Abstract
Chalcone isomerase, an enzyme in the isoflavonoid pathway in plants, catalyzes the cyclization of chalcone into (2S)-naringenin. Chalcone isomerase sequence family and three-dimensional fold appeared to be unique to plants and has been proposed as a plant-specific gene marker. Using sensitive methods of sequence comparison and fold recognition, we have identified genes homologous to chalcone isomerase in all completely sequenced fungi, in slime molds, and in many gammaproteobacteria. The residues directly involved in the enzyme’s catalytic function are among the best conserved across species, indicating that the newly discovered homologs are enzymatically active. At the same time, fungal and bacterial species that have chalcone isomerase-like genes tend to lack the orthologs of the upstream enzyme chalcone synthase, suggesting a novel variation of the pathway in these species.
Keywords: Isoflavonoid pathway, chalcone isomerase, comparative sequence analysis
On the heels of the complete genome sequencing, the projects of large-scale determination of three-dimensional structures representative of many protein families are making rapid progress (Burley and Bonanno 2002). One venue of exploring the increasingly dense protein structure space is to assess the usage of distinct spatial folds by different taxonomic lineages, aiming at the reconstruction of ancient fold repertoires and paths of their evolution (Orengo et al. 2001; Qian et al. 2001; Hegyi et al. 2002). Many spatial folds are ubiquitously found, and have apparently evolved prior to diversification of the major three domains of life, bacteria, archaea, and eukarya (Anantharaman et al. 2002; Aravind et al. 2002; Leipe et al. 2002). Other folds have limited distribution, most likely as a result of their emergence later in evolution, or lineage-specific gene losses (Aravind et al. 2000). Determination of the subsets of folds specific to a given lineage is of interest, because such subsets mark evolutionary transitions, and also because understanding of their functions may shed light on the biological differences between species that make use of these folds and those that lack them. Similar information can be obtained, at even larger scale, by analyzing the taxonomic distribution of conserved sequence families, whether or not their spatial structure is known.
Several differences in fold usage between bacteria and eukaryotes have been reported (Wolf et al. 1999; Lin and Gerstein 2000; Qian et al. 2001). In eukaryotes, certain lineage-specific expansions of gene families have been noticed, such as, for example, the LRR-NBS class of apoptosis signaling genes that has many dozens of members in each plant genome, but only a few genes in animal genomes, G protein-coupled receptors in animals (also found, at a limited scale, in fungi), or serine-threonine-tyrosine-like protein kinases in all species (Lespinet et al. 2002). Not much is known, however, about sequences and structures that would uniquely mark a major kingdom of multicellular eukaryotes—plants or animals. Chalcone isomerase (CHI; EC 5.5.1.6), an enzyme in the isoflavonoid pathway in plants (http://www.genome.ad.jp/dbget-bin/get_pathway?org_name=ot&mapno=00940), has been proposed as one such plant-specific gene marker, based on the apparent lack of similarity to any nonplant sequences or structures (Jez et al. 2000).
The isoflavonoid pathway in plants is the source of anthocyanins, which are used as floral pigments, as protectors against UV photodamage, and as second messengers for Rhizobium nodulation (Weisshaar and Jenkins 1998). Some anthocyanins exhibit antioxidant, antiasthmatic, antimalarial, antimicrobial, and other medically relevant properties, and have beneficial roles as components of human diet (Dixon and Steele 1999). Chalcone isomerase catalyzes the cyclization of chalcone (4,2′,4′,6′-tetrahydroxychalcone) and 6′-deoxychalcone (4,2′,4′-trihydroxychalcone), into, respectively, (2S)-naringenin (5,7,4′-trihydroxyflavanone), the key precursor of anthocyanins, and (2S)-5-deoxyflavanone (7,4′-dihydroxyflavanone). Both CHI substrates are synthesized by the upstream enzyme chalcone synthase (CHS, E.C. 2.3.1.74), in a multistep reaction that includes a series of additions of malonyl-CoA-derived acetate units to a phenylpropanoid precursor Coumaroyl-CoA (4-hydroxy-cinnamoyl-CoA).
