Abstract
Spontaneous conformational transition of the prion protein from an α-helical isoform to a β-sheet-rich isoform underlies the pathogenesis of sporadic prion diseases. To study the rate-limiting steps of spontaneous conversion, the formation of amyloid fibrils by the recombinant human PrP C-terminal fragment spanning residues 90–231 (recPrP) was monitored in the presence of urea. The kinetics of spontaneous fibril formation displayed sigmoidal behavior involving a lag phase. The shortest lag phase was observed at partially denaturing conditions, close to the concentration of urea corresponding to the middle point of unfolding. This result indicates that unfolding intermediates may be important for the conversion. To test whether unfolding intermediates are formed, we employed size-exclusion chromatography and circular dichroism spectroscopy to monitor urea denaturation of recPrP. Both techniques showed a single sigmoidal transition with very similar thermodynamic parameters of denaturation and that the transition can be described by a simple equilibrium between folded and denatured states. Detailed analyses of data, however, revealed that the dimensions of both the native and denatured species gradually increases with urea. Expansion of the native species is also accompanied by an increase in efficiency of the energy transfer from a single Trp residue to 1-anilinonaphthalene-8-sulfonate dye as measured by fluorescence. These data illustrate that thermodynamic character of the native ensemble changes gradually with environmental conditions. Such behavior is consistent with the thermodynamically variable model, and may be linked to the ability of PrP to adopt distinct abnormal conformations under pathologic conditions.
Keywords: prion protein, conformational transition, amyloid fibrils, unfolding intermediates, size exclusion chromatography
Spontaneous conversion of the normal cellular isoform of the prion protein, PrPC, into the abnormal pathological isoform, PrPSc, underlies sporadic forms of prion disease, including Creutzfeldt-Jakob disease in humans (Prusiner 2001). Sporadic Creutzfeldt-Jakob disease accounts for approximately 85% of all cases of prion disease. Elucidating the mechanism of the conversion of PrPC into PrPSc is important for the development of effective therapeutic strategies. The process of conversion requires a substantial conformational change: PrPC is a proteinase K-sensitive, α-helical monomer, whereas PrPSc is a proteinase K-resistant, aggregated β-sheet-rich multimer (Pan et al. 1993). At present, fundamental questions about the conformational transition remain unanswered. Why is the spontaneous conversion of PrPC into PrPSc so rare? What is the rate-limiting step in the conformational transition?
Very limited information about the initial stages of the conformational transition is available. In our current view, the early events of PrPSc formation involve a precursor species with a partially unstructured character (Cohen and Prusiner 1998). Substantial efforts have been devoted to identifying such species. Hydrogen-exchange experiments did not reveal any low-abundance states different from PrPC present under native conditions (Hosszu et al. 1999; Nicholson et al. 2002). However, kinetic folding intermediates of PrPC were suggested as possible candidates of a precursor. Whether or not such intermediates exist is controversial (Wildegger et al. 1999; Apetri and Surewicz 2002). Alternatively, PrPSc may be generated from equilibrium intermediates of PrPC unfolding. Earlier studies revealed that the folded α-helical recPrP (α-recPrP) undergoes a three-state, denaturant-induced or temperature-induced unfolding process under acidic conditions, showing an apparent equilibrium intermediate with a β-sheet-rich character (Swietnicki et al. 1997; Zhang et al. 1997; Hornemann and Glockshuber 1998). More detailed characterization of the denaturation profile demonstrated that the β-sheet-rich isoform is not an equilibrium intermediate in the unfolding of α-recPrP, and is formed only upon the oligomerization of recPrP (Baskakov et al. 2001; Morillas et al. 2001). At acidic pH, the β-sheet-rich isoform is thermodynamically more stable than the α-helical isoform (Baskakov et al. 2001). However, the β-sheet-rich isoform is less accessible kinetically and, for this reason, it may be misidentified as an equilibrium intermediate under partially denaturing conditions.
Up to now the biophysical nature of the transition state to PrPSc remains elusive. To approach this question we studied the kinetics of in vitro conversion of C-terminal fragment of recombinant human PrP that encompassed residues 90–231 (designated recPrP) to amyloid fibrils. We found that partially denaturing conditions favored fibril formation that indicates the importance of nonnative states for the conversion. Because the presence of unfolding intermediates still remains unclear, we investigated this question further and monitored urea-induced denaturation of α-recPrP using two techniques: circular dichroism (CD) and size-exclusion chromatography (SEC). Both SEC and CD showed very similar ΔG and m values, with no indication of unfolding intermediates. However, at low concentrations of urea, SEC and fluorescence energy-transfer studies demonstrated a change in thermodynamic character of the native ensemble. Our data indicate that the thermodynamic character of the native ensemble may gradually change as a function of environmental conditions. We speculate that such variable thermodynamic behavior of the native ensemble may be linked to the intrinsic ability of PrP to adopt different abnormal conformations under pathologic conditions.
Results
Does fibril formation occur through unfolding of α-recPrP?
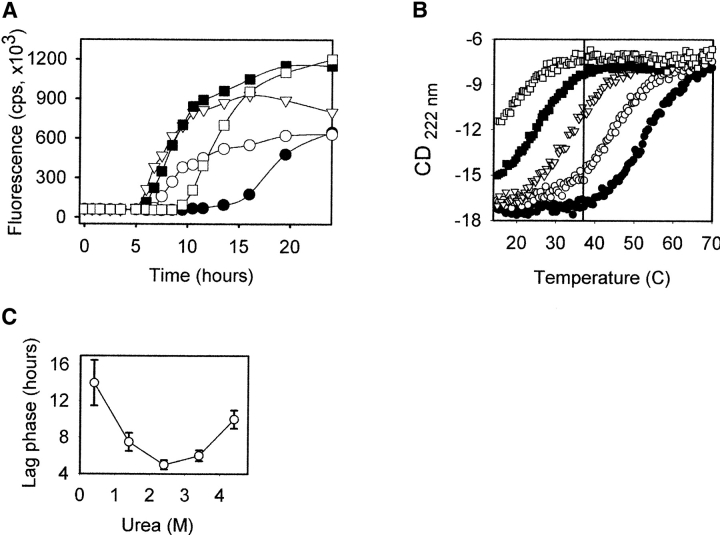
To test whether the conversion of the α-monomer to the amyloid fibrils requires unfolding we tested the effect of urea on the kinetics of fibril formation. Typical kinetics of spontaneous in vitro conversion of recPrP at 37°C showed a substantial lag-phase followed by a rapid increase of fibril accumulation as measured by Thioflavin T (ThT) fluorescence. The length of the lag phase varied substantially with urea concentration (Fig. 1A ▶). We observed that the shortest lag phase was in 2.4 M urea, while either the decrease in the urea concentration below 2.4 M or the increase above 2.4 M resulted in the longer lag phases (Fig. 1C ▶).
