Abstract
Dipeptidyl peptidase IV (DPPIV) is a member of the prolyl oligopeptidase family of serine proteases. DPPIV removes dipeptides from the N terminus of substrates, including many chemokines, neuropeptides, and peptide hormones. Specific inhibition of DPPIV is being investigated in human trials for the treatment of type II diabetes. To understand better the molecular determinants that underlie enzyme catalysis and substrate specificity, we report the crystal structures of DPPIV in the free form and in complex with the first 10 residues of the physiological substrate, Neuropeptide Y (residues 1–10; tNPY). The crystal structure of the free form of the enzyme reveals two potential channels through which substrates could access the active site—a so-called propeller opening, and side opening. The crystal structure of the DPPIV/tNPY complex suggests that bioactive peptides utilize the side opening unique to DPPIV to access the active site. Other structural features in the active site such as the presence of a Glu motif, a well-defined hydrophobic S1 subsite, and minimal long-range interactions explain the substrate recognition and binding properties of DPPIV. Moreover, in the DPPIV/tNPY complex structure, the peptide is not cleaved but trapped in a tetrahedral intermediate that occurs during catalysis. Conformational changes of S630 and H740 between DPPIV in its free form and in complex with tNPY were observed and contribute to the stabilization of the tetrahedral intermediate. Our results facilitate the design of potent, selective small molecule inhibitors of DPPIV that may yield compounds for the development of novel drugs to treat type II diabetes.
Keywords: Dipeptidyl peptidase IV, DPPIV, CD26, crystal structure, adenosine deaminase binding protein, serine protease, tetrahedral intermediate
The type II transmembrane serine protease, DPPIV, also known as CD26, or adenosine deaminase binding protein (ADAbp), is highly expressed on endothelial cells, differentiated epithelial cells and lymphocytes (Hegen et al. 1997; De Meester et al. 1999; Kahne et al. 1999). A soluble form of the enzyme was also found in plasma (Iwaki-Egawa et al. 1998; Durinx et al. 2000). As a dipeptidyl peptidase, DPPIV plays a major role in the regulation of physiological processes including immune, inflammatory, CNS, and endocrine functions. For example, DPPIV plays an important role in maintaining glucose homeostasis (Deacon et al. 1998; Balkan et al. 1999; Pauly et al. 1999; Drucker 2003). These studies reveal that DPPIV helps regulate plasma glucose levels by controlling the activity of the incretins glucagon-like peptide 1 (GLP-1) and glucose-dependent insulinotropic polypeptide (GIP). Inhibition of DPPIV in wild-type and diabetic mice leads to increased levels of unprocessed GLP-1 and GIP in the circulation, enhanced insulin secretion, and improved glucose tolerance. Selective inhibitors of DPPIV improve plasma glucose levels in human type II diabetics (Ahren et al. 2002). Independent of its dipeptidyl peptidase activity, DPPIV also binds adenosine deaminase (ADA; Morrison et al. 1993). This interaction has been shown to modulate immune function (Franco et al. 1998; Morimoto and Schlossman 1998).
The catalytic triad of DPPIV is composed of residues S630, D708, and H740, which are located within the last 140 residues of the C-terminal region (Ogata et al. 1992). The enzyme specifically removes dipeptides from the N terminus of peptide substrates that contain on average 30 residues and have a Pro or Ala in the penultimate position. In addition, a slow release has been observed for dipeptides composed of X-Ser or X-Gly (Bongers et al. 1992; De Meester et al. 1999; Hinke et al. 2000; Lambeir et al. 2002). Physiological peptides recognized by DPPIV that contain this specificity profile at their cleavage site include neuropeptides like neuropeptide Y, circulating peptide hormones like peptide YY, glucagon-like peptides (GLP)-1 and -2, gastric inhibitory peptides, as well as paracrine chemokines like RANTES (De Meester et al. 1999; Mentlein 1999). Catalytic efficiencies for the cleavage by DPPIV of different physiological substrates were determined by mass spectrometry-based protease assays (Lambeir et al. 2001a,b; Zhu et al. 2003). These studies demonstrated that residues surrounding the scissile bond mainly determine the substrate selectivity of DPPIV. However, there is supporting kinetic evidence that nonconserved residues along the entire length of the peptide are involved in long-range interactions that play a role in substrate binding and catalysis (Lambeir et al. 2001a,b, 2002; Zhu et al. 2003).
Crystal structures of DPPIV in complex with several small molecule inhibitors and substrates have been published (Engel et al. 2003; Hiramatsu et al. 2003; Oefner et al. 2003; Rasmussen et al. 2003; Thoma et al. 2003). However, the exact molecular determinants that contribute to the substrate specificity of DPPIV and how substrate peptides access the active site remains unclear. To help understand the function of DPPIV, we crystallized and solved the X-ray crystal structure of the enzyme in both its free form and in the presence of the first 10 residues of Neuropeptide Y. Neuropeptide Y is a physiological substrate of DPPIV widely distributed in the nervous system (Mentlein 1999), and involved in cardiovascular homeostasis and the regulation of insulin release (Ahren 2000; Ghersi et al. 2001). The catalytic efficiciency for N-terminal dipeptide cleavage of Neuropeptide Y by DPPIV is 3.0 × 106 M−1sec−1 (Mentlein et al. 1993). The DPPIV/tNPY structure provides direct evidence that the decapeptide accesses the active site through a side opening, unique to DPPIV, and not through the β-propeller opening. The latter mechanism was suggested for the closely related enzyme prolyl oligopeptidase (POP; Fulop et al. 1998, 2000). Our work also provides a detailed understanding of the molecular determinants that contribute to the substrate specificity of DPPIV. Moreover, in the DPPIV/tNPY crystal structure the peptide was trapped in a tetrahedral intermediate, and gives new insight into DPPIV enzyme catalysis. Earlier studies provided evidence for the existence of a tetrahedral intermediate, which was based on structural studies on complexes with small molecule “transition-state analog” inhibitors; ab initio quantum mechanics (QM), molecular mechanics (MM), and molecular dynamics (MD) simulations or combined time-resolved/pH jump crystallographic studies (Wilmouth et al. 2001; Topf et al. 2002a,b). Until now, no direct structural evidence of a single discrete intermediate formed between a physiological substrate and a serine protease has been published.
