Abstract
We expressed and characterized two sHsps, StHsp19.7 and StHsp14.0, from a thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. StHsp19.7 forms a filamentous structure consisting of spherical particles and lacks molecular chaperone activity. Fractionation of Sulfolobus extracts by size exclusion chromatography with immunoblotting indicates that StHsp19.7 exists as a filamentous structure in vivo. On the other hand, StHsp14.0 exists as a spherical oligomer like other sHsps. It showed molecular chaperone activity to protect thermophilic 3-isopropylmalate dehydrogenase (IPMDH) from thermal aggregation at 87°C. StHsp14.0 formed variable-sized complexes with denatured IPMDH at 90°C. Using StHsp14.0 labeled with fluorescence or biotin probe and magnetic separation, subunit exchanges between complexes were demonstrated. This is the first report on the filament formation of sHsp and also the high molecular chaperone activity of thermophilic archaeal sHsps.
Keywords: small heat shock protein, archaea, thermoacidophile, chaperone activity, oligomerization
All organisms express heat shock proteins (Hsps) to protect themselves from harm during times of stress. The major conserved families of these proteins have been shown to be involved in protein folding as molecular chaperones (Gething and Sambrook 1992; Hendrick and Hartl 1993; Parsell and Lindquist 1994). Five major families of Hsps have been identified (Buchner 1996). Among them, small heat shock proteins (sHsps) are ubiquitous in all types of organisms, including archaea, bacteria, and eukarya (Narberhaus 2002), and their expression is up-regulated significantly under conditions of cellular stress like heat shock (Horwitz 2000; Laksanalamai et al. 2001). In spite of a relatively low overall sequence homology among them, they were grouped together based on a conserved domain, the α-crystallin domain, which is named after the α-crystallin of the vertebrate eye lens (Caspers et al. 1995). The α-crystallin domain is preceded by a highly variable N-terminal region and is followed by a short, poorly conserved C-terminal extension (Leroux et al. 1997).
sHsps form large multimeric complexes of 9–40 subunits (12–42 kD in monomer size). To date, the three-dimensional crystal structures of MjHsp16.5, an sHsp from a hyperthermophilic archaeon, Methanococcus jannaschii, and Hsp16.9, an sHsp from wheat, have been determined. MjHsp16.5 forms a hollow spherical complex of 24 subunits (K.K. Kim et al. 1998), whereas Hsp16.9 exists as a dodecameric double disk (van Montfort et al. 2001). The crystal structures show that the α-crystallin domain is composed of β-strands and the two proteins use a similar dimer as a higher assembly building block but differ in their quaternary structure. In contrast, some members of the sHsp family like α-crystallin are remarkably polydispersed (Haley et al. 2000). The quaternary structure of sHsps is highly dynamic, which is often reflected by pronounced size heterogeneity or rapid subunit exchange (Bova et al. 2000, 2002). It was reported that oligomers of Hsp16.9 and Hsp26, an sHsp of Saccharomyces cerevisiae, underwent a reversible temperature-dependent dissociation into smaller species, and can also form high-molecular-weight complexes with a model substrate as a function of increased temperature (Haslbeck et al. 1999; van Montfort et al. 2001).
The sHsps endow thermotolerance to cells in vivo (van den IJssel et al. 1994; Plesofsky-Vig and Brambl 1995), and also protect some proteins from thermal aggregation and in some cases promote renaturation in vitro (Horwitz 1992; Jakob et al. 1993; Muchowski and Clark 1998). It has been suggested that sHsps bind to their substrate proteins at denaturing temperatures and form a stable complex with folding intermediates (Lee et al. 1997). As described above, the sHsp chaperone activity required the dissociation of the complex at heat shock temperatures (Haslbeck et al. 1999).
The molecular chaperone system of thermophilic archaea is very simple compared with that of other organisms. Among major chaperone families, only genes encoding Hsp70 and its cochaperones, group II chaperonin and sHsp, have been found in the genome sequences, and genes for Hsp104/ClpB and Hsp90 are absent (Bult et al. 1996; Kawarabayasi et al. 1998, 2001). Moreover, genes for the Hsp70/DnaK system were not found in the total genomic sequences of hyperthermophilic archaea. Therefore, it is believed that the sHsp plays the main role in the molecular chaperone system in hyperthermophilic archaea. To date, sHsps from two hyperthermophilic archaea, M. jannashii and Thermococcus sp. strain KS-1, have been characterized (R. Kim et al. 1998; Usui et al. 2001; Bova et al. 2002). Although they prevented thermal aggregation of some proteins in vitro, they were not very effective compared with other sHsps. Heat shock induction of sHsps in hyperthermophilic archaea was demonstrated in Thermococcus sp. strain KS-1 and Pyrococcus furiosus (Laksanalamai et al. 2001; Usui et al. 2001).
In this study, we characterized two sHsps, StHsp19.7 and StHsp14.0, from a thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7, which grows best in an acidic and aerobic conditions at ~80°C (Kawarabayasi et al. 2001; Suzuki et al. 2002). In studies of the molecular chaperone systems of S. tokodaii strain 7, the chaperonin subunits α and β were found to assemble into homo-oligomers, respectively, and arrest the spontaneous refolding of chemically denatured proteins in the same way as the purified Sulfolobus chaperonin (Nakamura et al. 1997; Yoshida et al. 1998). However, neither wild-type chaperonin nor recombinant homo-oligomers exhibited any ATP-dependent refolding activity. Recently, another chaperonin gene was found in the complete genomic sequence (Kawarabayasi et al. 2001).
We have found two open reading frames encoding sHsp, and expressed them in Escherichia coli. One of them, named StHsp19.7, formed a filamentous structure, and the other, StHsp14.0, exhibited efficient molecular chaperone activity at 87°C.
Results
Two sHsps exist in S. tokodaii strain 7
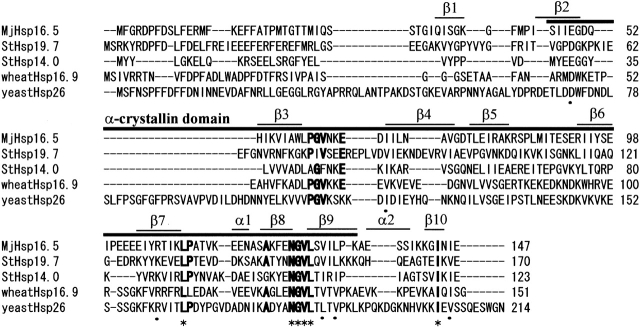
The deduced amino acid sequences of ST0555 and ST1653 of S. tokodaii strain 7 exhibited significant homology in the α-crystallin domain with MjHsp16.5 and some eukaryotic sHsps (Fig. 1 ▶). Among five residues (corresponding to G62, L111, A122, G127, and L129 of MjHSP16.5) highly conserved in sHsps (Narberhaus 2002), four exist in the amino acid sequences of ST0555 and ST1653. In addition, the secondary structure prediction showed that β-strands were also conserved in their putative α-crystallin domains (data not shown). Thus, it was concluded that they encode small heat shock proteins, which were named StHsp19.7 and StHsp14.0 by their deduced molecular masses of 19,717 D and 14,045 D, respectively.
Figure 1.
Amino acid sequence alignment of StHsp19.7 and StHsp14.0 with other sHsps. The sequences (with accession numbers) aligned are Hsp16.5 (AAB98273) of M. jannaschii, StHsp19.7 (BAB65551), StHsp14.0 (BAB66734), Hsp16.9 (S21600) of wheat, and Hsp26 (P15992) of S. cerevisiae. The alignment was performed by using CLUSTAL W. Asterisks indicate identical amino acids, and dots represent similar amino acids among all five sHsps compared. Boldface letters indicate amino acids that are conserved among at least four of the five sequences. The α-crystallin domain and the secondary structure assignment of MjHSP16.5 are shown on top of the alignment according to the crystal structure (K.K. Kim 1998).