Figure 1.
(A) Kinetic traces for recPrP (0.3 mg/mL) monitored by ThT fluorescence in the presence of 1 M GndHCl at pH 6.5 at 37°C as a function of concentration of urea: 0.4 M urea (filled circles); 1.4 M urea (empty circles); 2.4 M urea (empty triangles); 3.4 M urea (filled squares); 4.4 M urea (empty squares). The differences in ThT fluorescence observed at the final stage of the reaction (25 h) represent different yields of amyloid formation as recPrP undergoes nonspecific aggregation at low concentrations of urea. (B) The thermal denaturation of recPrP (0.2 mg/mL) as monitored by CD at 222 nm in the presence of 1 M GndHCl at pH 6.5 as a function of concentration of urea. The symbol definitions are the same as for A. The vertical bar at 37°C corresponds to the experimental conditions at which the kinetics of amyloid formation was monitored. recPrP displayed almost complete reversible denaturation behavior; minor hysteresis between the unfolding and the refolding curves (10%–20%) was observed after cooling down. (C) Dependence of the length of the lag phase of fibril formation on the concentration of urea. Data points represent the mean ± standard deviation for triplicate samples.
To determine the fraction of the native versus the unfolded state at each concentration of urea we monitored the temperature-induced denaturation using CD spectroscopy (Fig. 1B ▶). We found that recPrP was predominantly denatured in 4.4 M urea at 37°C, while the protein maintained its native state in 0.4 M urea at 37°C. The unfolding transition occurred between 1.4 M and 3.4 M urea, where both the native and the denatured states were abundant. The midpoint of the unfolding transition was close to the 2.4 M urea, in which the kinetics displayed the shortest lag phase. This experiment illustrates that the conversion to the amyloid fibrils is facilitated at partially denaturing conditions.
Using GndHCl instead of urea we found similar bell-shape dependence of the lag-phase on the concentration of the denaturant, in which the shortest lag-phase was observed at partially denaturing concentrations of GndHCl (data not shown). These data indicate that intermediate concentrations of the denaturants favor the conversion regardless of the chemical nature of the denaturant. The kinetics of fibril formation of murine recPrP 89–231 varied with urea concentration in a very similar way to that of human recPrP 90–231, illustrating that this property is not unique for human recPrP. Considering that the partially denaturing conditions facilitate fibril formation one may speculate that neither the native nor the unfolded states are important for the conversion to the fibrils, but rather, it is low abundant unfolding intermediate populated at partially denaturing conditions that leads to the fibrils.
Have any unfolding intermediates of α-recPrP been identified?
Earlier studies of unfolding intermediates of recPrP yielded conflicting results. The β-sheet-rich isoform of recPrP was originally proposed to be an unfolding intermediate in solvent- and temperature-induced equilibrium denaturation of α-recPrP at acidic pH (Swietnicki et al. 1997; Zhang et al. 1997; Hornemann and Glockshuber 1998). Recent studies demonstrated that (1) the β-isoform is not an equilibrium intermediate in the unfolding of α-recPrP, but is oligomeric and predominantly octameric; (2) recPrP can be folded selectively into either α-helical monomeric or β-sheet-rich oligomeric isoforms; (3) the conformational transition from α-recPrP into the β-oligomer is coupled with oligomerization; (4) the β-oligomer is not on the kinetic pathway to the fibrilar form (Baskakov et al. 2001, 2002). Because the β-oligomer is still referred to as an “unfolding intermediate” (Rezaei et al. 2002; Calzolai and Zahn 2003), it is very important to clarify this issue further.
We found that, unless specific precautious are taken, formation of minor amounts of the β-oligomer often accompanies the refolding of recPrP into the α-helical monomer. At acidic pH (<5.0), the β-oligomer is thermodynamically more stable than α-recPrP, but is less accessible kinetically (Baskakov et al. 2001). For this reason, the β-oligomer accumulates slowly in the process of storage and measurement. Numerous conditions favor the formation of the β-oligomer over α-recPrP: acidic pH, salt, intermediate to high concentrations of denaturants, elevated temperature, and high protein concentrations. For instance, specific conditions employed for solvent- and temperature-induced unfolding (denaturants and high temperature) may lead to the accumulation of substantial amounts of the β-oligomer through the course of an experiment. We found that recPrP assembles into the β-oligomer even under conditions normally considered to be highly denaturing (8 M urea, pH 5.0). Because the β-oligomer is frequently observed concomitantly with α-recPrP, a simple procedure for determining conformational heterogeneity of recPrP is needed.