Results
Structure and domain organization of DPPIV
The crystal structure of the extracellular domain (residues 39–766) of DPPIV was solved to a resolution of 2.1 Å. The structure consists of two domains: an N-terminal 8-bladed β-propeller domain (residues 61–495) and a C-terminal α/β hydrolase domain (Nardini and Dijkstra 1999; residues 39–55 and 497–766; Fig. 1 ▶). The propeller domain packs against the hydrolase domain, and the catalytic triad (S630, H740, and D708) is at the interface of the two domains. In vitro catalytic activity of recombinant DPPIV was measured. The catalytic efficiency for the cleavage of the fluorogenic substrate H-Ala-Pro-7-amido-4-trifluromethylcoumarin (Ala-Pro-AFC) by DPPIV is 5.2 × 106 M−1sec−1.
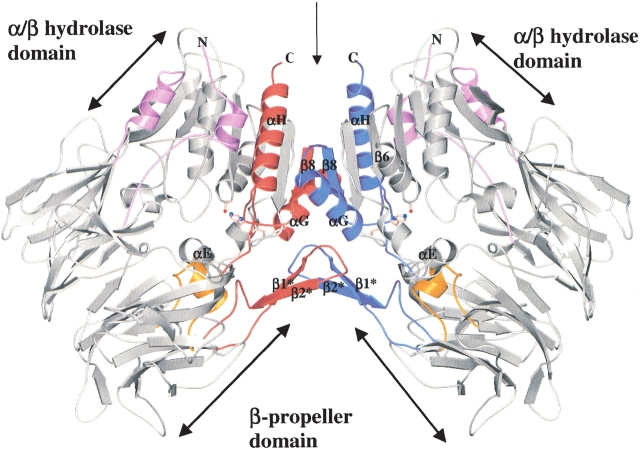
Figure 1.
Ribbon diagram showing overall structure of the DPPIV homodimer, viewed perpendicular to the twofold dyad axis. Secondary structural elements that are involved in dimer formation are represented in red and in blue. The active site residues are shown as ball-and-stick representations. The α-helix comprising residues E205 and E206 is indicated in gold. The figure was made using the programs MOLSCRIPT (Kraulis 1991) and Raster3D (Merrit and Bacon 1997).
The asymmetric unit is composed of two homodimers, the monomers of which are related by a twofold dyad axis (Fig. 1 ▶). This dimeric structure correlates with the biologically active form of DPPIV (Bednarczyk et al. 1991; De Meester et al. 1992; Gorrell et al. 2001; Ajami et al. 2003). The overall structures of the monomers are similar with root-mean-square deviations (RMSDs) from 0.64 Å to 0.98 Å for all heavy atoms and from 0.28 Å to 0.56 Å for the Cα atoms. The dimer interface buries a total of 2188 Å2 accessible surface area per monomer and comprises: (1) the last β-strand (β8) of the peptidase central β-sheet, (2) the last two α-helices (αG and αH), (3) the loop between β6 and αE, and (4) the antiparallel β-strand subdomain (β1* and β2*; Fig. 1 ▶). β8 mainly contains hydrophobic residues forming hydrophobic interactions at the center of the dimer interface. α-Helix H forms hydrogen bonds with the loop between αG and β8 in the other monomer. The antiparallel β-strand arm formed by β1* and β2* interacts with its related arm, αG, and the loop between β6 and αE in the other monomer.
The primary structure of DPPIV contains nine potential N-linked glycosylation sites: N85, N92, N150, N219, N229, N281, N321, N520, and N685. The first N-acetylglucosamine (NAG) sugar moiety is observed with clear electron density in all the nine predicted sites. Detailed structural and biochemical analysis revealed that the glycosylation of DPPIV is not important for catalytic activity, homodimer formation and ADA binding (Aertgeerts et al. 2004).
Substrate access to the active site
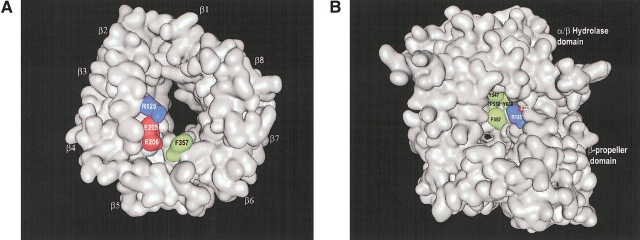
Bioactive peptides recognized by DPPIV could theoretically access the active site in two possible ways: through an opening in the propeller domain or via a side opening formed at the interface of the β-propeller and hydrolase domains (Fig. 2 ▶). The propeller opening is formed by the β-propeller domain, which is composed of an unusual eightfold repeat of blades. Each blade is composed of a four-strand antiparallel β-sheet. The β-propeller domain defines a funnel shaped, solvent-filled tunnel that extends from the β-propeller’s lower face to the active site. The lower face of the funnel, distal to the hydrolase domain, has a diameter of approximately 15 Å. The closing of the circle between the first and the last blade of propeller proteins has been termed “Velcro” (Neer and Smith 1996), and unlike most of the other known propeller proteins, the “Velcro” is not closed between the first and the last blades in the DPPIV structure. This is similar to the arrangement observed in POP (Fulop et al. 1998). The propeller opening connects to a larger side opening (~21 Å) formed at the interface of the β-propeller domain and the hydrolase domain. This oval-shaped cavity creates a second entrance to the active site (Fig. 2 ▶). To understand which entrance/exit pathway substrate peptides use to access the active site of DPPIV, we cocrystallized the enzyme with YPSKPDNPGE (tNPY), corresponding to the first 10 residues of the physiological substrate, Neuropeptide Y. (DPPIV used in the experiment contains a single mutation S716A. The catalytic efficiency of this mutant for cleavage of Ala-Pro-AFC is 41 × 106 M−1 sec−1, which is similar to the value measured for wild-type DPPIV. We also obtained crystals of wild-type DPPIV in complex with tNPY, but the crystals using DPPIV-S716A/tNPY diffracted to a higher resolution.) Clear continuous electron density was observed for the first six of the 10 residues of the peptide. Four of the six residues make molecular interactions (see below), with residues lining the side opening of DPPIV. No clear electron density was observed for the last four residues because they are solvent exposed and therefore not ordered in the structure. In conclusion, the crystal structure of the DPPIV/tNPY complex suggests that physiological substrates may employ the side opening of DPPIV to access the active site.
Figure 2.
(A) Surface representation of the β-propeller domain only, showing the propeller opening to the active site. The view was taken from the interface with the α/β-hydrolase domain and down the pseudo-eightfold axis. The four-strand antiparallel β-sheets of the eight blades are indicated (β1–β8). (B) Surface representation of whole DPPIV molecule, showing the side opening to the active site. Residues of DPPIV that make direct molecular interactions with tNPY are colored in both panels. Hydrophobic negatively charged and positively charged residues are shown in green, in red, and in blue, respectively. The figures were made with the program MOE (MOE, Chemical Computing Group).