The StHsp19.7 and StHsp14.0 genes were cloned in pET23a vectors and used for the expression of StHsp19.7 and StHsp14.0 in E. coli, respectively. The expressed proteins were confirmed to be StHsp19.7 and StHsp14.0 by N-terminal amino acid sequencing. StHsp19.7 has lost the N-terminal methionine.
StHsp19.7 exists as a filament and StHsp14.0 as a spherical complex
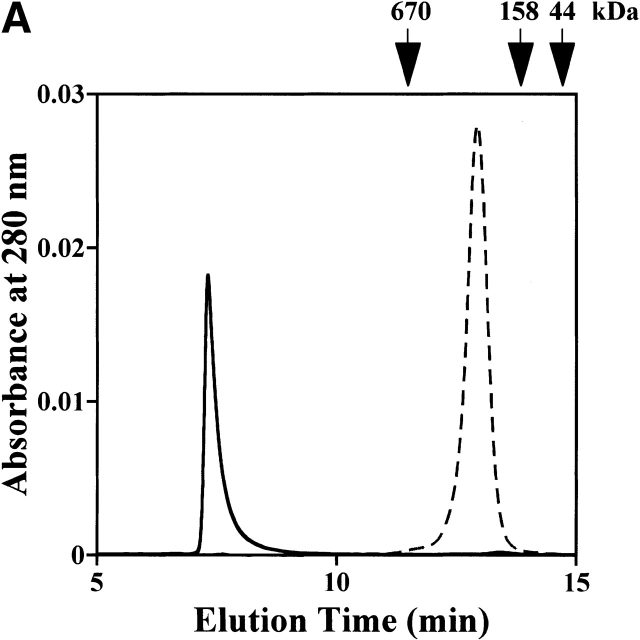
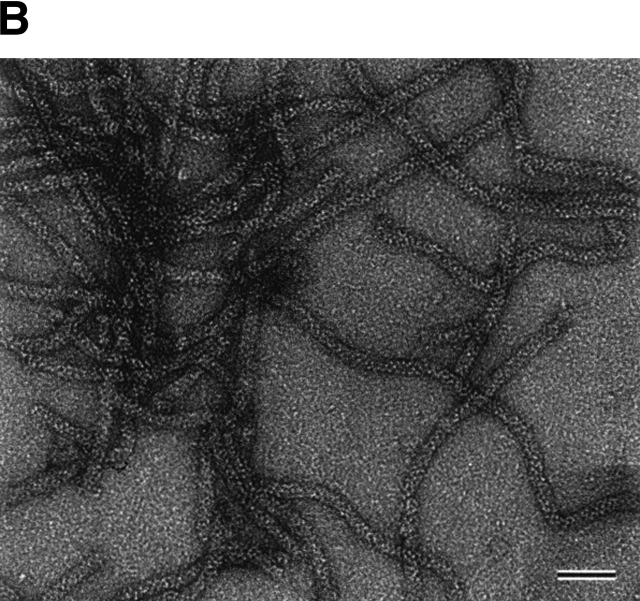
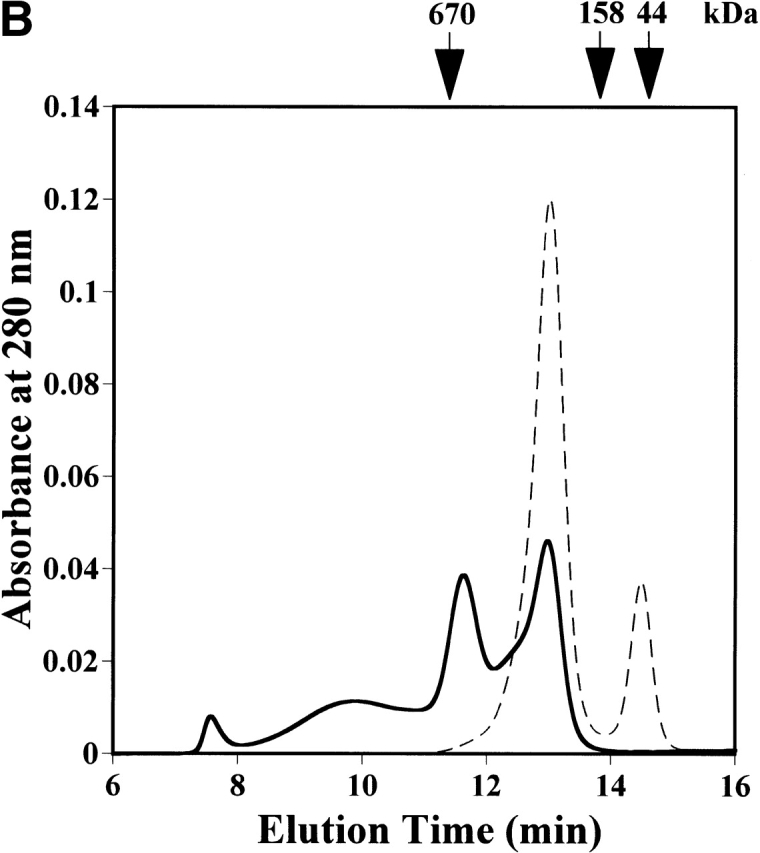
Size exclusion chromatography of the purified StHsp19.7 and StHsp14.0 revealed that they existed as oligomers (Fig. 2A ▶). The molecular mass of StHsp19.7 was larger than expected. Surprisingly, the electron microscopic image of negatively stained StHsp19.7 showed that it forms a filamentous structure (Fig. 2B ▶). The width of the filaments was measured to be 13–14 nm. Size exclusion chromatography and electron microscopic images showed that the StHsp19.7 filament was dissociated into spherical particles of 13–17 nm and shorter filaments by heat shock at 80°C, the optimal growth temperature of S. tokodaii strain 7 (Fig. 2C ▶).
Figure 2.
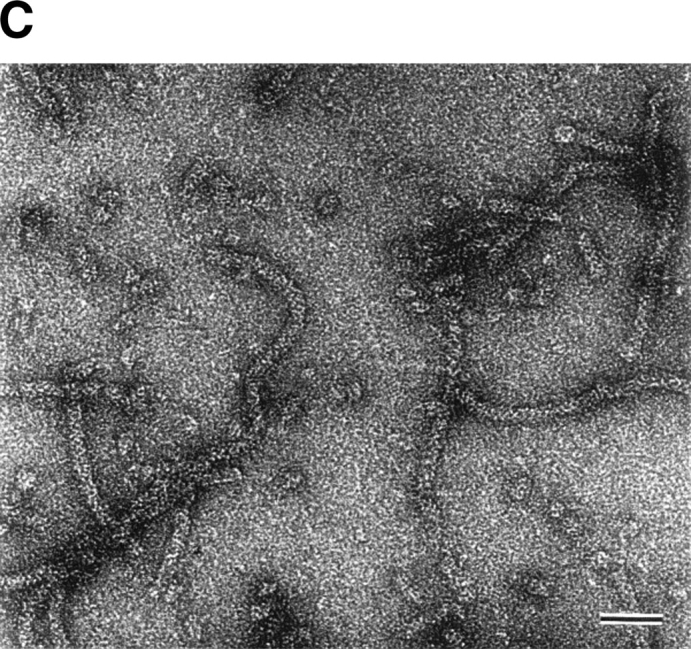
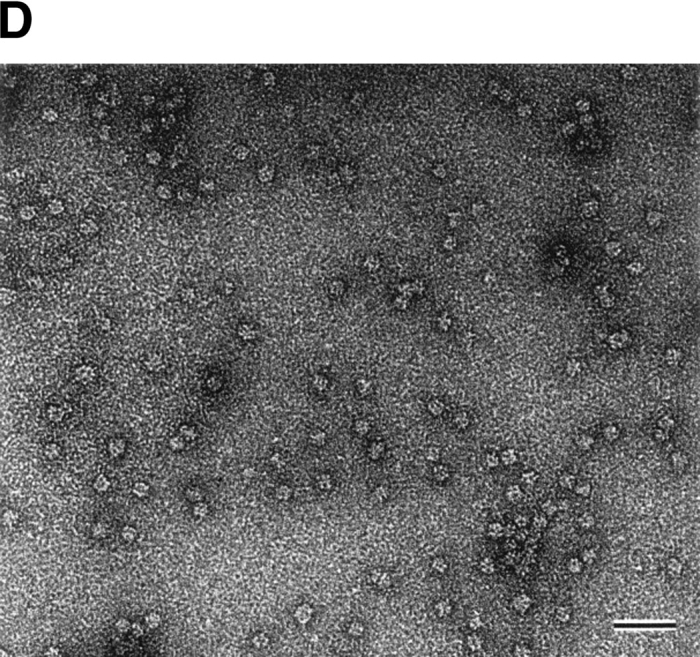
Quaternary structures of StHsp19.7 and StHsp14.0. (A) Size exclusion chromatography was performed using a TSKgel G4000SWXL column (Tosoh) as described in Materials and Methods. Purified StHsp19.7 (43.2 μM as monomer; solid line) and StHsp14.0 (43.2 μM as monomer; dotted line) elute as single peaks. The column was standardized with the following markers as indicated above the figure: thyroglobulin (670 kD); γ-globulin (158 kD); and ovalbumin (44 kD). (B) Negative-stain electron microscopic image of StHsp19.7 (7.1 μM). (C) Negative-stain electron microscopic image of StHsp19.7 (50.8 μM) kept at 80°C for 20 min, cooled on ice for 10 min, and centrifuged at 15,600g at 4°C for 10 min. The supernatant was diluted to 5.1 μM and then applied to grids as described in Materials and Methods. (D) Negative-stain electron microscopic image of StHsp14.0 (7.2 μM). Scale bar, 50 nm.