Determining conformational heterogeneity of refolded recPrP
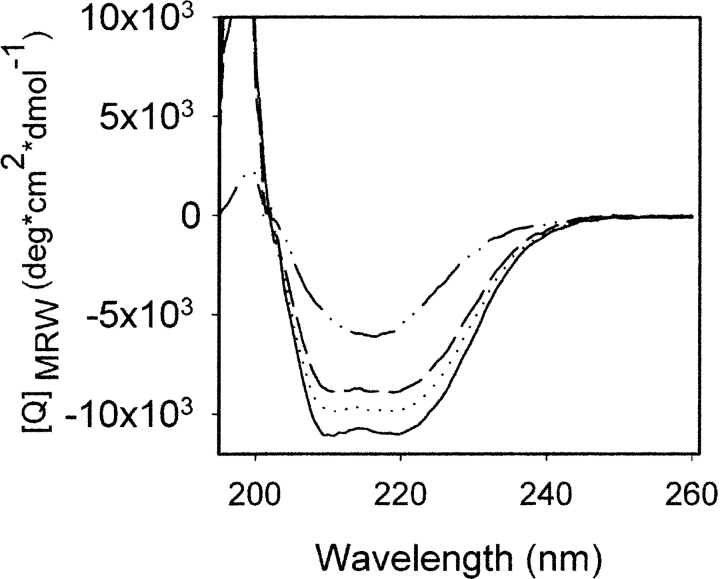
When the β-oligomer is formed, it remains assembled even in a low-salt buffer. Using CD spectroscopy only, it is easy to overlook the presence of minor amounts of the β-oligomer (<20%) in a preparation of α-recPrP. The CD spectra of recPrP samples obtained by mixing α-recPrP with the β-oligomer in 9 : 1 and 8 : 2 ratios are very similar to the spectrum of homogeneous α-recPrP (Fig. 2 ▶).
Figure 2.
Far-UV CD spectra of recPrP (0.25 mg/mL) refolded into either an α-helical monomer (solid line) or a β-sheet-rich oligomer (dotted/dashed line). Mixing of α-recPrP with β-recPrP in 9 : 1 (dotted line) and 8 : 2 (dashed line) ratios gave spectra similar to the spectrum of homogeneous α-recPrP. Spectra were recorded in 20 mM sodium acetate buffer (pH 5.0).
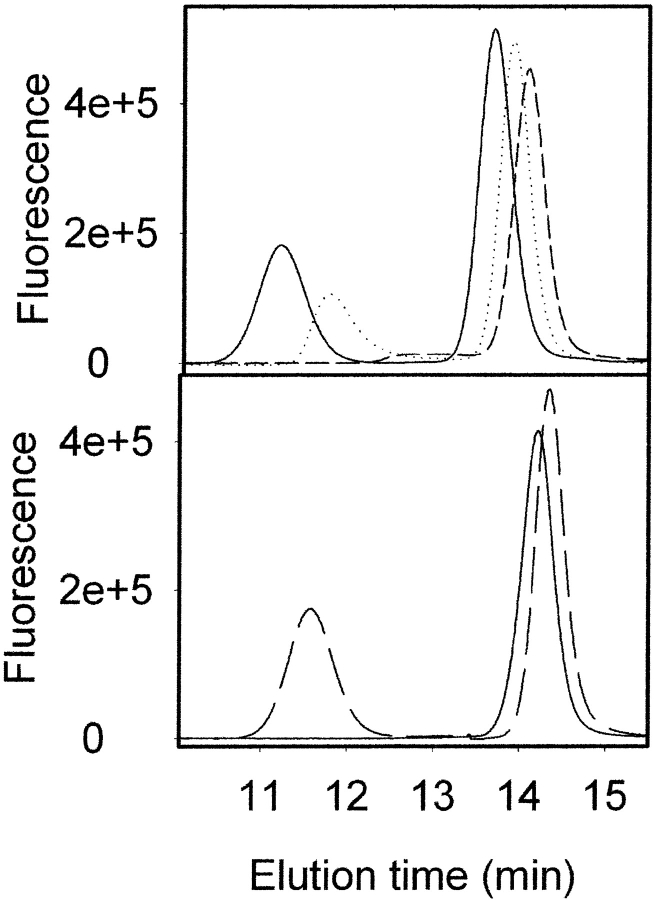
SEC provides an alternative method to determine conformational heterogeneity. As an oligomer, β-recPrP has a different elution volume from monomeric α-recPrP (Fig. 3 ▶). A potential difficulty of using SEC is the high propensity of the β-oligomer to adsorb to the column matrix even under conditions in which α-recPrP does not interact with the column matrix. A sample of recPrP containing 60% α-recPrP and 40% β-recPrP appears as a single peak, corresponding to the α-monomer when running buffer with 2 M urea and pH 5.0 is used (Fig. 3 ▶, bottom panel). However, the same sample is eluted as two peaks, corresponding to the α-monomer and the β-oligomer, with 2 M urea and pH 3.6. In addition to extremely acidic pH, high concentrations of urea (>6 M) at pH 5.0 also prevent adsorption of the β-oligomer to the column matrix. However, at concentrations of urea below 6 M, the β-oligomer peaks are characterized by a substantial increase in the elution time, marked asymmetry, and reduction of the peak area (Fig. 3 ▶, top panel). All of these features indicate an interaction of the β-oligomer with the column matrix. The β-oligomer is undetectable when the concentration of urea drops below 4.5 M at pH 5.0 (Fig. 4A ▶).
Figure 3.
Accessing the oligomerization state of recPrP using SEC. SEC profiles of recPrP containing 60% α-monomer and 40% β-oligomer were monitored (top) in the presence of different concentrations of urea in running buffer at pH 5.0: 4.0 M (dashed line), 4.75 M (dotted line), and 8 M (solid line); and (bottom) in the presence of 2 M urea at pH 3.6 (dashed line) and pH 5.0 (solid line).
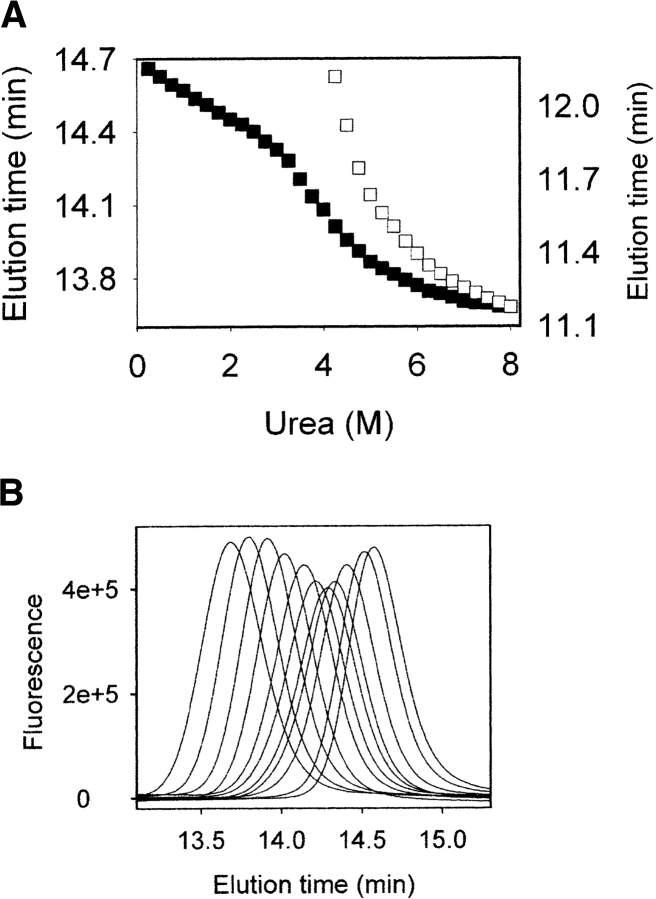
Figure 4.