Substrate specificity
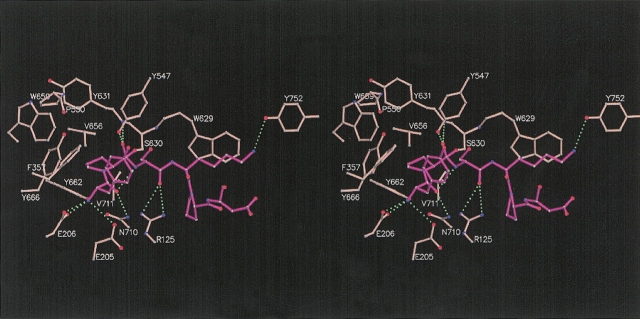
DPPIV cleaves the amide bond after the penultimate N-terminal residue (P1, according to Berger and Schechter 1970) of physiological peptides. Oligopeptide N termini are recognized by the negatively charged active site residues E205 and E206, and are anchored by hydrogen bond formation with the side chains of the two glutamates (Fig. 3 ▶). E205 and E206 reside on a short α-helix insertion (residues 200–206) protruding from the β-propeller domain and pointing toward the active site. The two glutamic acid residues are conformationally restrained by salt bridge formation and hydrogen bond interactions with residues R125, Y662, D663, and N710.
Figure 3.
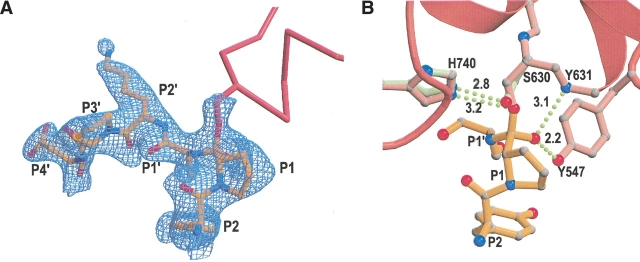
Stereo drawing of first six residues of Neuropeptide Y (magenta) and the underlying active site residues of DPPIV (pink) that make direct molecular interactions with the peptide. The peptide and selected DPPIV residues are shown as ball-and-stick representations. The peptide is not cleaved and trapped in a tetrahedral intermediate by which the carbonyl carbon is covalently linked to the active site S630. Hydrogen bonds are indicated as green dashed lines. The figure is made using the programs MOLSCRIPT (Kraulis 1991) and Raster3D (Merritt and Bacon 1997).
The best catalytic efficiencies for dipeptide cleavage by DPPIV was measured for peptides with a Pro or Ala at P1 (Lambeir et al. 2003; Leiting 2003). The well-defined hydrophobic S1 pocket lined by residues V656, Y631, Y662, W659, Y666, and V711 determines this specificity (Figs. 3 ▶, 4 ▶). The S2 pocket is hydrophobic and determined by the side chains of residues R125, F357, Y547, P550, Y631, and Y666. In this structure we observe two water molecules occupying this S2 pocket, and the P2 tyrosine is interacting with these waters and partially occupying the S2 site. The S1′ pocket is flat and not well defined, and the only interactions observed between the P1′ serine and the S1′ residues are nonspecific van der Waals interactions. The carbonyl oxygen of the P1′ serine makes a hydrogen bond with R125. The side chain of the P2′ lysine packs against the face of W629, completely occluding the tryptophane from solvent, and forms a hydrogen bond with the hydroxyl oxygen of Y752 (Fig. 3 ▶). Beyond P2′, no specific interactions are observed between the peptide and the underlying DPPIV residues.
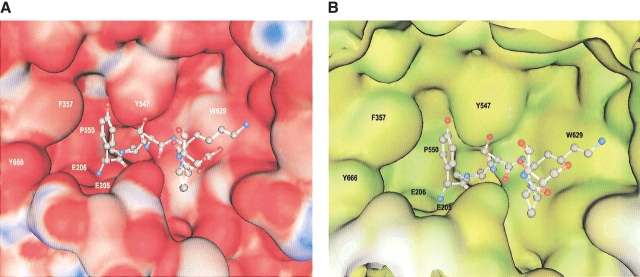
Figure 4.
Molecular surface representations showing the interaction of tNPY with DPPIV. Residues of the peptide are shown in ball-and-stick representations and DPPIV is shown as a solid surface. (A) Colors represent positive and negative electrostatic potential from blue (electropositive; white, neutral) to red (electronegative). (B) Colors represent hydrophobicity (green, polar; yellow, hydrophobic; white, exposed). The figures were made with the program MOE (MOE, Chemical Computing Group).
Ser 630 is located on “the nucleophilic elbow” formed by residues Gly-Trp-Ser630-Tyr-Gly. This sequence is essential for DPPIV activity (Ogata et al. 1992) and conserved in the α/β hydrolase family (Gly-X-Ser-X-Gly). The orientation of S630 is maintained by hydrogen bonds between the carbonyl oxygen of S630 and the amide of Y634, and the amide of S630 and the carbonyl oxygen of V653.
Tetrahedral intermediate
In the crystal structure of the DPPIV/tNPY complex, we observed that the peptide was not cleaved, but trapped in a tetrahedral intermediate (Fig. 5 ▶). As expected for tetrahedral intermediate formation, the Oγ atom of S630 was found in close contact (between 1.6–1.8 Å) with the carbonyl carbon of the scissile bond. The electron density map contoured at 3σ was continuous between the two atoms, and the electron density map contoured at 1σ was discontinuous between the Oγ atom of S630 and Nδ2 of H740. Comparison of this structure with a 2.1 Å structure of the free form of DPPIV shows that the hydroxyl group of the active site serine (S630) has moved significantly to optimally interact with the carbonyl carbon of the scissile bond (Fig. 5B ▶). In addition, the imidazole ring of H740 rotates by about 15° along the χ2 torsion (Fig. 5B ▶). The hydrogen bond distance between S630 and H740 in the native enzyme is 2.8 Å, whereas this distance changes to 3.2 Å in the transition state structure. The oxyanion is stabilized by hydrogen bond formation with the main chain amide of Y631 (~3.1 Å) and with the hydroxyl group of Y547 (~2.2 Å; Fig. 5B ▶). Formation of such a short, very strong, low-barrier hydrogen bond is expected in transition states, which stabilizes intermediates in enzymatic reactions and lowers the energy of transition states.
Figure 5.