StHsp14.0 eluted as a single peak, and the molecular mass was estimated to be ~220 kD (Fig. 2A ▶). This result implies that StHsp14.0 should exist as a uniform hexadecameric oligomer because the deduced molecular mass of the StHsp14.0 monomer was ~14.0 kD. However, because of the uncertainty of molecular weight predictions by size exclusion chromatography, more detailed studies would be necessary to determine the correct stoichiometry of the complex. Electron microscopic images of negatively stained StHsp14.0 showed that it exists as a spherical particle of 11–13 nm (Fig. 2D ▶). These results showed that StHsp14.0 has almost the same oligomeric structure as MjHsp16.5. Thus, we concluded that the StHsp14.0 complex also consists of 24 subunits based on analogy with other sHsps from hyperthermophilic archaea (K.K. Kim et al. 1998; Usui et al. 2001).
StHsp14.0 prevents the thermally induced aggregation of IPMDH
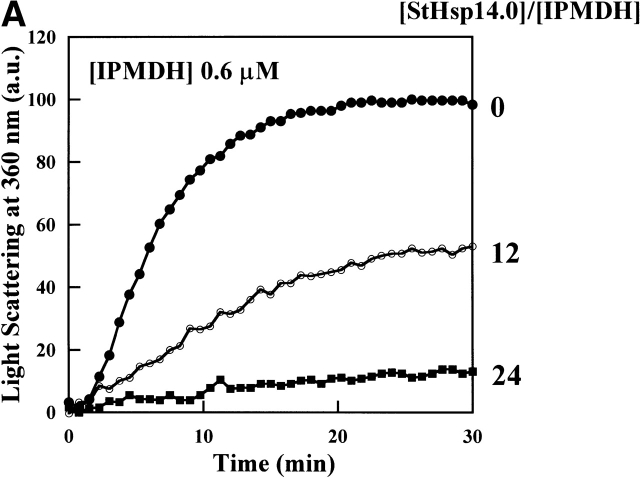
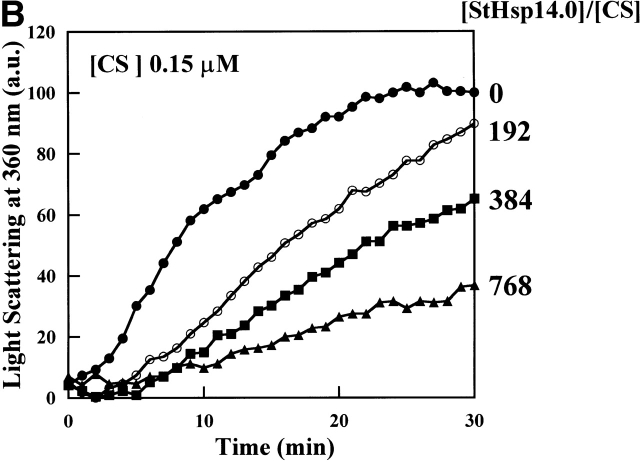
To examine if StHsp19.7 or StHsp14.0 possesses chaperone activity, their effects on the thermal aggregation of IPMDH from the thermophilic bacterium Thermus thermophilus HB8 were examined. IPMDH is a homodimer of the subunit with the molecular mass of ~37 kD (Imada et al. 1991). Although it is much more stable against thermal denaturation than mesophilic enzymes, it denatures to aggregate at 87°C as detected by light scattering at 360 nm. As shown in Figure 3A ▶, StHsp14.0 inhibited completely the thermal aggregation of IPMDH at 87°C at a 24 : 1 ratio of StHsp14.0 monomer to IPMDH monomer. Thus, the StHsp14.0 24-mer prevented the aggregation of IPMDH monomer at an almost equimolar ratio.
Figure 3.
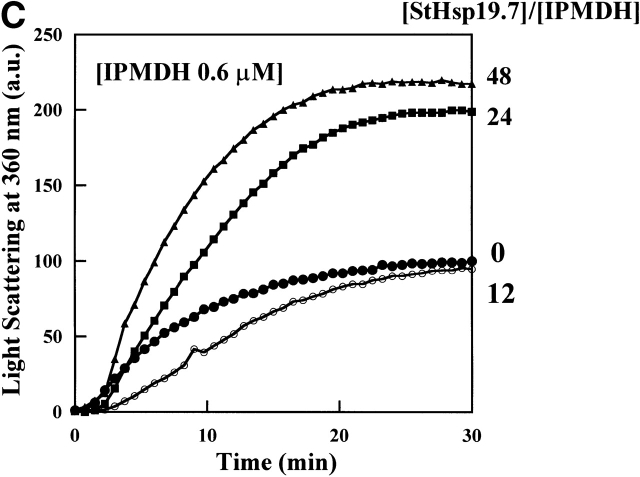
Thermal aggregation of IPMDH and CS was prevented by StHsp14.0. (A) IPMDH (0.6 μM as monomer) was incubated at 87°C in 50 mM Tris-HCl (pH 8.0) buffer, without or with StHsp14.0. (B) CS (0.15 μM as monomer) was incubated at 45°C in 50 mM Tris-HCl (pH 8.0) buffer, with or without StHsp14.0. (C) IPMDH (0.6 μM as monomer) was incubated at 87°C in 50 mM Tris-HCl (pH 8.0) buffer, without or with StHsp19.7. The aggregation of IPMDH or CS was monitored by measuring apparent light scattering at 360 nm with a spectrophotometer.
In previous reports, about 40–80-fold excess mole of MjHsp16.5 or T KS-1 sHsp 24-mer was required to prevent the thermal aggregation of the CS monomer at 40°–45°C (R. Kim et al. 1998; Usui et al. 2001). StHsp14.0 was also ineffective in suppressing the aggregation of the citrate synthase at 45°C, and a 32-fold excess of the StHsp14.0 24-mer over the CS monomer was required for complete suppression (Fig. 3B ▶). Therefore, sHsps of hyperthermophilic archaea are active around or over their optimal growth temperatures.
On the other hand, StHsp19.7 could not protect the thermal aggregation of IPMDH (Fig. 3C ▶). Because StHsp19.7 itself doesn’t aggregate at the temperature (data not shown), the aggregation of IPMDH should be enhanced in the presence of StHsp19.7.