(A) Dependence of the elution times of the monomer (filled squares, left axis) and the β-oligomer (empty squares, right axis) on urea concentration. The column was equilibrated with 20 mM sodium acetate buffer (pH 5.0) containing the same urea concentrations as samples of recPrP. (B) Elution profiles of monomeric recPrP in the presence of different urea concentrations: 1.0 M (the right-most peak), 1.75 M, 2.5 M, 3.0 M, 3.25 M, 3.5 M, 3.75 M, 4.25 M, 4.75 M, 5.75 M, and 8.0 M (the left-most peak).
Because formation of the β-oligomer is difficult to avoid and its detection is also difficult, special precautions are required. Using nonoptimal running buffer for SEC causes recPrP to appear as a homogeneous α-monomer, as judged by both SEC and CD, even though a substantial fraction of protein exists as the β-oligomer. Under other circumstances, the sample of recPrP containing the β-oligomer with minor amounts of the α-monomer may be confused with a β-monomer, because CD shows the sample as predominantly β-sheet-rich, whereas SEC detects only the monomeric species.
Test of unfolding intermediates
Whether equilibrium denaturation of α-recPrP exhibits a two- or three-state transition remains unclear. Appearance of the β-oligomer in the denaturation curve of α-recPrP in early experiments may obscure detection of true equilibrium intermediates. To investigate this question further, we monitored equilibrium denaturation of α-recPrP free from detectable amounts of the β-oligomer.
Typical elution profiles of α-recPrP monitored by SEC as a function of urea concentration show narrow chromatographic peaks at low urea concentrations, at which folded α-recPrP is the dominant species (Fig. 4B ▶). These peaks widen at the center of the transition and narrow again at high urea concentrations, at which the denatured state is the prevailing species. The α-recPrP undergoes interconversion between native and denatured states throughout the transition region, and the width of the peak reflects the rate of interconversion.
During denaturation, α-recPrP exhibits a single sigmoidal transition, when the elution time of the chromatographic peak is plotted against urea concentration (Fig. 4A ▶). However, the pre- and posttransition baselines have substantial slopes, which result from changes in the permeation properties of the column caused by increasing concentrations of urea. To adjust for the influence of the denaturant on the column matrix, the partition coefficient, Kd, is often used instead of the elution volume (Kd is defined in Materials and Methods; Baskakov and Bolen 1998; Baskakov et al. 2000). Kd takes into account the effects of denaturant on the column, and reflects only changes in the dimensions of the protein species. In a previous study, we showed that the 1/Kd parameter is proportional to the weighted average Stokes radius of rapidly interconverting protein species, and is a valid observable for quantitatively evaluating the thermodynamic parameters in solvent-induced denaturation of proteins (Baskakov and Bolen 1998).
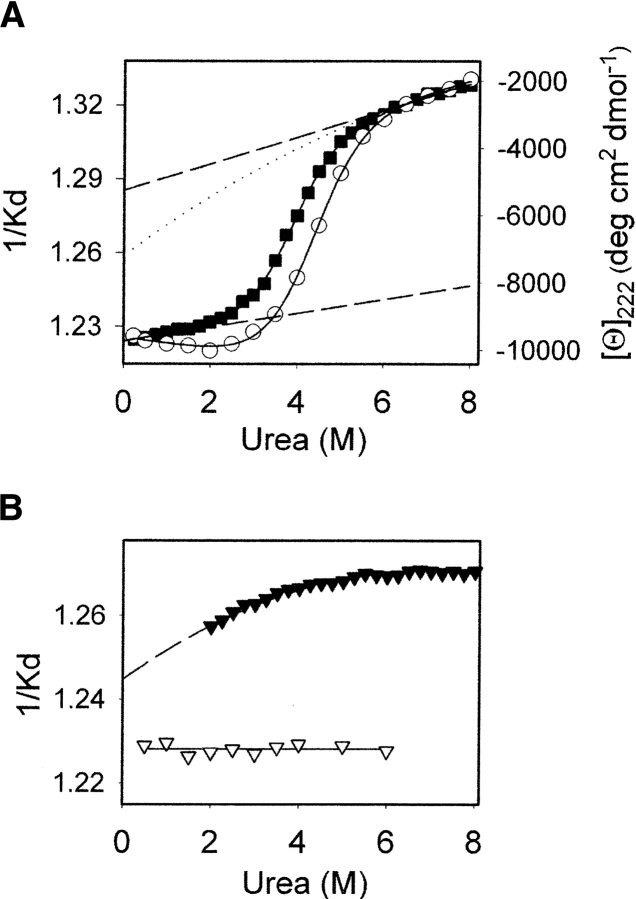
To test the validity of the two-state model of unfolding, we employed a superimposable test using two observable parameters of denaturation: mean residue ellipticity as measured by CD and the 1/Kd value derived from SEC (Fig. 5A ▶). Both CD and SEC techniques show a single sigmoidal transition in urea-induced denaturation. The ΔG0, m, and C1/2 values, three thermodynamic parameters of denaturation, determined from both CD and SEC, were nearly identical (Table 1). This indicates that the transition is well described by the simple equilibrium between the native and denatured states of α-recPrP.
Figure 5.