Schematic representations showing tetrahedral intermediate formation. (A) 2Fo−Fc electron density map contoured at 1σ of the first six residues of tNPY and of the active site serine (S630). The peptide and the side chain of S630 are shown as ball-and-stick representations, and part of the DPPIV molecule is represented as a pink ribbon diagram. (B) Schematic representation showing the difference in conformation and hydrogen bond formation of active site residues S630 and H740 between the free form (green) of the enzyme and the tetrahedral intermediate (pink). The first three residues of the peptide are shown in gold as ball-and-stick representations. Hydrogen bonds are represented as green dotted lines, and measured distances are indicated in angstroms. Part of the DPPIV molecule around the active site is represented in pink as a ribbon diagram. The figures were made using the programs MOLSCRIPT (Kraulis 1991) and Raster3D (Merrit and Bacon 1997) and XtalView (McRee 1999).
To verify our conclusion that the decapeptide was trapped in a tetrahedral intermediate, the peptide was omitted from the model, the active site S630 was changed to an alanine, and the structure was again refined using REFMAC (CCP 1994). The resultant electron density maps showed unambiguous density for the decapeptide, Oγ atom of S630 and the continuous electron density between the Oγ atom of S630 and the carbonyl carbon of the scissile bond. The asymmetric unit is composed of four independent DPPIV/tNPY complexes, and in all four structures, the peptide is trapped in the tetrahedral intermediate.
Discussion
DPPIV is an ectopeptidase implicated in the degradation of various peptides and hormones including glucagon family peptides, neuropeptides, and chemokines (Mentlein 1999). The enzyme selectively cleaves dipeptides at the N terminus when preferably Pro or Ala is present in the penultimate position. Several structural features that were observed from the crystal structure of the free form of the enzyme and the DPPIV/tNPY complex explain the substrate specificity of DPPIV and its mode of interaction with substrate peptides. Two channels give access to the active site: a propeller opening, and a side opening. Until now, no direct evidence has been described on which entrance/exit pathway is utilized by bioactive peptide substrates. No side opening was observed in the structure of POP, and a gating filter mechanism utilizing the propeller opening was proposed to explain its narrow substrate specificity (Fulop et al. 1998). Because the first and the last blade of the β-propeller domain are not closed, they might partially separate to facilitate substrate access of peptides to the active site. This hypothesis was subsequently proven by mutagenesis and kinetic data on POP (Fulop et al. 2000). DPPIV and POP have the same structural organization of the β-propeller domain, and DPPIV substrates could therefore theoretically utilize the central tunnel formed by this domain to access the active site. However, in contrast to the structure of POP, a second and larger side opening that facilitates access to the active site was observed in the structure of DPPIV. This opening is characterized by an oval shaped groove and is sterically the most favorable way to enter to and exit the active site. The DPPIV/tNPY structure provides direct evidence for peptides to employ this opening to access the active site of the enzyme. Six of the 10 residues of the substrate were ordered in the crystal structure, and the first four residues interact with underlying amino acids present in the side opening of DPPIV. The last four residues of the decapeptide are solvent exposed, and this observation provides further evidence that the propeller opening was not utilized, because based on the length and diameter of the tunnel, extensive interactions for all 10 residues would have been predicted.
The DPPIV/tNPY structure gives a detailed understanding of the molecular mechanisms that determine the interaction of the peptide with the residues present in the active site of DPPIV. The presence of two glutamates (E205 and E206) at the end of an α-helical segment that protrudes from the β-propeller domain into the active site of the enzyme determines the aminopeptidase function of DPPIV. Both residues are essential for enzyme activity (Abbott et al. 1999). The Glu motif is conserved in the DPPIV-like gene family, and was not found in the structure of POP. The Glu motif functions as a recognition site for the N terminus of peptide substrates, and anchors the substrate so that only dipeptides can be cleaved off. The hydrophobic S1 groove is shaped to optimally accommodate and interact with a Pro or Ala residue, and explains the strong preference of DPPIV for peptides with these amino acids in the penultimate position. The S2 subsite preferentially recognizes large hydrophobic and aromatic side chains. With respect to the shape and chemical composition of the S1′ subsite, we observed that most side chains can be modeled into this pocket; however, charged residues are not preferred due to possible unfavorable electrostatic interactions. Because of the π electron system of W629, the S2′ groove is well suited to accept large aliphatic side chains. We observed that residues P2–P2′ of the decapeptide are mainly recognized by DPPIV, and substrate recognition does not extend beyond P2′. This is in agreement with kinetic data measured in vitro on chemokines that are substrates for DPPIV (Lambeir et al. 2001a). However, the adenylyl cyclase-activating peptides, pituitary adenylate cyclase-activating polypeptide (PACAP)-27, and PACAP38 contain the same 27 amino acids at the N terminus, but PACAP38 was processed 15-fold more efficiently than PACAP27 (Lambeir et al. 2001b; Zhu et al. 2003). The only difference between the two peptides is a basic C-terminal extension present in PACAP38. These data suggest that secondary sites on DPPIV remote from the active site are important for substrate binding and catalysis.
In the crystal structure of the DPPIV/tNPY complex, the peptide is trapped in a tetrahedral intermediate that occurs during enzyme catalysis. From the electron density maps, we observed electron transfer between Oγ of S630 and the carbonyl carbon of the scissile bond; the measured distance was 1.75 ± 0.15 Å. The theoretical expected distance of this bond during the tetrahedral intermediate is ~1.4 Å (Topf et al. 2002a). This suggests that the intermediate is forming, but has not yet proceeded to completion. The oxyanion is stabilized by hydrogen bond formation with the main chain amide group of Y631 and the hydroxyl group of Y547. While we were writing our manuscript, Thoma et al. published the crystal structure of DPPIV in complex with diprotin A (Ile-Pro-Ile). The tripeptide was also trapped in a tetrahedral intermediate and its conformation is comparable to the first three residues of tNPY in our structure (Thoma et al. 2003).
The crystal structures of the free form of DPPIV and the DPPIV/tNPY complex suggest that physiological peptide substrates utilize the side opening, unique to DPPIV, to access the active site. The structures also provide a clear insight into the different molecular determinants that are responsible for the substrate specificity of DPPIV. Furthermore, in the DPPIV/tNPY complex, the decapeptide was trapped in a tetrahedral intermediate and gives thereby direct structural evidence for its existence and provides a detailed understanding in the molecular mechanisms that are utilized to stabilize the intermediate. The availability of the DPPIV/tNPY structure will assist in the rational design of highly specific and potent inhibitors that can be used to better understand the role of DPPIV, and as potential treatments for diabetes and related disorders.