CHS is a member of the thiolase superfamily, which includes enzymes involved in biosynthesis of various polyketide compounds. Phylogenetic analysis, corroborated by the biochemical data, defines several families of thiolases with distinct specificities and biological functions. In addition to CHS enzymes, the thiolase superfamily includes: β-ketoacyl-ACP synthases involved in lipid metabolism and broadly distributed in bacteria and eukarya; bacterial enzymes involved in polyketide antibiotic biosynthesis; hydroxymethylglutaryl-CoA synthases, catalyzing the first committed step in the mevalonate pathway of isoprenoid biosynthesis in eukaryotes, archaea, and a limited set of bacteria; and several families of bacterial, archaeal, and eukaryotic enzymes with unknown specificity (Smit and Mushegian 2000; Olsen et al. 2001). The CHS clade of the thiolase superfamily includes plant CHS enzymes with different specificities and a group of poorly characterized bacterial enzymes, found in at least five taxonomically distinct groups, alphaproteobacteria, gammaproteobacteria, bacilli, actinomycetales, and deinococcales (Table 1).
Table 1.
Occurrence of chalcone synthases and chalcone isomerases in completely sequenced or extensively covered genomes of bacteria and fungi
| Taxonomy | CHS | CHI |
| Bacteria | ||
| Actinomycetales | ||
| Mycobacterium tuberculosis H37Rv | 15608798a | none |
| Streptomyces coelicolor | 21225931a | none |
| Saccharopolyspora erythraea | 20384882 | none |
| Amycolatopsis mediterranei | 15131510 | none |
| Bacilli | ||
| Bacillus subtilis | 16079263 | none |
| Bacillus halodurans | 15613180 | none |
| Oceanobacillus iheyensis HTE831 | 23099203 | none |
| Deinococcales | ||
| Deinococcus radiodurans | 15807085 | none |
| Alphaproteobacteria | ||
| Magnetospirillum magnetotacticum | 23010812 | none |
| Rhodobacter sphaeroides | 22958576 | none |
| Rhodospirillum centenum | 5499727 | none |
| Rhizobium etli | 21492939 | none |
| Gammaproteobacteria | ||
| Pseudomonas fluorescens | 1163918 | none |
| Pseudomonas fluorescens PfO-1 | paralogs only | 23062459 |
| Pseudomonas putida KT2440 | paralogs only | 26989448 |
| Pseudomonas syringae pv. syringae B728a | paralogs only | 23471621 |
| Vibrio cholerae | paralogs only | 15641137 |
| Vibrio parahaemolyticus RIMD 2210633 | paralogs only | 28897899 |
| Pasteurella multocida | paralogs only | 15602689 |
| Haemophilus influenzae Rd | paralogs only | 16273308 |
| Ralstonia metallidurans | paralogs only | 22981147 |
| Shewanella oneidensis MR-1 | paralogs only | 24374786 |
| Eukaryotes | ||
| Fungi | ||
| Ascomycetes | ||
| Saccharomyces cerevisiae | paralogs only | 6321992, 6321993 |
| Schizosaccharomyces pombe | paralogs only | 19114577 |
| Neurospora crassa | paralogs only | 11359422 |
a This species contain a family of recently duplicated co-orthologs of CHS genes.
Broad distribution of CHS orthologs and universal presence of at least one CHS paralog in almost every cellular life form with a completely sequenced genome are thus in contrast with the apparently plant-specific nature of the downstream enzyme CHI. This raises interesting questions about evolution of isoflavonoid metabolism and of biochemical pathways in general. Can it be that only plants can isomerize chalcones? If so, does the presence of corresponding genes determine “plantness”? At what point in the evolution of life did the chalcone isomerase family and fold emerge, and is it possible to reconstruct any of its predecessors?