(A) Urea-induced equilibrium denaturation of α-recPrP is presented as change in 1/Kd (filled squares, left axis) and mean residue ellipticity (empty circles, right axis). The solid line represents the nonlinear least-squares fit of the data to a two-state model using linear extrapolation methods (Santoro and Bolen 1988). Dashed lines are linear extrapolations of (bottom line) the pretransition baseline to the posttransition region and (top line) the posttransition baseline to the pretransition region. Dotted line is an extrapolation of the posttransition baseline determined by fitting to a second-degree polynomial function. (B) 1/Kd values determined for the native state of chymotrypsinogen (empty triangles) and for the unfolded state of recPrP106 (filled triangles). Dashed line represents the fit of the data to a second-degree polynomial function.
Table 1.
Thermodynamic parameters for urea-induced unfolding of α-recPrP determined at pH
| Parameters used for fitting | ΔG (kcal/mole) | m (kcal/mole) | C1/2 (M) |
| Θa | −4.40 ± 0.25 | 1.03 ± 0.06 | 4.2 |
| 1/Kda | −4.44 ± 0.17 | 1.14 ± 0.05 | 3.9 |
| 1/Kdb | −4.57 ± 0.17 | 1.19 ± 0.05 | 3.8 |
a The thermodynamic parameters are results of nonlinear, least-squares best fits of the data using a linear extrapolation method (Santoro and Bolen 1988).
b Nonlinear fitting was performed using a second-degree polynomial for extrapolation of the baseline for the denaturated state.
Variable behavior of the native and denatured states
As we noted earlier, 1/Kd values in the pretransition region represent the dimensions of the native species and 1/Kd values in the posttransition region reflect dimensions of the denatured species, while the 1/Kd value within the transition region is the weighted average size of both native and denatured states. It is anticipated that 1/Kd values in the pre- and posttransition regions remain flat if the dimensions of the native and denatured species are independent of urea concentration. Contrary to this, both pre- and posttransition baselines show a positive slope, indicating that the native and denatured species increase as the concentration of urea increases (Fig. 5A ▶).
To determine whether the sloping baselines reflect changes in the physical properties of the protein species or a shift in parameters not related to the protein, we performed a control experiment monitoring 1/Kd values of chymotrypsinogen as a function of urea concentration, ranging from 0 M to 6 M. Chymotrypsinogen is highly resistant to urea denaturation, and is expected to remain in its native state over the range of urea concentrations used. Indeed, we found that 1/Kd values of chymotrypsinogen remain constant with all urea concentrations (Fig. 5B ▶), which indicates that 1/Kd values are unaffected by the urea-induced changes in the permeation properties of the column and that 1/Kd values represent only changes in the dimensions of the protein species. This result demonstrates that the volume of the native and denatured species of α-recPrP increases as urea concentrations increase.
In evaluating the unfolding transition by SEC, it is necessary to extrapolate the posttransition baseline toward 0 M urea. The linear extrapolation of 1/Kd values for the denatured state reveals considerable contraction of this state approaching 0 M urea (Fig. 5A ▶). To explore the validity of linear extrapolation, we used recPrP106, which is known to exist in the unfolded state even at low concentrations of denaturant (Baskakov et al. 2000). The unfolded state of recPrP106 expands with increasing urea concentration, and the 1/Kd value versus [urea] can be described by a second-degree polynomial (Fig. 5B ▶). These data demonstrate that a second-degree polynomial function is more appropriate than a linear function for accurate extrapolation of the dimensions of the denatured species.
When a second-degree polynomial is incorporated into the two-state model to represent the dependence of the denatured species on urea concentration, the extrapolation of the posttransition baseline to 0 M urea zero gives a very compact unfolded species (Fig. 5A ▶). A nonlinear least-squares analysis of the unfolding curve using a second-degree polynomial for the denatured state gives slightly higher ΔG and m values than those derived with linear extrapolation of the denatured state (Table 1). These thermodynamic parameters determined independently by CD and SEC are in good agreement.
Our data indicate that both the relative populations and physical properties of the native and denatured states of recPrP are dependent on denaturant concentrations. A reduction of the size of the denatured species with decreasing urea concentrations is consistent with the presence of residual structure in the denatured state, as was revealed by hydrogen-deuterium exchange experiments (Hosszu et al. 1999; Nicholson et al. 2002). Such “variable” behavior of the denatured state has been observed in other proteins, while the urea-induced change in properties of the native state is more unusual (Bolen and Yang 2000). To investigate this phenomenon in more detail, we employed ANS-dependent fluorescence.
Urea-induced change within the native state
The hydrophobic dye ANS has been used frequently to study conformational changes in proteins. ANS preferentially binds to hydrophobic protein surfaces. This binding is accompanied by a substantial increase in the fluorescence emission of the dye. It has been shown that ANS binds well to β-sheet-rich isoforms of recPrP, while its affinity to α-recPrP is relatively low (Safar et al. 1994; Baskakov et al. 2002). Despite low affinity, the fluorescence of ANS in the presence of α-recPrP is still much higher than its intrinsic fluorescence in the absence of protein (Fig. 6A ▶).
Figure 6.