StHsp14.0 binds to nonnative IPMDH and forms a large complex
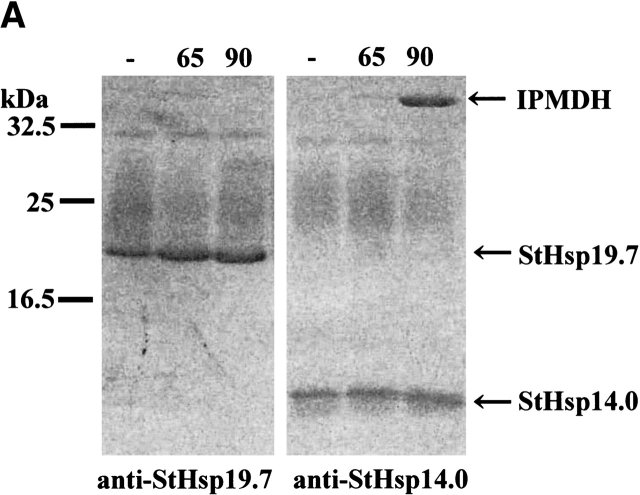
To examine whether Sulfolobus sHsps directly bind to IPMDH in a native or denatured state, immunoprecipitation from the mixture of StHsp19.7 or StHsp14.0 and IPMDH incubated at 65°C and 90°C was performed using anti-StHsp19.7 and anti-StHsp14.0 antibodies. StHsp19.7 bound to neither native IPMDH at 65°C, nor denatured IPMDH at 90°C, reflecting its lack of molecular chaperone activity (Fig. 4A ▶). IPMDH was precipitated with StHsp14.0 when preincubated at 90°C, but only StHsp14.0 was detected after incubation at 65°C (Fig. 4A ▶). Thus, it is confirmed that StHsp14.0 directly bound to denatured IPMDH to suppress thermal aggregation.
Figure 4.
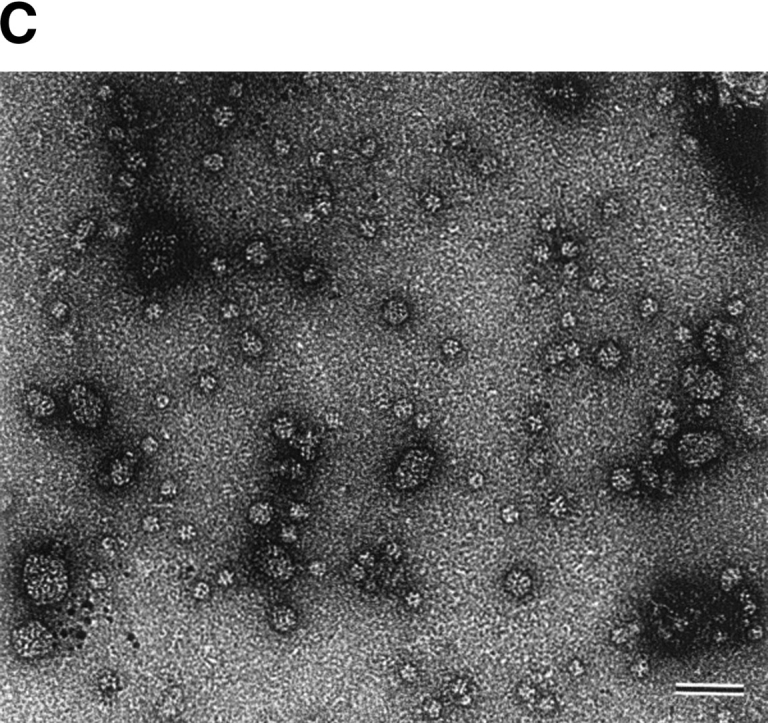
Complex formation between StHsp14.0 and IPMDH. (A) Immunoprecipitation of StHsp14.0 or StHsp19.7 and IPMDH. StHsp19.7 or StHsp14.0 and IPMDH were treated at 65°C or 90°C for 20 min, cooled on ice for 10 min, and then centrifuged at 15,600g at 4°C for 10 min. The supernatant was used for immunoprecipitation with IMMUNOchatcher, as described in Materials and Methods. Immunoprecipitation was performed with anti-StHsp19.7 or anti-StHsp14.0. Proteins were detected by SDS-PAGE using Coomassie brilliant blue. (B) Size exclusion chromatography was performed using a TSKgel G4000SWXL column (Tosoh) as described in Materials and Methods. StHsp14.0 and IPMDH were incubated at 90°C for 20 min, cooled on ice for 10 min (solid line), or not heat-treated (dotted line) and then centrifuged at 15,600g at 4°C for 10 min. The supernatant was applied to the column. The column was standardized with the markers as indicated in Figure 2A ▶. (C) Negative-stain electron microscopic images of complex between StHsp14.0 and IPMDH. StHsp14.0 (216.0 μM as monomer) and IPMDH (36.0 μM as monomer) were incubated at 90°C for 20 min, cooled on ice for 10 min, and then centrifuged at 15,600g 4°C for 10 min. The supernatant was applied to grids as described in Materials and Methods. Scale bar, 50 nm.
Using size exclusion chromatography and negative-stain electron microscopy, we examined the formation of complex between StHsp14.0 and nonnative IPMDH. When the mixture of StHsp14.0 and IPMDH was applied to the gel filtration column without heat treatment, two clearly separated peaks appeared (Fig. 4B ▶). The high-molecular-weight peak corresponds to the StHsp14.0 oligomer, and the low-molecular-weight peak corresponds to the IPMDH (dotted line). When StHsp14.0 was incubated with IPMDH at 90°C before loading, new broad and sharp peaks appeared at shorter elution times (Fig. 4B ▶, solid line).
Complex formation between StHsp14.0 and IPMDH at 90°C was also analyzed by electron microscopy. As shown in Figure 4C ▶, various sized particles, a spherical particle of 17–18 nm or a spread particle of 17–39 nm in diameter, larger than the spherical particles of the StHsp14.0 oligomer, were observed. Thus, StHsp14.0 likely binds to nonnative IPMDH to form complexes of variable size and shape. Eukaryotic sHsp also forms such complexes when incubated with substrate proteins under heat shock conditions (Ehrnsperger et al. 1997; Lee et al. 1997; Haslbeck et al. 1999).
Subunit exchange of StHsp14.0
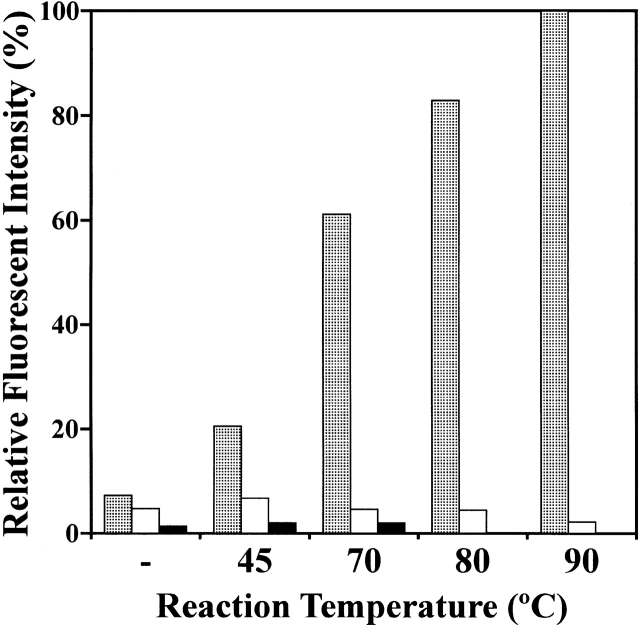
sHsp oligomers show a dynamic exchange of subunits at physiological temperatures, which is thought to be related to chaperone activity (Bova et al. 2000, 2002). To elucidate whether the StHsp14.0 oligomers also exchange subunits, we labeled StHsp14.0 with a fluorescent dye, Alexa Fluor 488, and biotin, independently. The labeling efficiencies were estimated to be 0.3 mole of Alexa Fluor 488 and 1.7 mole of biotin per StHsp14.0 subunit. The subunit exchange reaction was started by mixing equimolar amounts of fluorescence-labeled StHsp14.0 and biotin-labeled StHsp14.0. After incubation, streptavidin-linked magnetic beads were added to the reaction solution, and then StHsp14.0 complexes containing biotin-labeled sHsp subunits were recovered by magnetic separation. The recovered magnetic beads were suspended in buffer and used for fluorescence measurement. If StHsp14.0 oligomers exchange subunits, fluorescence would be detected because fluorescence-labeled subunits would be incorporated into complexes containing biotin-labeled complexes, which are captured by the streptavidin-linked magnetic beads. Although very little fluorescence was observed after the incubation at room temperature, the fluorescence of the captured fraction increased with the reaction temperature (Fig. 5 ▶, stippled bars). When the reaction temperature was increased to 90°C, a marked increase in fluorescent intensity was observed. The intensity was ~43% of the total fluorescent intensity of fluorescence-labeled StHsp14.0. At 45°C, the fluorescent intensity was ~20% of that at 90°C. This result corresponds with the low molecular chaperone activity of StHsp14.0 at 45°C (Fig. 3B ▶).
