aid in functional annotation of new HNHc proteins, biochemical and mutagenesis studies, and identification of targets for structural genomics initiatives. This work also suggests that the HNHc motif is present in a wider group than was suspected previously. Only 152 of the known HNHc proteins could be subclassified in the present work. However, as more proteins get added into the database, and with the commonality of the His-Me superfamily as suggested here, we suspect that this would soon be rectified.
An interesting observation during the course of this study was that almost all of the subsets defined here picked up McrA (an HNHc-containing protein, from the cryptic prophage e14, involved in restriction of nonglucosylated, hydroxymethylated DNA of T-even phages; SwissProt ID: P24200) on iterative HMMER searches, but just below the cutoff. A closer look at the alignments and the McrA sequence in the HNHc regions suggests that the differences between the various subsets can be based on certain distinct elements (Fig. 1 ▶). These include (1) the two cysteine double dyads and variations of these, which delimit the domain; (2) the first H of the HNHc, usually followed by a hydrophobic residue; (3) one to four glycines; (4) the N/D/Q residue of the HNHc, invariably followed by a hydrophobic residue; and (5) the final H/N of the HNHc. McrA and Subset 2 are at the opposite ends of the spectrum with regard to these features. Subset 2 is minimalist and has only the three elements H, N, N of the motif, whereas McrA has all the elements perfectly defined (Fig. 1 ▶). The different subsets seem to have arisen due to multiple divergences from a single McrA-like ancestor. Moreover, independent lineages and lack of a sufficient number of members might possibly account for the remaining HNHc sequences that could not be subclassified. This suggests that the number of subclasses could grow with more numbers of HNHc sequences being determined.
In at least two of the earlier studies on the HNHc group of proteins (with respect to Zn domains in group II introns), it has been suggested that the HNHc domain has a bacterial origin (Zimmerly et al. 2001). A large number of HNHc proteins are seen in viruses and in bacteriophages. The protein McrA that has all of the defining features of the eight subsets is actually a part of the cryptic prophage element e14 in Escherichia coli K-12. This is intriguing and suggests the possibility that the HNH domain could have originated in temperate bacteriophages. Such a suggestion would also be interesting in light of the hypothesis that these sequences are essentially selfish DNA, as temperate bacteriophages can act as very efficient transporters for such elements.
Electronic supplemental material
Domain organization and HNHc domain placement for each of the subsets.
Alignments for each of the eight subsets. The alignments were generated using CLUSTALw and displayed using BOXSHADE.
Clustering of the subsets.
Structural superposition data for the His-Me with the subsets.
Stereo view of the DNA docked on the models of the HNH region of the eight subsets.
Acknowledgments
We thank the anonymous reviewer for suggestions; Manalo Gouy for providing a Linux version for Njplot and modifying it to suit the requirements of the study; and Arun Krishnaswamy for python scripts. We thank Council Scientific Industrial Research (CSIR) for the fellowship for P.M. and K.M., and Department of Biotechnology (DBT) for the project funding to S.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material: See www.proteinscience.org
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03115604.
References
- Aravind, L., Makarova, K.S., and Koonin, E.V. 2000. Survey and summary: Holliday junction resolvases and related nucleases: Identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 28 3417–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y., Hsia, K., Doudeva, G., Chak, K., and Yuan, H.S. 2002. The crystal structure of the nuclease domain of Colicin E7 suggests a mechanism of binding to double-stranded DNA by the H-N-H endonucleases. J. Mol. Biol. 324 227–236. [DOI] [PubMed] [Google Scholar]
- Dalgaard, J.Z., Moser, M.J., Klar, A.J., Holley, W.R., Chatterjee, A., and Mian, I.S. 1997. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res. 25 4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya, A.E. 1994. Self-splicing group I and Group II introns encode homologous putative DNA endonucleases of a new family. Protein Sci. 3 1117–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin, N.V. 2001. Treble clef finger–a functionally diverse zinc-binding structural motif. Nucleic Acids Res. 29 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleanthous, C., Kuhlmann, U.C., Pommer, A.J., Ferguson, N., Radford, S.E., Moore, G.R., James, R., and Hemmings, A.M. 1999. Structural and mechanistic basis of the immunity toward endonuclease colicins. Nat. Struct. Biol. 6 243–252. [DOI] [PubMed] [Google Scholar]
- Miller, M.D., Cai, J., and Krause, K.L. 1999. The active site of Serratia endonuclease contains a conserved magnesium-water cluster. J. Mol. Biol. 288 975–987. [DOI] [PubMed] [Google Scholar]
- Perriere, G. and Gouy, M. 1996. WWW-query: An online retrieval system for biological sequence banks. Biochemie 78 364–369. [DOI] [PubMed] [Google Scholar]
- Raaijmakers, H., Vix, O., Toro, I., Golz, S., Kemper, B., and Suck, D. 1999. X-ray structure of T4 endonuclease VII: A DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture. EMBO J. 18 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, T.D. and Stephens, M.R. 1990. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shub, D.A., Goodrich-Blair, H., and Eddy, S.R. 1994. Amino acid sequence motif of group I intron encoded endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci. 19 402–404. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerly, S., Hausner, G., and Wu, X.C. 2001. Phylogenetic relationship among group II intron ORFs. Nucleic Acids Res. 29 1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]