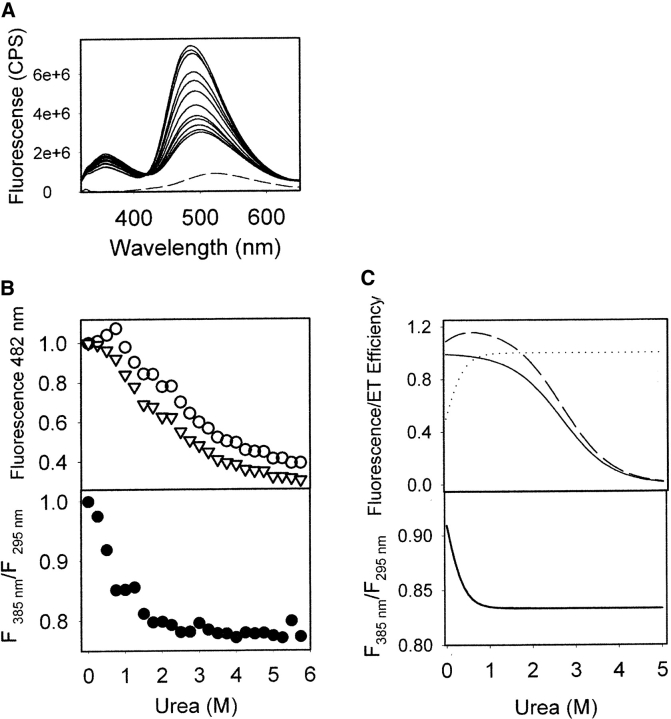
(A) Fluorescence spectra of ANS (100 μM) in the absence (dashed line) and in the presence of α-recPrP (0.15 mg/mL; solid lines) recorded at different concentrations of urea at 480 nm, from top to bottom: 0.5, 0.25, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 4.5, 5.0, 5.5, and 5.75 M upon excitation at 295 nm. (B)(Top) Urea-induced unfolding of α-recPrP monitored by ANS fluorescence upon excitation at 295 nm (empty circles) and at 385 nm (empty triangles). (Bottom) Normalized ratio of ANS fluorescence recorded upon excitation at 385 nm and at 295 nm. The emission wavelength was 482 nm. (C)(Top) The simulated change of ANS fluorescence upon excitation at 385 nm (solid line) and at 295 nm (dashed line). Dotted line represents change in the energy-transfer efficiency. (Bottom) The simulated dependence on urea concentration of the ratio of ANS fluorescence determined upon excitation at 385 nm and 295 nm.
To study the urea-induced change, we monitored ANS fluorescence emission upon excitation either at 295 nm or at 385 nm. Excitation at 385 nm with emission at 482 nm monitors the change in the environment of ANS, whereas 295 nm excites tryptophan (Trp) as well as ANS and emission at 482 nm therefore includes an energy transfer from Trp at PrP position 99 to ANS. Thus, the excitation protocol at 295 nm links the single Trp residue to ANS fluorescence emission.
We found that ANS fluorescence decreases as urea concentration increases (Fig. 6 ▶) with both excitation protocols (295 nm and 385 nm). Such changes reflect two processes: unfolding of α-recPrP and urea-induced dissociation of ANS from the protein. However, excitation at 295 nm showed a slight increase in fluorescence at low urea concentrations, followed by a decline at higher concentrations (Fig. 6B ▶). In addition to the dissociation of ANS and unfolding of the α-recPrP, the fluorescence profile seen upon excitation at 295 nm is believed to reflect a change in the energy-transfer efficiency from the Trp residue to ANS. The ratio of fluorescence intensities, measured upon excitations at 385 nm and 295 nm, decreases mostly within the pretransition region and remains stable in the transition and posttransition regions of the denaturation curve (Fig. 6B ▶). It is likely that such a change represents an increase in the efficiency of the energy transfer from the Trp residue to ANS. To test whether this is a possible explanation, we performed simulations of fluorescence intensity in the donor–acceptor system, as described in Materials and Methods.
Our simulations demonstrate that the fluorescence of the acceptor may increase slightly at low urea concentrations if the protocol for excitation of both donor and acceptor is used (Fig. 6C ▶). This observation can be attributed to an increase in the efficiency of the energy transfer that dominates at low concentrations of urea. However, at a higher urea concentration, the fluorescence of the acceptor gradually decreases due to protein unfolding and ANS dissociation. The simulated ratio of estimated fluorescence intensities from excitation at 385 nm and 295 nm decreases as urea concentration increases. Agreement between our experimental data and simulations indicates that a decrease in the fluorescence ratio most likely represents increased efficiency of energy transfer. Remarkably, this ratio increases mostly within the pretransition region, which is consistent with rearrangement within the native state of α-recPrP. This urea-induced change is not detectable using far-UV CD, and is barely seen by near-UV CD (data not shown).
Discussion
Sporadic conversion of PrPC into PrPSc is an extremely rare event (Cohen and Prusiner 1998). The mechanism of the conversion and factors affecting its probability are poorly understood. Recent biophysical studies of recPrP in vitro revealed that the protein can adopt multiple nonnative β-sheet-rich isoforms (Lu and Chang 2001; Baskakov et al. 2002). Whether any of these isoforms are biologically relevant is currently under investigation. Our recent study shows that the amyloid fibrils of recPrP have similar epitope presentation as PrPSc indicating their common overall architecture (Baskakov et al. 2002). Furthermore, we found that the in vitro conversion of recPrP into fibrils mimics species barrier (submitted to publication), an essential attribute of prion propagation. To the extent that these tests capture important features of prion propagation, fibril formation may be considered as a more relevant model system to study in vitro than the conversion to the β-oligomer.
Regardless of which abnormal isoform possesses infectivity and/or toxicity, the ability of PrP to form distinct, abnormal conformers reflects the complexity of the energetic landscape of folding, as well as high conformational plasticity. These peculiar features are believed to underlie the molecular mechanism of prion diseases. One of the more intriguing issues is how PrP can form different nonnative isoforms in the absence of covalent modification and without any templates.
It has been postulated that an unfolding intermediate of PrPC may be a precursor to PrPSc (Swietnicki et al. 1997; Hornemann and Glockshuber 1998). Because of several technical difficulties, in particular, the confusion of the β-oligomer with an unfolding intermediate, the existence of a true equilibrium intermediate has remained questionable. Here, we have presented a reliable procedure that allows probing of the structural heterogeneity of refolded α-recPrP and detection of residual amounts of the β-oligomer that might be present. When recPrP is refolded into a homogeneous α-monomer, no unfolding intermediates are detected by either CD or SEC. Furthermore, both techniques revealed almost identical thermodynamic parameters of denaturation, which argues against the formation of an unfolding intermediate.