Figure 5.
Effect of temperature on subunit exchange of StHsp14.0. The subunit exchange reaction was performed with or without incubation at 45°C, 70°C, 80°C, or 90°C for 20 min. The reaction solution contained fluorescence-labeled StHsp14.0 and biotin-labeled StHsp14.0 (stippled bars), biotin-labeled StHsp14.0 and free fluorescence label (white bars), or fluorescence-labeled StHsp14.0 and free biotin label (black bars). The fluorescence intensity of the magnetic beads fractions was measured as described in Materials and Methods. The strongest intensity equals 100%.
To unequivocally show that the florescence of the magnetic beads fraction is caused only by an exchange of subunits between oligomers, we performed an additional control experiment. We mixed excess unreacted free fluorescence label with biotin-labeled StHsp14.0 or excess unreacted free biotin label with fluorescence-labeled StHsp14.0, and performed the same experiment as described above. We did not observe any remarkable fluorescence increase in either experiment (Fig. 5 ▶).
Native StHsp19.7 exists as filaments or spherical particles
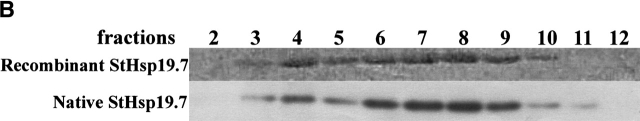
To investigate whether StHsp19.7 forms filamentous complexes in vivo, the cellular lysate of S. tokodaii strain 7 was fractionated by size exclusion chromatography, and then aliquots of the fractions were analyzed by SDS-PAGE with immunoblotting using anti-StHsp19.7 antibody. Recombinant StHsp19.7 was also size fractionated after being incubated at 80°C, and analyzed by SDS-PAGE with Coomassie brilliant blue staining.
The elution profile of recombinant StHsp19.7 shows two broad peaks, which correspond to short filaments and spherical particles as shown in Figure 2C ▶ (Fig. 6A ▶). SDS-PAGE analyses of the fractions showed that StHsp19.7 in the cellular lysate eluted in almost the same manner as recombinant StHsp19.7 (Fig. 6B ▶). The results indicate that native StHsp19.7 may exist as filaments and spherical particles in the cells.
Figure 6.
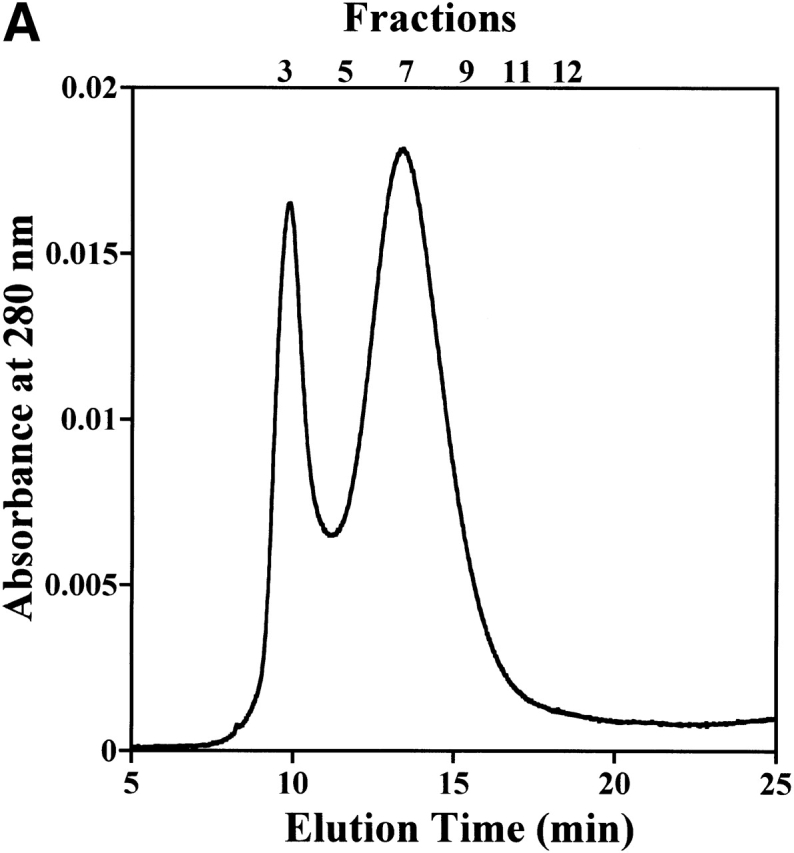
Filament formation of StHsp19.7 in S. tokodaii strain 7. (A) Size exclusion chromatography was performed using a Bio-Prep SE-1000/17 (Bio-Rad) as described in Materials and Methods. Recombinant StHsp19.7 (45.7 μM as monomer) was incubated at 80°C for 20 min and cooled on ice for 10 min. After centrifugation at 15,600g at 4°C for 10 min, the supernatant was applied to the column. (B) Fractions of size exclusion chromatography of recombinant StHsp19.7 and S. tokodaii strain 7 lysate were separated by SDS-PAGE, and analyzed by Coomassie blue staining (upper) and immunoblotting with anti-StHsp19.7 antibody (lower), respectively.
Discussion
There are two sHsp genes in the genomic sequence of the thermoacidophilic crenarchaeon S. tokodaii strain 7. We have expressed both of them, StHsp19.7 and StHsp14.0, in E. coli and performed biochemical characterizations. Electron microscopic images showed that StHsp19.7 exists as a filament of spherical complexes (Fig. 2B ▶). This is the first report on the formation of filaments by sHsps. It was reported that chaperonin from the cognate archaeon Sulfolobus shibatae formed filaments in the presence of Mg2+ and nucleotides (Trent et al. 1997). It is proposed that chaperonin filaments exist in vivo and play a cytoskeletal role in archaea. We have shown that StHsp19.7 exists as filaments and spherical particles in the cell, by size exclusion chromatography with immunoblotting (Fig. 6A,B ▶). Although more detailed study is necessary, StHsp19.7 might exist and function as filaments in vivo.
StHsp14.0 prevented thermal aggregation of IPMDH from T. thermophilus HB8 at 87°C (Fig. 3A ▶). This result shows that StHsp14.0 has molecular chaperone activity similar to other sHsps. However, the chaperone activity was low when the assay was carried out at lower temperature, ~45°C (Fig 3B ▶). This indicates that the chaperone activity of hyperthermophilic sHsp is activated only at high temperature. In contrast, other molecular chaperones from hyperthermophilic archaea, group II chaperonin, and prefoldin could protect CS from porcine heart at 45°C with high efficiency (Iizuka et al. 2001; Okochi et al. 2002). We have shown that an exchange of subunits occurs between StHsp14.0 complexes and the efficiency of the exchange is highly dependent on temperature (Fig. 5 ▶). Recently, subunit exchange of a hyperthermophilic archaeal sHsp, MjHsp16.5, was also reported (Bova et al. 2002). The eukaryotic sHsp complex dissociates into small species at the heat shock temperature (Haslbeck et al. 1999; van Montfort et al. 2001). The dissociation of the complex is likely to be required for the molecular chaperone activity of sHsp, and recently it was reported that dynamic change during oligomerization is essential to the function of sHsp in vivo (Giese and Vierling 2002).