Results and Discussion
We searched the nonredundant protein sequence database at the National Center for Biotechnology Information using CHI family members as queries and the PSI-BLAST program (Altschul et al. 1997), setting the threshold for inclusion into the probabilistic model at random match probability E = 0.01, and trying both composition-corrected and -uncorrected statistics (−t option).
Using the sequence from Medicago sativa (GenBank ID 116134) as the query, after detecting many plant CHI-like enzymes, we found, at the fourth iteration, uncharacterized proteins from Neurospora crassa (gi 28923131) with an E-value of 2 × 10−11 and, at the fifth iteration, the ortholog from Saccharomyces cerevisiae (gi 6321992; YHR198c) with an E-value of 6 × 10−44. Homologs from other fungi followed, as well as the uncharacterized protein from bacterium Ralstonia metallidurans (gi 22981147, E-value 0.079; seventh iteration). When these sequences were used as queries in further database searches, we detected statistically significant similarities to a group of proteins from several bacteria. For instance, the homolog from Shewanella oneidensis (gi 24374786), which was not found in the PSI-BLAST search with CHI as the query, was identified after it received an E-value of 4 × 10−51 in the second iteration of the PSI-BLAST with the Ralstonia protein. Statistically significant matches to the members of the extended CHI family were also observed in the unfinished genomes of slime mold Dictyostelium, of Physcomitrella moss (http://www.moss.leeds.ac.uk/blast.html), and of several bacteria and fungi. Every completely sequenced fungal genome contains at least one CHI-like gene (baker’s yeast has two tandemly duplicated genes on chromosome VIII), and many free-living and parasitic gammaproteobateria also have one (Table 1, Fig. 1 ▶).
Figure 1.
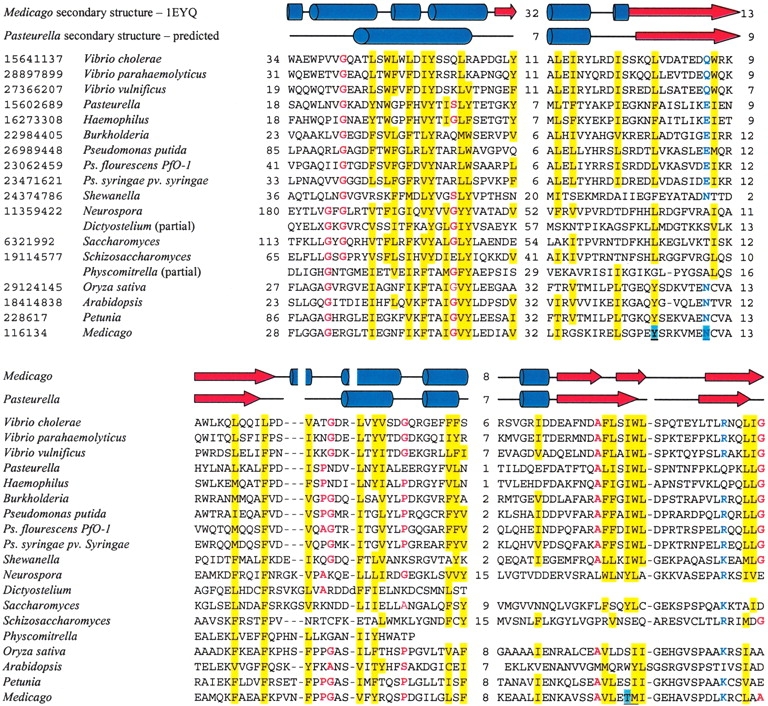
Multiple sequence alignment of CHI-like proteins, constructed with the MACAW program (Schuler et al. 1991). GenBank identifiers are shown before each sequence. Other numbers indicate distance, in amino acid residues, from the N termini and between conserved blocks. In the secondary structure line, red rectangles indicate α-helices and blue rectangles indicate β-strands. In the predicted secondary structure lane, the consensus of the methods included in the META-PP server (Eyrich and Rost 2003) is shown. Yellow shading indicates conserved bulky hydrophobic residues (I,L,V,M,F,Y, and W), red type indicates residues with small side chain (A, G, and S) and structure-breaking P, and blue type indicates conservation of acidic/amide residues (D,E,N, and Q) and basic residues (K and R). Blue shading in the Medicago sequence indicates the residues directly implicated in catalysis.