Materials and methods
Protein expression and purification
The cDNA encoding human DPPIV was isolated by PCR from spleen cDNA (Clontech) and the extracellular domain (residues 39–766) was cloned into the SmaI site of a modified pFastBacHTb vector (Invitrogen). The final construct contains a baculovirus gp67 signal peptide followed by a His6 tag fused to the coding sequence corresponding to residues 39–766 of DPPIV. Recombinant baculovirus was generated by transposition using the Bac-to-Bac system (Gibco-BRL). Large-scale production of recombinant protein was performed by infection of Trichoplusia ni (Hi5) insect cells (Gibco-BRL) for 48 h in 5-L Wave Bioreactors (Wave Biotech). The secreted glycosylated recombinant protein was isolated from the cell culture medium by diafiltration using crossflow ultrafiltration followed by passage over a nickel chelate resin (Binding buffer: 25 mM Tris [pH 7.9], 400 mM NaCl). The column was washed overnight (0.2 mL/min) with 50 mM K2HPO4 (pH 7.9); 400 mM NaCl; 20 mM Imidazole-HCl, and 0.25 mM TCEP followed by five column volumes (1 mL/min) of 50 mM Tris HCl (pH 7.9), 400 mM NaCl and 0.25 mM TCEP. Protein bound was eluted with four column volumes of 50 mM Tris-HCl (pH 7.9), 400 mM NaCl, 200 mM imidazole-HCl, and 0.25 mM TCEP. To remove oligomeric forms, the sample was further purified over a size-exclusion column (BioSep SEC S3000, 300 × 21.2 mm, Phenomenex) equilibrated with 25 mM Tris (pH 7.6); 150 mM NaCl; 0.25 mM TCEP; and 1 mM EDTA. A yield of 16 mg/L culture (3 × 106 cells/mL) was obtained. DPPIV substituted with Se-Met was produced as described above with the following modifications: 16 h following infection the Hi5-infected insect cells were centrifuged at 500 × g for 15 min and the pelleted cells resuspended in an equal volume of protein-free methionine-free ESF-921 medium (Expression Systems). Following 4 h of further growth Se-Met (Acros) was added to a final concentration of 50 mg/L. The medium from the infected culture was harvested 64 h following viral infection. The Se-Met-substituted protein was purified as described above. The incorporation of Se-Met was estimated to be in the region of 30%–40%. The complex of DPPIV with a decapeptide (YPSKPDNPGE; tNPY) (custom synthesized by Biopeptide Co., LLC) that corresponds to the first 10 amino acids of Neuropeptide Y was formed at pH 7.6 (25 mM Tris, 250 mM NaCl, 0.25 mM TCEP, 1 mM EDTA) by incubation for 30 min at room temperature using a 10-fold molar excess of tNPY (final concentration 1 mM) over DPPIV (final concentration 0.1 mM).
Determination of catalytic activity
The determination of the catalytic constants of DPPIV and DPPIV-S716A for dipeptide cleavage was performed using a fluorescent assay. Enzyme (0.1 nM) was mixed with 0.4–400 μM of Ala-Pro-AFC (Bachem) in 20 mM Tris (pH 7.4) 20 mM KCl, 0.1 mg/mL BSA, and 1% DMSO in a 96-well half-area plate and monitored kinetically at Ex400 nm and Em505 nm using Molecular Devices SpectraMax Gemini. Assays were performed in duplicate for each sample. MDL data analysis toolbox was used for analysis of Michaelis-Menten kinetics.
Crystallization and data collection
Wild-type DPPIV, Se-Met DPPIV, and the DPPIV/tNPY complex were crystallized at 4°C using Syrrx’s automated Nanovolume CrystallisationTM technology (Hosfield et al. 2003). In all cases, the reservoir solution was 20% PEG MME 2000, 100 mM Bicine (pH 8.0–8.5). Thick plate-shaped crystals appeared in about 5 days, which grew to about 0.5 mm in longest dimension and varying width and thickness. For X-ray data collection, crystals were flash-frozen at 100 K using 25% v/v ethylene glycol as a cryoprotectant. Data were collected at Advanced Light Source (ALS) and Stanford Synchrotron Laboratory (SSRL) beam lines and processed with both HKL2000 programs and MOSFLM (Otwinowski and Minor 1997; Leslie et al. 2002). For heavy atom derivatization, the native crystals were soaked in varying concentrations of heavy atom solutions made in synthetic mother liquor. Extensive screening of a large number of heavy atom-soaked crystals resulted in two useful isomorphous derivatives: di-μ-iodobis(ethylenediamine)diplatinum (PIP), and ethylmercurithiosalicylate (EMTS). In addition, a three wavelength multiple wavelength anomalous diffraction (MAD) data set was collected using Se-Met crystals. The data were processed using MOSFLM of CCP4 (1994). The data collection statistics are listed in Table 1.
Table 1.
Heavy atom and Se-Met data statistics for human DPPIV
| Native | Se-Met | EMTSa | PIPb | |||
| Unit cell parameters | ||||||
| a (Å) | 121.8 | 121.9 | 122.2 | 121.4 | ||
| b (Å) | 124.1 | 123.0 | 123.1 | 121.7 | ||
| c (Å) | 144.5 | 145.0 | 145.8 | 144.2 | ||
| β (°) | 114.7 | 114.9 | 114.9 | 114.8 | ||
| Wavelength (Å) | 1.0 | 0.97893 | 0.97913 | 0.91837 | 1.00720 | 1.0721 |
| Resolution (Å) | 2.1 | 2.8 | 3.0 | 3.0 | 2.8 | 3.0 |
| Total observations | 996,059 | 1,323,896 | 656,502 | 646,108 | 914,648 | 582,365 |
| Unique reflections | 218,087 | 92,320 | 91,599 | 91,218 | 96,270 | 76,748 |
| Completeness (%) | 96.4 (95.0) | 96.9 (96.0) | 95.8 (94.0) | 95.4 (93.0) | 99.8 (100) | 99.8 (98.3) |
| Rsymm (I) | 0.062 (0.524) | 0.168 (0.701) | 0.164 (697) | 0.170 (0.652) | 0.204 (0.716) | 0.119 (0.639) |
| l/σ (I) | 19.8 (2.3) | 15.0 (2.4) | 10.4 (2.7) | 8.4 (2.2) | 12.4 (1.6) | 16.6 (2.5) |
| Number of sites | 56 | 8 | 16 | |||
| Phasing power (ano) | 1.043 | 0.46 | 0.61 | 1.01 | 0.98 | |
| Phasing power (iso) | 0.50 | 0.38 | 2.83 | 1.35 | ||
| Figure of merit | 0.385 (acentric), 0.345 (centric) | |||||
| NCS correlation | 0.44 (initial) 0.81 (final) | |||||
a Ethylmercurithiosalicylate (EMTS).
b Di-μ-iodobis(ethylenediamine)diplatinum (PIP).