Therefore, we speculate that the dissociation of the StHsp14.0 complex into suboligomeric species at elevated temperatures is indispensable to its molecular chaperone activity. Although we have tried to detect dissociated StHsp14.0 suboligomeric complexes by gel filtration chromatography after heat shock treatment, we could not detect any oligomers with slower retention times (data not shown). This result indicates that suboligomeric complexes of StHsp14.0 immediately reform into a complex after heat shock in the absence of unfolded proteins.
StHsp14.0 forms large complexes with denatured proteins at elevated temperatures (Fig 4B,C ▶). Similar to the case for other sHsps (Ehrnsperger et al. 1997; Lee et al. 1997), the association between StHsp14.0 with IPMDH was not specific, and the complex existed as a more irregular shape as observed in the Hsp26–CS complex (Haslbeck et al. 1999). We propose that dissociated StHsp14.0 with exposed hydrophobic regions binds randomly to intermediate-state IPMDH during heat shock.
We showed that the thermoacidophilic archaeal sHsp StHsp14.0 had chaperone properties at temperatures higher than the optimal growth temperature like the sHsps of other organisms. This study supports the prediction that the heat-induced dissociation of the StHsp14.0 oligomer would result in a smaller species that initiates the interaction with substrate, followed by reassembly into a larger StHsp14.0–substrate complex. The molecular chaperone system of thermophilic archaea is very simple compared with that of other organisms. In the S. tokodaii strain 7 genome sequence, among major chaperone families, only genes encoding Hsp60s, three group II chaperonin subunits, and two sHsps have been found, but genes for other principal chaperones, Hsp70/DnaK, Hsp104/ClpB, and Hsp90, are absent (Kawarabayasi et al. 2001). Therefore, this report indicated that StHsp14.0 plays the main role in the molecular chaperone system in S. tokodaii strain 7 rather than bacteria and eukarya. However, the mechanism of sHsp chaperone activity is still vague. StHsp14.0 may be useful for understanding the relationships between the molecular structure and function of sHsps from not only archaea but also others.
Materials and methods
Strains, enzymes, and reagents
The thermoacidophilic crenarchaeon Sulfolobus tokodaii strain 7 was obtained from Tairo Oshima of Tokyo University of Pharmacy and Life Science. E. coli BL21(DE3) harboring the plasmids for expression of 3-isopropylmalate dehydrogenase (IPMDH) from the thermophilic bacterium Thermus thermophilus HB8 was a gift from Akihiko Yamagishi of Tokyo University of Pharmacy and Life Science. Ex Taq DNA polymerase and other reagents for gene manipulation were obtained from TAKARA Bio Inc. Citrate synthase (CS) from porcine heart was purchased from Sigma Chemical Co.
Plasmid constructs for Sulfolobus sHsps expression
ORFs ST0555 and ST1653 of the thermoacidophilic crenarchaeon S. tokodaii strain 7 were searched for sequence homology with other sHsps. The proteins encoded by theses genes are named StHsp19.7 and StHsp14.0, respectively. Full-length StHsp19.7 and StHsp14.0 genes were amplified from the total DNA of S. tokodaii strain 7 by PCR using primers, 5′-CCA-TAT-GTC-AAG-GAAATA-TAG-A-3′, 5′-GGG-ATC-CCT-ACT-CAA-CCT-TAA-TTTC-3′; and 5′-CCA-TAT-GTA-TTA-TTT-AGG-AAA-AG-3′, 5′-GGG-ATC-CTC-ATT-CTA-TCT-TTA-TAA-CA-3′; and then subcloned into the pT7Blue T vectors, respectively. After sequence confirmation, the genes were excised with NdeI and BamHI, and then introduced into the NdeI/BamHI site of pET23a to construct pSTSH1E and pSTSH2E, respectively.
Purification of StHsp19.7 and StHsp14.0
E. coli BL21(DE3) transformed with pSTSH1E was grown at 37°C in Luria-Bertani medium containing 100 μg/mL ampicillin. The cells were harvested by centrifugation at 4600g at 4°C for 10 min. The harvested cells were resuspended in buffer A (50 mM Tris-HCl at pH 8.0, 0.1 mM EDTA, and 1 mM dithiothreitol) and lysed by sonication. The suspension of disrupted cells was centrifuged at 25,000g at 4°C for 30 min, and the supernatant was heated at 75°C for 30 min. The denatured protein was removed by centrifugation at 25,000g at 4°C for 30 min. The supernatant was applied to a DEAE-Toyopearl anion exchange column (Tosoh) equilibrated with buffer A. Proteins were eluted with a linear gradient of 0–500 mM NaCl in buffer A. Fractions containing StHsp19.7 were pooled, concentrated by ultrafiltration (Centriprep 10, Millipore), and then applied to a HiLoad 26/60 Superdex 200pg size exclusion column (Amersham Biosciences) equilibrated with buffer B (50 mM Tris-HCl at pH 8.0, 1 mM dithiothreitol, and 150 mM NaCl). Fractions containing StHsp19.7 were collected and then concentrated by ultrafiltration (Centriprep 10, Millipore).
E. coli BL21(DE3), transformed with pSTSH2E, was grown, and StHsp14.0 was purified as described above.
Purification of IPMDH
E. coli BL21(DE3) cells harboring the plasmids for expression of IPMDH from the thermophilic bacterium T. thermophilus HB8 were grown at 37°C in Luria-Bertani medium containing 100 μg/mL ampicillin. The cells were harvested by centrifugation at 4600g at 4°C for 10 min. The harvested cells were resuspended in buffer A and lysed by sonication. The suspension of disrupted cells was centrifuged at 25,000g at 4°C for 30 min, and the supernatant was heated at 65°C for 30 min. The denatured protein was removed by centrifugation at 25,000g at 4°C for 30 min. The supernatant was precipitated by the addition of ammonium sulfate to a final concentration of 20%. After centrifugation at 14,000g at 4°C for 30 min, the supernatant was applied to a Butyl-Toyopearl hydrophobic interaction column (Tosoh) equilibrated with buffer A containing 20% ammonium sulfate. Proteins were eluted with a linear gradient of 20%–0% ammonium sulfate in buffer A. Fractions containing IPMDH were concentrated and desalted by ultrafiltration (Centriprep 10, Millipore), and then applied to a UNO Q6 anion exchange column (Bio-Rad) equilibrated with buffer A. Proteins were eluted with a linear gradient of 0–500 mM NaCl in buffer A. Fractions containing IPMDH were collected and then concentrated by ultrafiltration (Centriprep 10).
N-terminal amino acid sequencing
The purified StHsp19.7 or StHsp14.0 was subjected to SDS polyacrylamide gel electrophoresis (SDS-PAGE), and transferred onto polyvinylidine difluoride membranes. The membranes were stained with Coomassie brilliant blue R-250. The bands corresponding to StHsp19.7 and StHsp14.0 were cut out, and analyzed with a 492 Procise Protein sequencing System (Applied Biosystems).
Size exclusion chromatography of sHsps
Size exclusion chromatography was performed using a TSKgel G4000SWXL (Tosoh) with buffer (50 mM Tris-HCl at pH 7.3, 150 mM NaCl, and 1 mM dithiothreitol) at a flow rate of 0.8 mL/min with absorbance monitored at 280 nm. Gel filtration standard protein (Bio-Rad) was used to calibrate the column. To examine the formation of complex, a mixture of StHsp14.0 (216.0 μM as monomer) and IPMDH (36.0 μM as monomer) was incubated at 90°C for 20 min, cooled on ice for 10 min, and centrifuged at 15,600g at 4°C for 10 min, and then the supernatant was applied to TSKgel G4000SWXL (Tosoh).
Electron microscopy
The protein solutions were applied individually onto specimen grids covered with thin carbon support films, which had been made hydrophilic by an ion spatter device (JEOL HDT-400), then negatively stained with 1% uranyl acetate for 30 sec. The images were taken on Kodak electron image films (SO-163) under low electron dose conditions at a magnification of 50,000× in an electron microscope (Philips Tecnai F20) operated at 120 kV.