To cross-validate sequence and structure similarity, we queried the fold prediction metaserver (Ginalski et al. 2003) with bacterial and fungal proteins detected by PSI-BLAST and analyzed the identity of the best matches and their 3D Jury consensus prediction scores. A CHI was ranked as the best-fitting fold against all bacterial and yeast sequences, with similarity scores from 90 to 150, typical of true positives. The highest score for a structure not belonging to the CHI fold (first false positive) was always below 30.
Multiple sequence alignment of plant, fungal, and bacterial proteins with CHI-like fold (Fig. 1 ▶) revealed several consecutive blocks of evolutionary conservation, closely corresponding to the secondary structure elements of plant CHI and readily explainable based on knowledge of the catalytic mechanism (Jez and Noel 2002; Jez et al. 2002).
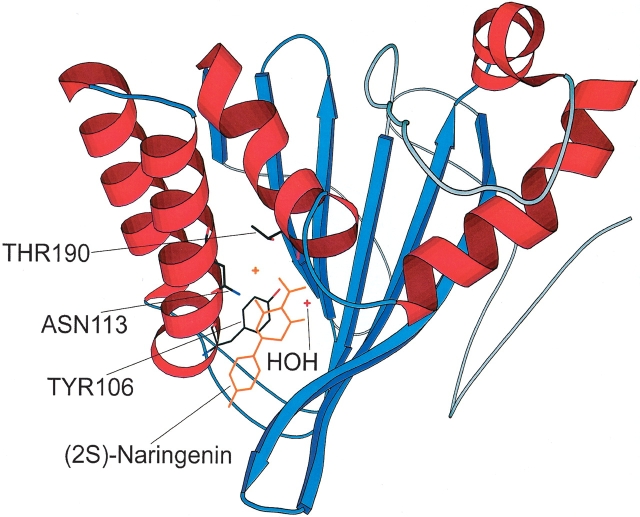
In plant CHI, the isomerization seems to occur via polarization of bound water by the side chain of Tyr 106, followed by water-mediated deprotonation of the 2′-hydroxyl group of chalcone, formation of an oxyanion, and intramolecular attack on the α,β unsaturated double bond (Jez and Noel 2002). In the (2S)-naringenin-binding cleft, a hydrogen bond network links the substrate, a bound water molecule, and five side chains, including Tyr 106. Other interactions between the enzyme and the substrate are predominantly hydrophobic, involving many side chains on the walls of the cleft, although hydrogen bonds from Asn 113 and Thr 190 are also involved (Jez et al. 2002). Analysis of the multiple alignment (Figs. 1 ▶ and 2 ▶) indicates that most of the catalytic core, including the complete substrate-binding cleft, is well conserved in CHI-like proteins from fungi and bacteria. In particular, the set of β-strands forming the large sheet and two α helices on the other side of the cleft are preserved. Asn 113 is among the best conserved residues in the family, and Thr 190 is either conserved or replaced by similarly sized residues, suggesting how hydrogen bonds may be formed between the CHI-like molecules and their substrates. In contrast, Tyr 106 is replaced in CHI-like enzymes by similarly bulky residues that, however, lack a hydroxyl group. Given that the change of Tyr 106 into phenylalanine in alfalfa CHI results in 70-fold reduction in cyclization rate of 6′-deoxychalcone, which is still a 105-fold enhancement over the spontaneous reaction rate, and that maximal reaction rate seems to require the conditions when substantial fraction of the substrate is polarized in solution (Jez and Noel 2002), it is likely that CHI-like proteins retain enzymatic activity even in the absence of conserved tyrosine.