Structure determination
For wild-type DPPIV, experimental phases were derived by a combination of multiple isomorphous replacement with anomalous scattering (MIRAS) and MAD methods using the heavy atom derivatives (PIP and EMTS) and Se-Met derivative. A highly redundant peak data set collected at Pt absorption edge was used to locate the 16 sites in the asymmetric unit with the program SHELXD (Sheldrick 1998). The 16-Pt sites were refined using the program SHARP (de La Fortelle and Bricogne 1997), and initial protein phases were calculated. The phases that resulted from these derivatives were further improved by solvent flattening and fourfold crystallographic averaging using the program DM (CCP 1994). The heavy atom phases obtained as described above were successful in locating all the 56 Se positions when crossphased on to the Se absorption edge data set and eight Hg sites of the EMTS derivative. These Se and Hg sites were included along with the Pt sites for refinement in SHARP. The protein phases calculated where improved by solvent flattening and noncrystallographic symmetry averaging using the program DM (CCP 1994). The resulting electron density maps were of very good quality, and were used for chain tracing and model building using XtalView (McRee 1999).
The structure of the DPPIV/tNPY complex was determined by molecular replacement using AMORE (CCP 1994) using the crystal structure of the free form of DPPIV as a search model. The structure was refined using the program REFMAC (CCP 1994). Several cycles of model building with XtalView (McRee 1999) and refinement were performed for improving the quality of the model. Refinement statistics are presented in Table 2.
Table 2.
Data collection and refinement statistics for wild-type DPPIV and DPPIV/tNPY complex
| Wild-type DPPIV | DPPIV/tNPY complex | |
| Crystals | ||
| Space group | P21 | P21 |
| Cell dimension | a = 121.8 Å; b = 124.1 Å; c = 144.5 Å; β = 114.7° | a = 122.5 Å; b = 122.5 Å; c = 145.3 Å; β = 114.9° |
| Solvent content (%) | 55.0 | 55.0 |
| Data processing statistics | ||
| Wavelength (Å) | 1.0 | 1.01 |
| Resolution range (Å) | 20.0/2.1 | 42.2/2.3 |
| Total reflections | 996,059 | 311,524 |
| Unique reflections | 218,087 | 153,023 |
| I/σ(I) | 19.8 (2.3) | 8.4 (1.4) |
| Completeness (%) | 96.4 (95.0) | 93.4 (93.4) |
| Multiplicity | 4.6 | 2.0 |
| Rmerge | 0.062 (0.524) | 0.054 (0.531) |
| Refinement statistics | ||
| Number of protein/sugar/water residues | 2914/37/1678 | 2932/25/1249 |
| Reflection in working/free set | 17,551/4355 | 144,953/8070 |
| Rfactor/Rfree | 21.8/24.9 | 23.4/26.8 |
| Average B-factor (Å2) | 21.9 | 47.2 |
| r.m.s. deviation of Bonds (Å)/angle (°) from ideality | 0.009/1.46 | 0.010/1.55 |
| Ramachandran plot | ||
| Residues in most favor region (%) | 88.5 | 84.8 |
| Additionally allowed region (%) | 11.1 | 14.4 |
| Generously allowed region (%) | 0.3 | 0.6 |
| Disallowed region (%) | 0.2 | 0.2a |
a The catalytic serine situates in a so-called “nucleophile elbow” and is in disallowed region of the Ramachandran plot.
Acknowledgments
We thank Gyorgy Snell for X-ray data collection; Lihong Shi for kinetic data analysis; Andy Jennings for the preparation of Figure 2 ▶; and Keith Wilson, David Weitz, Robert Wijnands, and Steve Kaldor for careful reading of the manuscript and helpful suggestions. This work is based on research conducted at the Advanced Light Source (ALS) and at the Stanford Synchrotron Radiation Laboratory (SSRL). ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the U.S. Department of Energy (DOE) under Contract No. DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory. SSRL is funded by the DOE Office of Basic Energy Sciences. SSRL beam lines are supported by the NIH, National Center for Research Resources, Biomedical Technology Program, and by the DOE Office of Biological and Environmental Research. We thank the staff at ALS and at SSRL for their excellent support in the use of the beam lines.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
DPPIV, dipeptidyl peptidase IV
NPY, Neuropeptide Y
tNPY, N-terminal decapeptide (residues 1–10) of Neuropeptide Y
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03460604.
