Abstract
Ribosomal protein L27 is located near the peptidyltransferase center at the interface of ribosomal subunits, and is important for ribosomal assembly and function. We report the crystal structure of ribosomal protein L27 from Thermus thermophilus HB8, which was determined by the multiwavelength anomalous dispersion method and refined to an R-factor of 19.7% (Rfree = 23.6%) at 2.8 Å resolution. The overall fold is an all β-sheet hybrid. It consists of two sets of four-stranded β-sheets formed around a well-defined hydrophobic core, with a highly positive charge on the protein surface. The structure of ribosomal protein L27 from T. thermophilus HB8 in the RNA-free form is investigated, and its functional roles in the ribosomal subunit are discussed.
Keywords: ribosomal protein L27, protein–RNA interactions, ribosome, Thermus thermophilus HB8, crystal structure
The location of ribosomal protein L27 in the Escherichia coli 50S ribosomal subunit has been probed by immuno-electron microscopy, which revealed its position to be on the interface of the subunits (Lotti et al. 1987). A series of biochemical and genetic approaches, including cross-linking studies, showed that protein L27 resides near the peptidyltransferase center, which is the functionally essential region of the ribosome (Traut et al. 1986; Stöffler-Meilicke and Stöffler 1990; Noller 1991). Notably, protein L27 in the 70S ribosome was most efficiently labeled by the 3′-end of tRNA, in either the A-site or the P-site, suggesting that it must be a central element for tRNA binding (Noller 1991; Wower et al. 2000).
Although the high-resolution crystallographic structure of the 50S subunit from Haloarcula marismortui was solved, this particle lacks a homologous counterpart to the bacterial L27 protein (Ban et al. 2000). The ribosomal protein L21e occupies the position equivalent to that of protein L27. In 2001, the crystal structure of the Thermus thermophilus 70S ribosome was determined at 5.5 Å, and some electron density was ascribed to protein L27, but the protein has not yet been fitted to the density (Yusupov et al. 2001). The exact location of L27 was determined in the 3.1 Å crystal structure of the 50S ribosomal subunit from Deinococcus radiodurans, using the Cα atom coordinates (PDB entry 1NKW, chain U; Harms et al. 2001).
To understand the functional importance of ribosomal proteins, detailed structural information is required. In this work, the crystal structure of the ribosomal protein L27 from T. thermophilus HB8 was determined by the multi-wavelength anomalous dispersion (MAD) method (Hendrickson 1991). The structural characteristics of the protein L27 in the RNA-free form were investigated, and its functional roles in the ribosomal subunit are discussed.
Results and Discussion
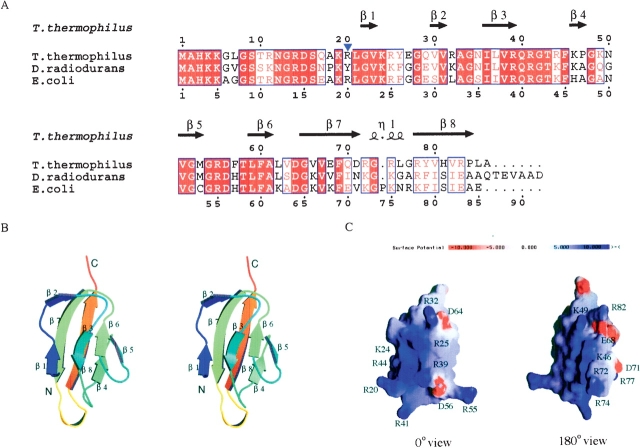
The crystal structure was determined by the MAD method, as described in Materials and Methods. Crystallographic statistics are shown in Table 1. There are four molecules in the crystallographic asymmetric unit. They are essentially identical, with a root-mean-square deviation (RSMD) of ~0.41 Å for the main-chain atoms. The N-terminal region (residues 1–19) of each monomer lacks interpretable electron densities because of disorder, and was excluded from the model. The overall fold of the globular domain of the monomers can be described as a β-barrel-sandwich hybrid (Fig. 1B ▶). It consists of two sets of four-stranded β-sheets formed around a well-defined hydrophobic core. One β-sheet is composed of the short antiparallel β-strands, β1, β3, β6, and β5. The other is formed by the long antiparallel β-strands of β7 and β8 and the short strands of β2 and β4. The electrostatic surface potential reveals a highly positively charged area (Fig. 1C ▶). The salt-bridges of Arg 32–Asp 64 and Arg 39–Asp 56 stabilize the β-sheets of β2–β7 and β3–β5, respectively. The β7–β8 sheets are stabilized by the salt-bridges of Glu 68–Arg 82 and Asp 71–Arg 77. Other basic residues contribute to the extensive positively charged cluster on the protein surface, which should be important for protein L27 to fit in the ribosome and to bind efficiently with rRNA.