Figure 2.
Mapping of sequence conservation onto the spatial structure of chalcone isomerase from alfalfa (PDB ID 1EYQ). Conserved elements in Figure 1 ▶ are represented in ribbon form; nonconserved elements, by a thin gray wire.
The least conserved sequence segments tend to be located far away from the substrate-binding pocket (Fig. 2 ▶). In particular, the distal loops connecting core elements in bacterial and fungal proteins could not be confidently aligned to the plant sequences, and the pair of solvent-exposed β-hairpins, as well as the bundle of short α helices at the tip of the CHI molecule, are also not conserved and may be missing altogether in bacteria. The distal α-helical region contains several residues involved in dimerization of plant CHI, and their poor conservation in bacteria suggests that bacterial CHI-like proteins are unlikely to form similar dimers.
The origin of CHI fold remains unclear. We were not able to detect any distant relatives of CHI-like superfamily members among the proteins with currently known structures, using a variety of structure comparison methods. The natural substrates of fungal and bacterial CHI-like enzymes are unknown. Analysis of gene content in completely sequenced genomes (Table 1) shows that orthologs of plant CHS and plant CHI tend to exist in these species to the exclusion of one another, and that, most likely, there is no plantlike pathway of naringenin biosynthesis in fungi and bacteria. Whenever a CHS ortholog (presumably, producing a chalconelike compound) is found in bacteria, there is no isomerase, and whenever a chalcone isomerase-like enzyme is found, there is no CHS, so that the substrate of isomerase-like enzyme is unlikely to be chalcone. Thus, an isoflavonoid-like compound appears to be produced and self-cyclicized in fungi and some bacteria, but its chemical identity and biological role remain to be investigated.
We have shown that the CHI-like fold and sequence family is not restricted to flowering plants, but is found in mosses (a deep clade of higher plants), in fungi and early-branching mycetozoa, and in one division (gamma) of proteobacteria. Plants, animals, and fungi are thought to have diverged about 1.6 Ga ago (Hedges and Kumar 2003), whereas the last common ancestor of gammaproteobacteria may have lived more recently, perhaps 0.7 Ga ago (May et al. 2001). The CHI-like enzymes may have emerged in early eukaryotes (failing to establish themselves in animals) and could have been acquired by proteobacteria in just one act of interkingdom gene transfer. Results of phylogenetic analysis are compatible with such a scenario, in that plant, fungal, and bacterial CHI-like enzymes form three compact clades distant from each other, indicating ancient divergence and/or rapid evolution (data not shown). Other evolutionary scenarios, such as, for example, early bacterial origin and symbiotic acquisition of CHI by a eukaryotic lineage that preceded the divergence of plants and fungi, would involve gene losses in some eukaryotes and in multiple bacterial lineages. Sequencing of simple eukaryotes and additional bacteria will be required to time the emergence and track the routes of dissemination of the chalcone isomerase-like fold in various kingdoms of life.
In conclusion, the distal part of the isoflavonoid biosynthesis pathway, leading to the anthocyanin precursor (2S)-naringenin, appears to be plant-specific from the chemical point of view. At the genome level, however, this biochemical innovation was enabled by the recruitment and co-adaptation of enzymes from preexisting broadly distributed families and folds.