References
- Abbott, C.A., McCaughan, G.W., and Gorrell, M.D. 1999. Two highly conserved glutamic acid residues in the predicted β propeller domain of dipeptidyl peptidase IV are required for its enzyme activity. FEBS Lett. 458 278–284. [DOI] [PubMed] [Google Scholar]
- Aertgeerts, K., Ye, S., Shi, L., Prasad, S.G., Witmer, D., Chi, E., Sang, B.-C., Wijnands, R.A., Webb, D.R., and Swanson, R.V. 2004. N-linked glycosylation of dipeptidyl peptidase IV (CD26): Effects on enzyme activity, homodimer formation, and adenosine deaminase binding. Protein Sci. 13 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahren, B. 2000. Autonomic regulation of islet hormone secretion—Implications for health and disease. Diabetologia 43 393–410. [DOI] [PubMed] [Google Scholar]
- Ahren, B., Simonsson, E., Larsson, H., Landin-Olsson, M., Torgeirsson, H., Jansson, P.A., Sandqvist, M., Bavenholm, P., Efendic, S., Eriksson, J.W., et al. 2002. Inhibition of dipeptidyl peptidase IV improves metabolic control over a 4-week study period in type 2 diabetes. Diabetes Care 25 869–875. [DOI] [PubMed] [Google Scholar]
- Ajami, K., Abbott, C.A., Obradovic, M., Gysbers, V., Kahne, T., McCaughan, G.W., and Gorrell, M.D. 2003. Structural requirements for catalysis, expression, and dimerization in the CD26/DPIV gene family. Biochemistry 42 694–701. [DOI] [PubMed] [Google Scholar]
- Balkan, B., Kwasnik, L., Miserendino, R., Holst, J.J., and Li, X. 1999. Inhibition of dipeptidyl peptidase IV with NVP-DPP728 increases plasma GLP-1 (7–36 amide) concentrations and improves oral glucose tolerance in obese Zucker rats. Diabetologia 42 1324–1331. [DOI] [PubMed] [Google Scholar]
- Bednarczyk, J.L., Carroll, S.M., Marin, C., and McIntyre, B.W. 1991. Triggering of the proteinase dipeptidyl peptidase IV (CD26) amplifies human T lymphocyte proliferation. J. Cell. Biochem. 46 206–218. [DOI] [PubMed] [Google Scholar]
- Berger, A. and Schechter, I. 1970. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 257 249–264. [DOI] [PubMed] [Google Scholar]
- Bongers, J., Lambros, T., Ahmad, M., and Heimer, E.P. 1992. Kinetics of dipeptidyl peptidase IV proteolysis of growth hormone-releasing factor and analogs. Biochim. Biophys. Acta 1122 147–153. [DOI] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project 4). 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50 760–763. [DOI] [PubMed] [Google Scholar]
- Deacon, C.F., Hughes, T.E., and Holst, J.J. 1998. Dipeptidyl peptidase IV inhibition potentiates the insulinotropic effect of glucagon-like peptide 1 in the anesthetized pig. Diabetes 47 764–769. [DOI] [PubMed] [Google Scholar]
- de La Fortelle, E. and Bricogne, G. 1997. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276 472–494. [DOI] [PubMed] [Google Scholar]
- De Meester, I., Vanhoof, G., Hendriks, D., Demuth, H.U., Yaron, A., and Scharpe, S. 1992. Characterization of dipeptidyl peptidase IV (CD26) from human lymphocytes. Clin. Chim. Acta 210 23–34. [DOI] [PubMed] [Google Scholar]
- De Meester, I., Korom, S., Van Damme, J., and Scharpe, S. 1999. CD26, let it cut or cut it down. Immunol. Today 20 367–375. [DOI] [PubMed] [Google Scholar]
- Drucker, D.J. 2003. Therapeutic potential of dipeptidyl peptidase IV inhibitors for the treatment of type 2 diabetes. Expert Opin. Investig. Drugs 12 87–100. [DOI] [PubMed] [Google Scholar]
- Durinx, C., Lambeir, A.M., Bosmans, E., Falmagne, J.B., Berghmans, R., Haemers, A., Scharpe, S., and De Meester, I. 2000. Molecular characterization of dipeptidyl peptidase activity in serum: Soluble CD26/dipeptidyl peptidase IV is responsible for the release of X-Pro dipeptides. Eur. J. Biochem. 267 5608–5613. [DOI] [PubMed] [Google Scholar]
- Engel, M., Hoffmann, T., Wagner, L., Wermann, M., Heiser, U., Kiefersauer, R., Huber, R., Bode, W., Demuth, H.U., and Brandstetter, H. 2003. The crystal structure of dipeptidyl peptidase IV (CD26) reveals its functional regulation and enzymatic mechanism. Proc. Natl. Acad. Sci. 100 5063–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco, R., Valenzuela, A., Lluis, C., and Blanco, J. 1998. Enzymatic and extraenzymatic role of ecto-adenosine deaminase in lymphocytes. Immunol. Rev. 161 27–42. [DOI] [PubMed] [Google Scholar]
- Fulop, V., Bocskei, Z., and Polgar, L. 1998. Prolyl oligopeptidase: An unusual β-propeller domain regulates proteolysis. Cell 94 161–170. [DOI] [PubMed] [Google Scholar]
- Fulop, V., Szeltner, Z., and Polgar, L. 2000. Catalysis of serine oligopeptidases is controlled by a gating filter mechanism. EMBO Rep. 1 277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghersi, G., Chen, W., Lee, E.W., and Zukowska, Z. 2001. Critical role of dipeptidyl peptidase IV in neuropeptide Y-mediated endothelial cell migration in response to wounding. Peptides 22 453–458. [DOI] [PubMed] [Google Scholar]
- Gorrell, M.D., Gysbers, V., and McCaughan, G.W. 2001. CD26: A multifunctional integral membrane and secreted protein of activated lymphocytes. Scan. J. Immunol. 54 249–264. [DOI] [PubMed] [Google Scholar]
- Hegen, M., Kameoka, J., Dong, R.P., Morimoto, C., and Schlossman, S.F. 1997. Structure of CD26 (dipeptidyl peptidase IV) and function in human T cell activation. Adv. Exp. Med. Biol. 421 109–116. [DOI] [PubMed] [Google Scholar]
- Hinke, S.A., Pospisilik, J.A., Demuth, H.U., Mannhart, S., Kuhn-Wache, K., Hoffmann, T., Nishimura, E., Pederson, R.A., and McIntosh, C.H. 2000. Dipeptidyl peptidase IV (DPIV/CD26) degradation of glucagon. Characterization of glucagon degradation products and DPIV-resistant analogs. J. Biol. Chem. 275 3827–3834. [DOI] [PubMed] [Google Scholar]
- Hiramatsu, H., Kyono, K., Higashiyama, Y., Fukushima, C., Shima, H., Sugiyama, S., Inaka, K., Yamamoto, A., and Shimizu, R. 2003. The structure and function of human dipeptidyl peptidase IV, possessing a unique eight-bladed β-propeller fold. Biochem. Biophys. Res. Commun. 302 849–854. [DOI] [PubMed] [Google Scholar]
- Hosfield, D., Palan, J., Hilgers, M., Scheibe, D., McRee, D.E., and Stevens, R.C. 2003. A fully integrated protein crystallization platform for small-molecule drug discovery. J. Struct. Biol. 142 207–217. [DOI] [PubMed] [Google Scholar]