Our current studies demonstrate that thermodynamic characters of the native and denatured states of recPrP are variable. Thus, the native ensemble expands slightly with increasing urea concentration, as monitored by SEC. The increase in the dimensions of the native species is accompanied by an increase in the efficiency of energy transfer from a single Trp residue to ANS. Because this rearrangement in the native species is not evident by far-UV CD, we speculate that the physical nature of these changes is the loss of some tertiary contacts, which confers higher flexibility while keeping secondary structure essentially intact. Our observation is in agreement with pressure-induced denaturation experiments, which revealed changes occurring within the native species upon increasing pressure (Kuwata et al. 2002). Our data are also consistent with dynamic light-scattering experiments, which demonstrated that the dimension of the native state varies slightly with pH (Sokolowski et al. 2003). The denatured ensemble of recPrP also displays changes as a function of urea concentration, as judged by SEC. The size of the denatured species diminishes as urea concentration decreases, which is in agreement with hydrogen-deuterium exchange experiments (Nicholson et al. 2002).
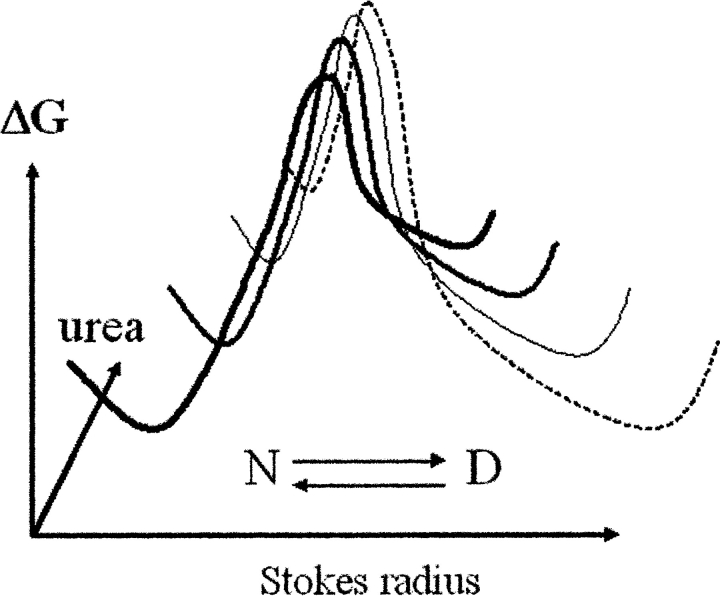
The gradual change we observed within the native and denatured species is consistent with the thermodynamically variable model, which postulates that the thermodynamic characters of the native and/or denatured ensemble change continuously as a function of the solvent environment (Fig. 7 ▶) (Yang et al. 1999; Bolen and Yang 2000). The gradual change of the thermodynamic character of the native ensemble does not contradict to the absence of conformational changes detected by CD. Other proteins that display a thermodynamically variable behavior of the native ensembles did not show any changes of secondary structure within the native ensemble either (Bolen and Yang 2000).
Figure 7.
Free-energy diagram illustrating the thermodynamically variable character of native and denatured states of recPrP. Not only do the relative populations of the native (N) and the denatured (D) states change as a function of urea concentration, but the physical properties of both states are variable depending on solvent conditions.
With no unfolding intermediates having been identified, one may speculate that variable thermodynamic behavior is linked to the diversity of misfolding pathways for PrP. Gradual environment-dependent change in the physical properties of the native and denatured ensemble may bias the adoption of particular pathways of misfolding under a different solution environment. As we observed previously, the formation of the β-oligomer is favored at pH below 5.5, whereas optimal pH for the conversion into amyloid fibrils is between 5.0 and 7.2 (Baskakov et al. 2002). Interestingly, both abnormal isoforms, the β-oligomer and the amyloid form, coexist at pH 4.0–6.0. These pH values are close to the slightly acidic environment of lysosomes and endosomes, where the conformational transition into pathological PrPSc is believed to occur (McKinley et al. 1990; Arnold et al. 1995). Our present findings indicate that changes in environmental conditions may have important implications in determining the misfolding pathways of PrP in vivo and the formation of distinct abnormal isoforms.
Materials and methods
Protein expression, purification, and refolding
The expression and purification of human recPrP and recombinant mouse PrP of 106 residues (recPrP106) were performed as described previously (Mehlhorn et al. 1996). The purified protein was seen to be a single pure species with an intact disulfide bond as confirmed by SDS-PAGE and electrospray mass spectrometry. The β-oligomer and α-monomer isoforms of recPrP were prepared as described previously (Baskakov et al. 2001).
Formation of amyloid fibrils in vitro
The oxidized form of recPrP spontaneously converts into the amyloid fibrils upon continuous shaking at 37°C in phosphate buffer, pH 6.5, in 1 M GndHCl and various concentrations of urea (Baskakov et al. 2002). To monitor the kinetics of fibril formation, 7-μL aliquots were withdrawn and diluted 40-fold into 10 mM sodium-acetate buffer (pH 5.5) to a final concentration of recPrP 7.5 μg/mL. After addition of Thioflavine T (ThT; Molecular Probes) to a final concentration of 10 μM, fluorescence spectra were recorded in 0.4-cm rectangular cuvettes with excitation at 445 nm on a FluoroMax-3 fluorimeter (Jobin Yvon); both excitation and emission slits were 3 nm. ThT bound to the fibrils showed typical fluorescence spectra with the emission maximum at 482 nm.
Circular dichroism and thermal denaturation
CD spectra of recPrP (0.25 mg/mL) were recorded in a 0.1-cm cuvette with a J-810 CD spectrometer (Jasco) scanning at 20 nm/min, with a bandwidth of 1 nm and data spacing of 0.5 nm. Each spectrum represents the average of three individual scans after subtracting the background spectra. To monitor the unfolding curve, recPrP was diluted to various concentrations of urea in 20 mM sodium acetate (pH 5.0) and incubated at room temperature for 30 min.
The thermal denaturations were recorded with a J-810 CD spectrometer interfaced with a temperature control unit at 222 nm with a bandwidth of 1 nm. The concentration of protein was 0.2 mg/mL in a 0.1-cm civette and 0.04 mg/mL in a 0.5-cm cuvette. The samples were equilibrated at 15°C for 20 min following by increases of temperature with a constant rate 1°C/min, where the temperature in a cuvette was monitored during each experiment using microprobe. The reference CD spectra were collected at 15°C before and after each experiment.