Thermal aggregation measurements
The effect of StHsp19.7 or StHsp14.0 on the thermally induced aggregation of IPMDH (0.6 μM) from T. thermophilus HB8 and citrate synthase (0.15 μM) from porcine heart in 50 mM Tris-HCl (pH 8.0) buffer was monitored by light scattering at 360 nm using a UV-1600PC spectrophotometer (Shimadzu Biotech) equipped with a thermostatted cell compartment preheated to 87°C or 45°C, respectively.
Culture conditions for S. tokodaii strain 7
S. tokodaii strain 7 was grown in the medium at 80°C aerobically. This medium contains 1.3 g of (NH4)2SO4; 0.28 g of KH2PO4; 0.25 g of MgSO4 • 7 H2O; 0.07 g of CaCl2 • 2 H2O; 0.02 g of FeCl3 • 6 H2O; 1.8 mg of MnCl2 • 4 H2O; 4.5 mg of Na2B4O7 • 10 H2O; 0.22 mg of ZnSO4 • 7 H2O; 0.05 mg of CuCl2 • 2 H2O; 0.03 mg of Na2MoO4 • 2 H2O; 0.03 mg of VOSO4 • 2 H2O; 0.01 mg of CoSO4; 1.0 g of yeast extract; 1.0 g of casamino acid; and 1 L of deionized water (Brock et al. 1972). The pH was adjusted to 3.0 with H2SO4. For Western blot analysis, S. tokodaii strain 7 cells were introduced into 500 mL of the above medium and incubated at 80°C for 3 d.
Antibody preparation, immunoprecipitation, Western blot analysis
Rabbit polyclonal antibodies against the recombinant StHsp19.7 or StHsp14.0 were raised. The IgG fractions were purified using an affinity column (Affi-Gel Protein A, Bio-Rad).
Immunoprecipitation experiments were performed using IMMUNOcatcher (CytoSignal) according to the supplier’s instructions. StHsp19.7 or StHsp14.0 and IPMDH were treated at 65°C or 90°C in buffer B for 20 min, cooled on ice for 10 min, and centrifuged at 15,600g at 4°C for 10 min. Then, 10 μL of protein A purified anti-StHsp19.7 or StHsp14.0 antibody was added to 200 μL of the supernatant sample and incubated at room temperature for 1 h. Protein A/G resin (10 μL) was added to the mixture and incubated for 30 min. Proteins bound to the resin were collected by centrifugation, washed, and resuspended in SDS-PAGE sample solution. After incubation for 15 min, bound proteins were collected by centrifugation and then analyzed by SDS-PAGE.
The S. tokodaii strain 7 cells grown at 80°C were harvested by centrifugation at 4600g at 4°C for 10 min. The harvested cells were resuspended in buffer A and lysed by sonication. The suspension of disrupted cells was centrifuged at 25,000g at 4°C for 30 min. The supernatant was applied to a Bio-Prep SE-1000/17 size exclusion column (Bio-Rad) equilibrated with buffer B, and eluted at a flow rate of 0.5 mL/min. Fractions were separated by SDS-PAGE using 15% gels and transferred to a polyvinylidine difluoride membrane with a semidry blotter (Nippon Eido). The membrane was used for immunoblotting with anti-StHsp19.7 antibody. Detection was performed using a horseradish peroxidase-linked protein A, and reactive bands were visualized with Enhanced Chemiluminescence (ECL) Detection Reagents (Amersham Biosciences).
Labeling of StHsp14.0 with fluorescent dye or biotin
The labeling of StHsp14.0 with fluorescent dye was performed using an Alexa Fluor 488 Protein Labeling Kit (Molecular Probes), according to the manufacture’s instructions. StHsp14.0 was diluted to 2 mg/mL in buffer C (50 mM sodium phosphate at pH 7.5, and 150 mM NaCl), and then sodium bicarbonate was added to the solution (final concentration 100 mM). The solution was transferred to a vial of Alexa Fluor 488 reactive dye. The reaction was performed at room temperature for 1 h in the dark with continuous stirring. Covalently bound probes were separated from free, unreacted probes using the column supplied with the kit. Labeling efficiency was determined by measuring the absorbance of the solution at 280 nm and 494 nm.
The labeling of StHsp14.0 with biotin probe was performed using an EZ-Link Sulfo-NHS-Biotinylation Kit (PIERCE Biotechnology) according to the instructions supplied. StHsp14.0 was diluted to 2 mg/mL in buffer C. Then 1.3 mg of Sulfo-NHS-Biotin was added to the solution. The reaction was run at room temperature for 30 min. Covalently bound probe was separated from free, unreacted probes using the D-Salt Dextran Desalting Column MWCO 5000 supplied with the kit. Biotin incorporation, moles biotin per mole protein, was determined by HABA assay as recommended in the instructions (Green 1965, 1975).
Measurements of subunits exchange by magnetic separation and fluorescence
To initiate the subunit exchange reaction, 3.6 μM each of fluorescence-labeled and biotin-labeled StHsp14.0 monomer in buffer C was mixed together in a reaction tube at various temperatures for 20 min, and cooled on ice. As a control experiment, biotin-labeled StHsp14.0 (3.6 μM) and free fluorescence label (4.6 times the fluorescence intensity of fluorescence-StHsp14.0; 3.6 μM) or fluorescence-labeled StHsp14.0 (3.6 μM) and free biotin label (2.6 times the absorbance at 500 nm in the HABA assay [Green 19652.6 times the absorbance at 500 nm in the HABA assay [Green 1975] of biotin-labeled StHsp14.0; 3.6 μM) in buffer C were mixed together in a reaction tube. Magnetic beads covalently coupled with streptavidin (Dynabeads M-280 Streptavidin, DYNAL) in an equivalent volume were added to the reaction solution. The suspension was kept on ice for 5 min with occasional pipetting. The magnetic beads fraction was separated from the solution fraction using a magnet, buffer C was added, and the mixture was transferred to a microtiter plate. The fraction was subsequently excited at 532 nm, and the fluorescence emission cut off below 526 nm by a filter was measured using a Typhoon 8600 (Amersham Bioscience).
Other methods
Proteins were analyzed by SDS-PAGE (Laemmli 1970). The separated proteins were visualized by staining with Coomassie brilliant blue R-250. The protein concentration was measured by the Bradford method with a protein assay kit (Bio-Rad) using bovine serum albumin as the standard (Bradford 1976).
Acknowledgments
The work reported here is a part of the 21st Century COE (Center of Excellence) program of “Future Nano-Materials” research and education project, which is financially supported by the Ministry of Education, Science, Sports, Culture, and Technology through Tokyo University of Agriculture & Technology. This work was also supported by grants-in-aid for scientific research on priority areas (13033008 and 14037216, 15032212), and a grant of the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Science, Sports and Culture of Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
sHsp, small heat shock protein
IPMDH, 3-isopropylmalate dehydrogenase from Thermus thermophilus HB8
SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis
CS, citrate synthase from porcine heart
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03264204.