Table 1.
X-ray data collection and refinement statistics
| X-ray data | Native | Peak | Edge | High remote | Low remote |
| Space group | P43 | P43 | P43 | P43 | P43 |
| Unit cell (Å) | |||||
| a = b | 48.543 | 49.108 | 49.106 | 49.092 | 49.101 |
| c | 139.120 | 139.323 | 139.325 | 139.324 | 139.326 |
| Resolution (Å) | 2.8 | 3.0 | 3.0 | 3.0 | 3.0 |
| Wavelength (Å) | 0.9000 | 0.9785 | 0.9793 | 0.9600 | 0.9845 |
| No. of observations | 59,602 | 54,165 | 54,613 | 53,997 | 54,022 |
| No. of unique reflections | 7947 (766)d | 6526 (653) | 6580 (667) | 6585 (656) | 6588 (653) |
| Completeness (%) | 99.7% (97.3%) | 99.1% (98.9%) | 99.1% (98.8%) | 99.1% (98.8%) | 99.1% (98.6%) |
| I/σ(I) | 27.8 (3.6) | 17.1 (6.4) | 18.0 (5.3) | 17.1 (5.3) | 18.5 (5.1) |
| Rsyma | 0.074 (0.430) | 0.110 (0.319) | 0.103 (0.331) | 0.103 (0.333) | 0.105 (0.340) |
| Refinement data | |||||
| Rcyst (%)b | 19.7 | ||||
| Rfree (%)c | 23.6 | ||||
| No. of protein atoms | 529 | ||||
| No. of water molecules | 18 | ||||
| Average B factor (Å) | 52.3 | ||||
| RMSD from standard stereochemistry | |||||
| Bond lengths (Å) | 0.009 | ||||
| Bond angles (°) | 1.0 | ||||
| Dihedral angles (°) | 25.2 | ||||
| Improper angles (°) | 0.70 | ||||
| Ramachandran plot statistics | |||||
| Most favored regions (%) | 88.0 | ||||
| Additional allowed regions (%) | 11.5 | ||||
| Generously allowed regions (%) | 0.5 | ||||
| Disallowed regions (%) | 0.0 | ||||
aRsym = |∑I − 〈I〉/∑I|, where I is the observed integrated intensity, 〈I〉 is the average integrated intensity obtained from multiple measurements, and the summation is overall observed reflections.
bRcryst = |Fobs – Fcalc|/Fobs.
cRfree calculated with randomly selected reflections (10%).
d Numbers in parentheses represent values in the highest resolution shell (native 2.80–2.90 Å, SeMet 3.00–3.11 Å).
Figure 1.
(A) Alignment of the ribosomal protein L27 sequences from T. thermophilus HB8, D. radiodurans, and E. coli. The multiple alignment was achieved with CLUSTAL X (Thompson et al. 1997). The secondary structural elements, determined on the basis of the X-ray structure of T. thermophilus protein L27, are indicated with arrows for β-strands and a coil for a 310 helix at the top of the alignment. The identical amino acid residues are shown in white letters highlighted in red, whereas the similar residues are shown in red letters. (B) Stereo structure of the T. thermophilus protein L27. The stereo views were prepared by the programs MOLSCRIPT (Kraulis et al. 1991) and RASTER3D (Merritt and Bacon 1997). (C) Electrostatic potential surface of the T. thermophilus ribosomal protein L27. The molecular surface presentation was generated by GRASP (Nicholls et al. 1991).