- Iwaki-Egawa, S., Watanabe, Y., Kikuya, Y., and Fujimoto, Y. 1998. Dipeptidyl peptidase IV from human serum: Purification, characterization, and N-terminal amino acid sequence. J. Biochem. 124 428–433. [DOI] [PubMed] [Google Scholar]
- Kahne, T., Lendeckel, U., Wrenger, S., Neubert, K., Ansorge, S., and Reinhold, D. 1999. Dipeptidyl peptidase IV: A cell surface peptidase involved in regulating T cell growth (review). Int. J. Mol. Med. 4 3–15. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Lambeir, A.M., Proost, P., Durinx, C., Bal, G., Senten, K., Augustyns, K., Scharpe, S., Van Damme, J., and De Meester, I. 2001a. Kinetic investigation of chemokine truncation by CD26/dipeptidyl peptidase IV reveals a striking selectivity within the chemokine family. J. Biol. Chem. 276 29839–29845. [DOI] [PubMed] [Google Scholar]
- Lambeir, A.M., Durinx, C., Proost, P., Van Damme, J., Scharpe, S., and De Meester, I. 2001b. Kinetic study of the processing by dipeptidyl-peptidase IV/CD26 of neuropeptides involved in pancreatic insulin secretion. FEBS Lett. 507 327–330. [DOI] [PubMed] [Google Scholar]
- Lambeir, A.M., Proost, P., Scharpe, S., and De Meester, I. 2002. A kinetic study of glucagon-like peptide-1 and glucagon-like peptide-2 truncation by dipeptidyl peptidase IV, in vitro. Biochem. Pharmacol. 64 1753–1756. [DOI] [PubMed] [Google Scholar]
- Lambeir, A.M., Durinx, C., Scharpe, S., and De Meester, I. 2003. Dipeptidyl-peptidase IV from bench to bedside: An update on structural properties, functions, and clinical aspects of the enzyme DPP IV. Crit. Rev. Clin. Lab. Sci. 40 209–294. [DOI] [PubMed] [Google Scholar]
- Leiting, B., Pryor, K.D., Wu, J.K., Marsilio, F., Patel, R.A., Craik, C.S., Ellman, J.A., Cummings, R.T., and Thornberry, N.A. 2003. Catalytic properties and inhibition of proline-specific dipeptidyl peptidases II, IV and VII. Biochem J. 371 525–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, A.G., Powell, H.R., Winter, G., Svensson, O., Spruce, D., McSweeney, S., Love, D., Kinder, S., Duke, E., and Nave, C. 2002. Automation of the collection and processing of X-ray diffraction data—A generic approach. Acta Crystallogr. D Biol. Crystallogr. 58 1924–1928. [DOI] [PubMed] [Google Scholar]
- McRee, D.E. 1999. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125 156–165. [DOI] [PubMed] [Google Scholar]
- Mentlein, R. 1999. Dipeptidyl-peptidase IV (CD26)—Role in the inactivation of regulatory peptides. Regul. Pept. 85 9–24. [DOI] [PubMed] [Google Scholar]
- Mentlein, R., Dahms, P., Grandt, D., and Kruger, R. 1993. Proteolytic processing of neuropeptide Y and peptide YY by dipeptidyl peptidase IV. Regul. Pept. 49 133–144. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A. and Bacon, D.J. 1997. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 277 505–524 [DOI] [PubMed] [Google Scholar]
- Morimoto, C. and Schlossman, S.F. 1998. The structure and function of CD26 in the T-cell immune response. Immunol. Rev. 161 55–70. [DOI] [PubMed] [Google Scholar]
- Morrison, M.E., Vijayasaradhi, S., Engelstein, D., Albino, A.P., and Houghton, A.N. 1993. A marker for neoplastic progression of human melanocytes is a cell surface ectopeptidase. J. Exp. Med. 177 1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardini, M. and Dijkstra, B.W. 1999. α/β hydrolase fold enzymes: The family keeps growing. Curr. Opin. Struct. Biol. 9 732–737. [DOI] [PubMed] [Google Scholar]
- Neer, E.J. and Smith, T.F. 1996. G protein heterodimers: New structures propel new questions. Cell 84 175–178. [DOI] [PubMed] [Google Scholar]
- Oefner, C., D’Arcy, A., Mac Sweeney, A., Pierau, S., Gardiner, R., and Dale, G.E. 2003. High-resolution structure of human apo dipeptidyl peptidase IV/CD26 and its complex with 1-[([2-[(5-iodopyridin-2-yl)amino]-ethyl] amino)-acetyl]-2-cyano-(S)-pyrrolidine. Acta Crystallogr. D Biol. Crystallogr. 59 1206–1212. [DOI] [PubMed] [Google Scholar]
- Ogata, S., Misumi, Y., Tsuji, E., Takami, N., Oda, K., and Ikehara, Y. 1992. Identification of the active site residues in dipeptidyl peptidase IV by affinity labeling and site-directed mutagenesis. Biochemistry 31 2582–2587. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Pauly, R.P., Demuth, H.U., Rosche, F., Schmidt, J., White, H.A., Lynn, F., McIntosh, C.H., and Pederson, R.A. 1999. Improved glucose tolerance in rats treated with the dipeptidyl peptidase IV (CD26) inhibitor Ile-thiazolidide. Metabolism 48 385–389. [DOI] [PubMed] [Google Scholar]
- Rasmussen, H.B., Branner, S., Wiberg, F.C., and Wagtmann, N. 2003. Crystal structure of human dipeptidyl peptidase IV/CD26 in complex with a substrate analog. Nat. Struct. Biol. 10 19–25. [DOI] [PubMed] [Google Scholar]
- Sheldrick, G. 1998. Direct methods for solving macromolecular structures. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Thoma, R., Loffler, B., Stihle, M., Huber, W., Ruf, A., and Hennig, M. 2003. Structural basis of proline-specific exopeptidase activity as observed in human dipeptidyl peptidase-IV. Structure 11 947–959. [DOI] [PubMed] [Google Scholar]
- Topf, M., Varnai, P., Schofield, C.J., and Richards, W.G. 2002a. Molecular dynamics simulations of the acyl-enzyme and the tetrahedral intermediate in the deacylation step of serine proteases. Proteins 47 357–369. [DOI] [PubMed] [Google Scholar]
- Topf, M., Varnai, P., and Richards, W.G. 2002b. Ab initio QM/MM dynamics simulation of the tetrahedral intermediate of serine proteases: Insights into the active site hydrogen-bonding network. J. Am. Chem. Soc. 124 14780–14788. [DOI] [PubMed] [Google Scholar]
- Wilmouth, R.C., Edman, K., Neutze, R., Wright, P.A., Clifton, I.J., Schneider, T.R., Schofield, C.J., and Hajdu, J. 2001. X-ray snapshots of serine protease catalysis reveal a tetrahedral intermediate. Nat. Struct. Biol. 8 689–694. [DOI] [PubMed] [Google Scholar]
- Zhu, L., Tamvakopoulos, C., Xie, D., Dragovic, J., Shen, X., Fenyk-Melody, J.E., Schmidt, K., Bagchi, A., Griffin, P.R., Thornberry, N.A., et al. 2003. The role of dipeptidyl peptidase IV in the cleavage of glucagon family peptides: In vivo metabolism of pituitary adenylate cyclase activating polypeptide-(1–38). J. Biol. Chem. 278 22418–22423. [DOI] [PubMed] [Google Scholar]