References
- Bova, M.P., McHaourab, H.S., Han, Y., and Fung, B.K. 2000. Subunit exchange of small heat shock proteins. Analysis of oligomer formation of αA-crystallin and Hsp27 by fluorescence resonance energy transfer and site-directed truncations. J. Biol. Chem. 275 1035–1042. [DOI] [PubMed] [Google Scholar]
- Bova, M.P., Huang, Q., Ding, L., and Horwitz, J. 2002. Subunit exchange, conformational stability, and chaperone-like function of the small heat shock protein 16.5 from Methanococcus jannaschii. J. Biol. Chem. 277 38468–38475. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Brock, T.D., Brock, K.M., Belly, R.T., and Weiss, R.L. 1972. Sulfolobus: A new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 84 54–68. [DOI] [PubMed] [Google Scholar]
- Buchner, J. 1996. Supervising the fold: Functional principles of molecular chaperones. FASEB J. 10 10–19. [PubMed] [Google Scholar]
- Bult, C.J., White, O., Olsen, G.J., Zhou, L., Fleischmann, R.D., Sutton, G.G., Blake, J.A., FitzGerald, L.M., Clayton, R.A., Gocayne, J.D., et al. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273 1058–1073. [DOI] [PubMed] [Google Scholar]
- Caspers, G.J., Leunissen, J.A., and de Jong, W.W. 1995. The expanding small heat shock protein family, and structure predictions of the conserved “α-crystallin domain.” J. Mol. Evol. 40 238–248. [DOI] [PubMed] [Google Scholar]
- Ehrnsperger, M., Graber, S., Gaestel, M., and Buchner, J. 1997. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 16 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething, M.J. and Sambrook, J. 1992. Protein folding in the cell. Nature 355 33–45. [DOI] [PubMed] [Google Scholar]
- Giese, K.C. and Vierling, E. 2002. Changes in oligomerization are essential for the chaperone activity of a small heat shock protein in vivo and in vitro. J. Biol. Chem. 277 46310–46318. [DOI] [PubMed] [Google Scholar]
- Green, N.M. 1965. A spectrophotometric assay for avidin and biotin based on binding of dyes by avidin. Biochem. J. 94 23c–24c. [DOI] [PubMed] [Google Scholar]
- ———. 1975. Avidin. Adv. Protein Chem. 29 85–133. [DOI] [PubMed] [Google Scholar]
- Haley, D.A., Bova, M.P., Huang, Q.L., McHaourab, H.S., and Stewart, P.L. 2000. Small heat shock protein structures reveal a continuum from symmetric to variable assemblies. J. Mol. Biol. 298 261–272. [DOI] [PubMed] [Google Scholar]
- Haslbeck, M., Walke, S., Stromer, T., Ehrnsperger, M., White, H.E., Chen, S., Saibil, H.R., and Buchner, J. 1999. Hsp26: A temperature-regulated chaperone. EMBO J. 18 6744–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick, J.P. and Hartl, F.U. 1993. Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem. 62 349–384. [DOI] [PubMed] [Google Scholar]
- Horwitz, J. 1992. α-Crystallin can function as a molecular chaperone. Proc. Natl. Acad. Sci. 89 10449–10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. The function of α-crystallin in vision. Semin. Cell Dev. Biol. 11 53–60. [DOI] [PubMed] [Google Scholar]
- Iizuka, R., Yoshida, T., Maruyama, T., Shomura, Y., Miki, K., and Yohda, M. 2001. Glycine at the 65th position plays an essential role in ATP-dependent protein folding by Archael group II chaperonin. Biochem. Biophys. Res. Commun. 289 1118–1124. [DOI] [PubMed] [Google Scholar]
- Imada, K., Sato, M., Tanaka, N., Katsube, Y., Matsuura, Y., and Oshima, T. 1991. Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 Å resolution. J. Mol. Biol. 222 725–738. [DOI] [PubMed] [Google Scholar]
- Jakob, U., Gaestel, M., Engel, K., and Buchner, J. 1993. Small heat shock proteins are molecular chaperones. J. Biol. Chem. 268 1517–1520. [PubMed] [Google Scholar]
- Kawarabayasi, Y., Sawada, M., Horikawa, H., Haikawa, Y., Hino, Y., Yamamoto, S., Sekine, M., Baba, S., Kosugi, H., Hosoyama, A., et al. 1998. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5 55–76. [DOI] [PubMed] [Google Scholar]
- Kawarabayasi, Y., Hino, Y., Horikawa, H., Jin-no, K., Takahashi, M., Sekine, M., Baba, S., Ankai, A., Kosugi, H., Hosoyama, A., et al. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Res. 8 123–140. [DOI] [PubMed] [Google Scholar]
- Kim, K.K., Kim, R., and Kim, S.H. 1998. Crystal structure of a small heat shock protein. Nature 394 595–599. [DOI] [PubMed] [Google Scholar]
- Kim, R., Kim, K.K., Yokota, H., and Kim, S.H. 1998. Small heat shock protein of Methanococcus jannaschii, a hyperthermophile. Proc. Natl. Acad. Sci. 95 9129–9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Laksanalamai, P., Maeder, D.L., and Robb, F.T. 2001. Regulation and mechanism of action of the small heat shock protein from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183 5198–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, G.J., Roseman, A.M., Saibil, H.R., and Vierling, E. 1997. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 16 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux, M.R., Melki, R., Gordon, B., Batelier, G., and Candido, E.P. 1997. Structure–function studies on small heat shock protein oligomeric assembly and interaction with unfolded polypeptides. J. Biol. Chem. 272 24646–24656. [DOI] [PubMed] [Google Scholar]
- Muchowski, P.J. and Clark, J.I. 1998. ATP-enhanced molecular chaperone functions of the small heat shock protein human αB crystallin. Proc. Natl. Acad. Sci. 95 1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, N., Taguchi, H., Ishii, N., Yoshida, M., Suzuki, M., Endo, I., Miura, K., and Yohda, M. 1997. Purification and molecular cloning of the group II chaperonin from the acidothermophilic archaeon, Sulfolobus sp. strain 7. Biochem. Biophys. Res. Commun. 236 727–732. [DOI] [PubMed] [Google Scholar]
- Narberhaus, F. 2002. α-Crystallin-type heat shock proteins: Socializing minichaperones in the context of a multichaperone network. Microbiol. Mol. Biol. Rev. 66 64–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okochi, M., Yoshida, T., Maruyama, T., Kawarabayasi, Y., Kikuchi, H., and Yohda, M. 2002. Pyrococcus prefoldin stabilizes protein-folding intermediates and transfers them to chaperonins for correct folding. Biochem. Biophys. Res. Commun. 291 769–774. [DOI] [PubMed] [Google Scholar]
- Parsell, D.A. and Lindquist, S. 1994. Heat shock proteins and stress tolerance. In The biology of heat shock proteins and molecular chaperones (eds. R.I. Morimoto et al.), pp. 457–494. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Plesofsky-Vig, N. and Brambl, R. 1995. Disruption of the gene for hsp30, an α-crystallin-related heat shock protein of Neurospora crassa, causes defects in thermotolerance. Proc. Natl. Acad. Sci. 92 5032–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., Iwasaki, T., Uzawa, T., Hara, K., Nemoto, N., Kon, T., Ueki, T., Yamagishi, A., and Oshima, T. 2002. Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. strain 7), a new member of the genus Sulfolobus isolated from Beppu Hot Springs, Japan. Extremophiles 6 39–44. [DOI] [PubMed] [Google Scholar]
- Trent, J.D., Kagawa, H.K., Yaoi, T., Olle, E., and Zaluzec, N.J. 1997. Chaperonin filaments: The archaeal cytoskeleton? Proc. Natl. Acad. Sci. 94 5383–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui, K., Yoshida, T., Maruyama, T., and Yohda, M. 2001. Small heat shock protein of a hyperthermophilic Archaeum, Thermococcus sp. strain KS-1, exists as a spherical 24 mer and its expression is highly induced under heat-stress conditions. J. Biosci. Bioeng. 92 161–166. [DOI] [PubMed] [Google Scholar]
- van den IJssel, P.R., Overkamp, P., Knauf, U., Gaestel, M., and de Jong, W.W. 1994. α A-crystallin confers cellular thermoresistance. FEBS Lett. 355 54–56. [DOI] [PubMed] [Google Scholar]
- van Montfort, R.L.M., Basha, E., Friedrich, K.L., Slingsby, C., and Vierling, E. 2001. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat. Struct. Biol. 8 1025–1030. [DOI] [PubMed] [Google Scholar]
- Yoshida, T., Yohda, M., Suzuki, M., Yazaki, K., Miura, K., and Endo, I. 1998. Characterization of homo-oligomeric complexes of α and β chaperonin subunits from the acidothermophilic archaeon, Sulfolobus sp. strain 7. Biochem. Biophys. Res. Commun. 242 640–647. [DOI] [PubMed] [Google Scholar]