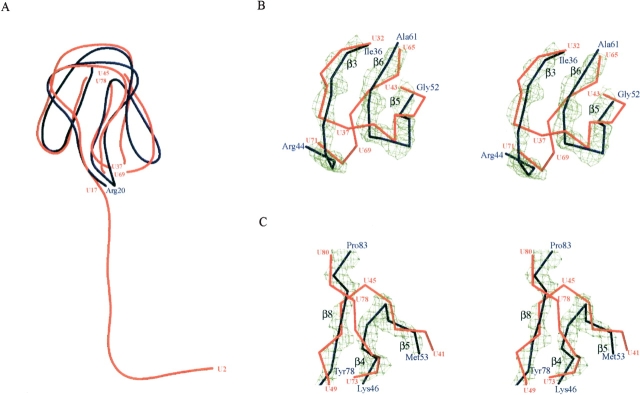
The structure of ribosomal protein L27 in this study is very similar to the globular domain in the D. radiodurans 50S ribosomal subunit as shown in Figure 2A ▶ (1NKW; Harms et al. 2001). It is consistent with the results from previous studies, which showed that the conformations of ribosomal proteins in the ribosome are similar to their monomer forms in isolation (Moore and Steiz 2003). The N-terminal region (residues 1–19) was disordered in the RNA-free form, probably because of the flexibility, although the N terminus of the D. radiodurans protein L27 was shown to be the extended tail. Two connecting regions of the β-strands are different, whereas the number and the locations of the β-strands are almost the same (Fig. 2 B,C ▶). The chain-tracing order of some strands was reversed in the structural model of D. radiodurans protein L27. The better resolution of the monomer form would achieve the accurate structure determination of protein L27.
Figure 2.
Superposition of ribosomal protein L27 backbone representations from T. thermophilus (blue) and D. radiodurans (red). The blue residue numbers are for T. thermophilus and the red numbers are for D. radiodurans. (A) Overall structures of protein L27. Two pairs of Cα atoms (U37, U69, and U45, U78) were removed from the D. radiodurans protein L27. (B,C) Stereo showing the structural differences between the proteins L27 from T. thermophilus and D. radiodurans. The 2Fobs – Fcalc maps of the main-chain atoms of β8–β4–β5 (B) and β3–β6–β5 (C) are contoured at 2σ. The electron density surface was prepared by the program CONSCRIPT (Lawrence and Bourke 2000).
A single, short 310 helix between β7 and β8 exhibits the characteristic positively charged structure containing two Arg residues, and the corresponding Lys residues in the E. coli counterpart have been shown to cross-link with U2334 in the 23S rRNA (Thiede 1998). In E. coli, the end of strand β7, Val 67 to Lys 72, was efficiently affinity-labeled by antibiotics for the peptidyltransferase (Bischof et al. 1995). Considering these results with the present structural information, this short 310 helix would play a key role in the peptidyltransferase reaction (Wower et al. 1989; Kirillov et al. 2002). Superposition of the T. thermophilus protein L27 structure on the D. radiodurans 50S model shows that the N-terminal tail starts from Lys 19 toward the opposite direction of the 310 helix (Fig. 2 ▶). This N-terminal tail would involve the assembly of ribosome and the tRNA positioning (Schwartz et al. 1983; Podkowinski and Gornicki 1989; Osswald et al. 1995). The crystal structure of protein L27 from T. thermophilus HB8 reported here provides detailed structural information and will be useful in future ribosome studies.