Acknowledgments
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03395404.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman, V., Koonin, E.V., and Aravind, L. 2002. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 30 1427–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., Watanabe, H., Lipman, D.J., and Koonin, E.V. 2000. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl. Acad. Sci. 97 11319–11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., Anantharaman, V., and Koonin, E.V. 2002. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: Implications for protein evolution in the RNA world. Proteins 48 1–14. [DOI] [PubMed] [Google Scholar]
- Burley, S.K. and Bonanno, J.B. 2002. Structuring the universe of proteins. Annu. Rev. Genomics Hum. Genet. 3 243–262. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A. and Steele, C.L. 1999. Flavonoids and isoflavonoids—A gold mine for metabolic engineering. Trends Plant Sci. 4 394–400. [DOI] [PubMed] [Google Scholar]
- Eyrich, V.A. and Rost, B. 2003. META-PP: Single interface to crucial prediction servers. Nucleic Acids Res. 31 3308–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginalski, K., Elofsson, A., Fischer, D., and Rychlewski, L. 2003. 3D-Jury: A simple approach to improve protein structure predictions. Bioinformatics 19 1015–1018. [DOI] [PubMed] [Google Scholar]
- Hedges, S.B. and Kumar, S. 2003. Genomic clocks and evolutionary timescales. Trends Genet. 19 200–206. [DOI] [PubMed] [Google Scholar]
- Hegyi, H., Lin, J., Greenbaum, D., and Gerstein, M. 2002. Structural genomics analysis: Characteristics of atypical, common, and horizontally transferred folds. Proteins 47 126–141. [DOI] [PubMed] [Google Scholar]
- Jez, J.M. and Noel, J.P. 2002. Reaction mechanism of chalcone isomerase. pH dependence, diffusion control, and product binding differences. J. Biol. Chem. 277 1361–1369. [DOI] [PubMed] [Google Scholar]
- Jez, J.M., Bowman, M.E., Dixon, R.A., and Noel, J.P. 2000. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat. Struct. Biol. 7 786–791. [DOI] [PubMed] [Google Scholar]
- Jez, J.M., Bowman, M.E., and Noel, J.P. 2002. Role of hydrogen bonds in the reaction mechanism of chalcone isomerase. Biochemistry 41 5168–5176. [DOI] [PubMed] [Google Scholar]
- Leipe, D.D., Wolf, Y.I., Koonin, E.V., and Aravind, L. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317 41–72. [DOI] [PubMed] [Google Scholar]
- Lespinet, O., Wolf, Y.I., Koonin, E.V., and Aravind, L. 2002. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 12 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. and Gerstein, M. 2000. Whole-genome trees based on the occurrence of folds and orthologs: Implications for comparing genomes on different levels. Genome Res. 10 808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, B.J., Zhang, Q., Li, L.L., Paustian, M.L., Whittam, T.S., and Kapur, V. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. 98 3460–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, J.G., Kadziola, A., von Wettstein-Knowles, P., Siggaard-Andersen, M., and Larsen, S. 2001. Structures of β-ketoacyl-acyl carrier protein synthase I complexed with fatty acids elucidate its catalytic machinery. Structure 9 233–243. [DOI] [PubMed] [Google Scholar]
- Orengo, C.A., Sillitoe, I., Reeves, G., and Pearl, F.M. 2001. Review: What can structural classifications reveal about protein evolution? J. Struct. Biol. 134 145–165. [DOI] [PubMed] [Google Scholar]
- Qian, J., Luscombe, N.M., and Gerstein, M. 2001. Protein family and fold occurrence in genomes: Power-law behaviour and evolutionary model. J. Mol. Biol. 313 673–681. [DOI] [PubMed] [Google Scholar]
- Schuler, G.D., Altschul, S.F., and Lipman, D.J. 1991. A workbench for multiple alignment construction and analysis. Proteins 9 180–190. [DOI] [PubMed] [Google Scholar]
- Smit, A. and Mushegian, A. 2000. Biosynthesis of isoprenoids via mevalonate in Archaea: The lost pathway. Genome Res. 10 1468–1484. [DOI] [PubMed] [Google Scholar]
- Weisshaar, B. and Jenkins, G.I. 1998. Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1 251–257. [DOI] [PubMed] [Google Scholar]
- Wolf, Y.I., Brenner, S.E., Bash, P.A., and Koonin, E.V. 1999. Distribution of protein folds in the three superkingdoms of life. Genome Res. 9 17–26. [PubMed] [Google Scholar]