Materials and methods
Protein expression, purification, and crystallization
The gene encoding ribosomal protein L27 was amplified by PCR from T. thermophilus HB8 genomic DNA and cloned into the pET26b vector (Novagen). Recombinant protein L27 was overex-pressed in the E. coli strain BL21 (DE3) or B834 (DE3). Cell lysate was subjected to heat treatment for 30 min at 70°C, and a series of cation-exchange chromatography and Superdex 75 columns (Amersham Biosciences) was carried out. The estimated yield was 4.5 mg of purified protein per liter of culture.
Both of the native and SeMet crystals were obtained in drops composed of 2 μL of protein solution (6 mg/mL, 0.4 M NaCl, 1 mM DTT, 20 mM Tris at pH 8.0) and 2 μL of reservoir solution (0.1 M ammonium acetate, 25% polyethylene glycol 4000, 0.05 M tri-sodium citrate dehydrate at pH 6.0) at 30°C. The addition of Anapoe 35 (Hampton Research) improved the SeMet crystal.
Structure determination
The MAD data were collected from a single SeMet-derivative crystal at four wavelengths to 3.0 Å resolution and were used to determine the phases. The structure was refined to an R-factor of 19.7% (Rfree = 23.6%) at 2.8 Å resolution using the native data. The native data were collected at RIKEN beamline BL44B2 at SPring-8 (Adachi et al. 2001), and the MAD data were collected at RIKEN beamline BL26B1 at SPring-8 (Yamamoto et al. 2002). All data were processed using the HKL2000 and SCALEPACK programs (Otwinowski and Minor 1997). General handling of the scaled data was carried out with programs in the CCP4 suite (CCP4 1994). The positions of the Se atoms and the initial phases were determined using the program SOLVE (Terwilliger and Berendzen 1999), and the phases were improved with RESOLVE (Terwilliger 2001), giving an overall figure of merit (FOM) of 0.69. The model building was done by the program O (Jones et al. 1991) and refined by the program CNS (Brunger et al. 1998). The NCS restraints were released in the final stage of the refinements. The coordinates have been deposited in the Protein Data Bank (PDB entry: 1V8Q). Coordinate superpositionings were done by the programs LSQKAB (Kabsch 1976) and LSQMAN (Kleywegt and Jones 1997).
Acknowledgments
We are grateful to N. Kamiya, H. Naitow, Y. Kawano, T. Hikima, and M. Yamamoto, and to H. Nakajima and T. Matsu for their assistance in our data collection at SPring-8 beam lines. We also thank T. Kaminishi for his technical assistance. This work was supported by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analysis, Ministry of Education, Culture, Sports, Science and Technology of Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04864904.
References
- Adachi, S., Oguchi, T., Tanida, H., Park, S.Y., Shimizu, H., Miyatake, H., Kamiya, N., Shiro, Y., Inoue, Y., Ueki, T., et al. 2001. The RIKEN structural biology beamline II (BL44B2) at the SPring-8. Nucl. Instrum. Methods Phys. Res. A 467–468 711–714. [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289 905–920. [DOI] [PubMed] [Google Scholar]
- Bischof, O., Urlaub, H., Kruft, V., and Wittmann-Liebold, B. 1995. Peptide environment of the peptidyl transferase center from E. coli 70S ribosomes as determined by thermoaffinity labeling with dihydrospiramycin. J. Biol. Chem. 270 23060–23064. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., Delano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50 760–763. [DOI] [PubMed] [Google Scholar]
- Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F., and Yonath, A. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107 679–688. [DOI] [PubMed] [Google Scholar]
- Hendrickson, W.A. 1991. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science 254 51–58. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. 1976. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A 32 922–923. [Google Scholar]
- Kirillov, S.V., Wower, J., Hixson, S.S., and Zimmermann, R.A. 2002. Transit of tRNA through the Escherichia coli ribosome: Cross-linking of the 3′ end of tRNA to ribosomal proteins at the P and E sites. FEBS Lett. 514 60–66. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G.J. and Jones, T.A. 1997. Detecting folding motifs and similarities in protein structures. Methods Enzymol. 277 525–545. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Lawrence, M. and Bourke, B., 2000. CONSCRIPT: A program for generating electron density isosurfaces for presentation in protein crystallography. J. Appl. Crystallogr. 33 990–991. [Google Scholar]
- Lotti, M., Stöffler-Meilicke, M., and Stöffler, G. 1987. Localization of ribosomal protein L27 at the peptidyl transferase centre of the 50 S subunit, as determined by immuno-electron microscopy. Mol. Gen. Genet. 210 498–503. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A. and Bacon, D.J. 1997. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 277 505–524. [DOI] [PubMed] [Google Scholar]
- Moore, P.B. and Steitz, T.A. 2003. The structural basis of large ribosomal subunit function. Annu. Rev. Biochem. 72 813–850. [DOI] [PubMed] [Google Scholar]
- Nicholls, A., Sharp, K.A., and Honig, B. 1991. Protein folding and association: Insight from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11 281–296. [DOI] [PubMed] [Google Scholar]
- Noller, H.F. 1991. Ribosomal RNA and translation. Annu. Rev. Biochem. 60 191–277. [DOI] [PubMed] [Google Scholar]
- Osswald, M., Doring, T., and Brimacombe, R. 1995. The ribosomal neighbourhood of the central fold of tRNA: Cross-links from position 47 of tRNA located at the A, P or E site. Nucleic Acids Res. 23 4635–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Podkowinski, J. and Gornicki, P. 1989. Ribosomal proteins S7 and L1 are located close to the decoding site of E. coli ribosome—Affinity labeling studies with modified tRNAs carrying photoreactive probes attached adjacent to the 3′-end of the anticodon. Nucleic Acids Res. 11 8767–8782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, I., Vincent, M., Strycharz, W.A., and Kahan, L. 1983. Photochemical cross-linking of translation initiation factor 3 to Escherichia coli 50S ribosomal subunits. Biochemistry 22 1483–1489. [DOI] [PubMed] [Google Scholar]
- Stöffler-Meilicke, M. and Stöffler G. 1990. Topography of the ribosomal proteins from Escherichia coli within the intact subunits as determined by immunoelectron microscopy and protein–protein cross-linking. In The ribosome: Structure, function and evolution (eds. W.E. Hill et al.), pp. 123–133. American Society for Microbiology, Washington, DC.
- Terwilliger, T.C. 2001. Map-likelihood phasing. Acta Crystallogr. D 57 1763–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C. and Berendzen, J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D 55 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiede, B. 1998. Precise determination of RNA–protein contact sites in the 50 S ribosomal subunit of Escherichia coli. Biochem. J. 334 39–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL-X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut, R.R., Tewari, D.S., Sommer, A., Gavino, G.R., Olson, H.M., and Glitz, D. 1986. Protein topography of ribosomal functional domains: Effects of monoclonal antibodies to different epitopes in Escherichia coli protein L7/L12 on ribosome function and structure. In Structure, function and genetics of ribosomes (eds. B. Hardesty and G. Kramer), pp. 286–308. Springer Verlag, Berlin.
- Wower, J., Hixson, S.S., and Zimmermann, R.A. 1989. Labeling the peptidyltransferase center of the Escherichia coli ribosome with photoreactive tRNA (Phe) derivatives containing azidoadenosine at the 3′ end of the acceptor arm: A model of the tRNA–ribosome complex. Proc. Natl. Acad. Soc. 86 5232–5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower, J., Kirillov, S.V., Wower, I.K., Guven, S., Hixson, S.S., and Zimmermann, R.A. 2000. Transit of tRNA through the Escherichia coli ribosome. J. Biol. Chem. 275 37887–37894. [DOI] [PubMed] [Google Scholar]
- Yamamoto, M., Kumasaka, T., Ueno, G., Ida, K., Kanda, H., Miyano, M., and Ishikawa, T. 2002. RIKEN Structural Genomics Beamlines at SPring-8. Acta Crystallogr. A 58 C302. [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292 883–896. [DOI] [PubMed] [Google Scholar]