Size-exclusion chromatography
The experiments were carried out on a 300× 7.8 mm Toyopearl TSK3000SWxl HPLC gel filtration column (Tosoh Corporation). Protein samples (0.2 mg/mL) were incubated before injection for 30 to 120 min in column buffer (20 mM sodium acetate buffer, pH 5.0) containing different concentrations of urea. The column was operated using flow rate 0.8 mL/min and with mechanical injection within a fully automated HPLC system (Shimadzu) that allowed the elution volume to be repeated to within 0.005 μL. Partition coefficients (Kd) were calculated by Kd = (Ve − Vo)/Vi, in which Ve is the elution volume of the protein sample, Vo is the void volume, represented by the elution volume of Dextran Blue, and Vi is the difference in elution volume between aminobenzoic acid and Dextran Blue.
ANS fluorescence and data simulation
The fluorescence spectra of ANS were recorded in 0.4-cm rectangular cuvettes with excitations either at 295 nm or 385 nm (an excitation slit of 3 nm and an emission slit of 5 nm) on a FluoroMax-3 fluorimeter. Samples of α-recPrP (0.15 mg/mL) were incubated in the presence of 100 μM ANS at different concentrations of urea in 20 mM sodium acetate buffer (pH 5.0) for 30 min before measurement.
To explain rationally observed changes in fluorescence, we provide a simplified model. In this model, we consider that the fluorescent signal of ANS is determined by the relative fraction of ANS bound to the native state of protein and the possible energy transfer between the tryptophan residue (donor, D) and protein-bound ANS (acceptor, A). In this donor–acceptor system, when one wavelength (385 nm) exclusively excites the acceptor and a second wavelength (295 nm) excites both the donor and acceptor, the fluorescence signal from the acceptor can be described as follows:
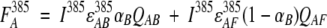 |
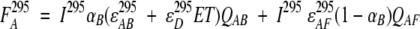 |
The fluorescence signal from the donor will be:
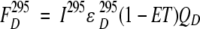 |
in which I295 and I385 denote the excitation intensity at 295 nm and 385 nm, respectively; 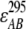 ,
, 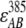 ,
, 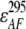 , and
, and 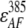 represent the acceptor extinction coefficient for bound (B) and free (F) form at 295 nm and 385 nm, as indicated by the respective superscript values;
represent the acceptor extinction coefficient for bound (B) and free (F) form at 295 nm and 385 nm, as indicated by the respective superscript values; 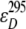 is the extinction coefficient of the donor at 295 nm; αB is the fraction of the bound acceptor; QAB and QAF are quantum yields of bound and free acceptor, respectively; QD represents quantum yield of the donor; and ET is the efficiency of energy transfer. In the case of ANS fluorescence, the product of the quantum yield and the extinction coefficient of free ANS is much smaller than that of the bound dye:
is the extinction coefficient of the donor at 295 nm; αB is the fraction of the bound acceptor; QAB and QAF are quantum yields of bound and free acceptor, respectively; QD represents quantum yield of the donor; and ET is the efficiency of energy transfer. In the case of ANS fluorescence, the product of the quantum yield and the extinction coefficient of free ANS is much smaller than that of the bound dye:
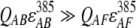 |
We assume that both the fraction of ANS bound to the native state and the efficiency of energy transfer change with urea concentration. To simulate the urea-induced change in the fraction of bound ANS, we used the Henderson-Hasselbalch equation:
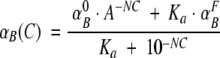 |
in which 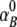 and
and 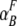 are the initial and final fractions of bound ANS, respectively; Ka represents the equilibrium association constant for ANS; C is the concentration of urea; and N is an arbitrary constant. Similarly, the urea-dependent change in the energy-transfer efficiency can be described by:
are the initial and final fractions of bound ANS, respectively; Ka represents the equilibrium association constant for ANS; C is the concentration of urea; and N is an arbitrary constant. Similarly, the urea-dependent change in the energy-transfer efficiency can be described by:
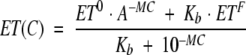 |
in which ET0 and ETF are the efficiencies of energy transfer at initial and final conformational states, respectively; Kb represents equilibrium constants between the two states; C is the concentration of urea; and M is an arbitrary constant. The simulation mimics experimentally observed ratios of ANS fluorescence when the parameters N, M, Ka, and Kb were fixed.
Acknowledgments
We thank Dr. Joseph P.Y. Kao for critical reading of the manuscript. This work was supported by NIH grants AG022116 to I.V.B., and AG02132 and AG10770, as well as by a gift from the G. Harold and Leila Y. Mathers Charitable Foundation to S.B.P. and G.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
1 PrPC, cellular isoform of the prion protein
PrPSc, disease-associated isoform of the prion protein
recPrP, recombinant prion protein
α-recPrP, α-helical monomeric form of recPrP
ANS, 1-anilinonaphthalene-8-sulfonate
CD, circular dichroism
recPrP106, recombinant prion protein of 106 residues
ThT, Tioflavin T
GndHCl, guanidine hydrochloride
SEC, size-exclusion chromatography
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03457204.
References
- Apetri, A.C. and Surewicz, W.K. 2002. Kinetic intermediate in the folding of human prion protein. J. Biol. Chem. 277 44589–44592. [DOI] [PubMed] [Google Scholar]
- Arnold, J.E., Tipler, C., Laszlo, L., Hope, J., Landon, M., and Mayer, R.J. 1995. The abnormal isoform of the prion protein accumulates in late-endosome-like organelles in scrapie-infected mouse brain. J. Pathol. 176 403–411. [DOI] [PubMed] [Google Scholar]
- Baskakov, I.V. and Bolen, D.W. 1998. Monitoring the sizes of denatured ensembles of Staphylococcal nuclease proteins: Implications regarding m values, intermediates, and thermodynamics. Biochemistry 37 18010–18017. [DOI] [PubMed] [Google Scholar]
- Baskakov, I.V., Aagaard, C., Mehlhorn, I., Wille, H., Groth, D., Baldwin, M.A., Prusiner, S.B., and Cohen, F.E. 2000. Self-assembly of recombinant prion protein of 106 residues. Biochemistry 39 2792–2804. [DOI] [PubMed] [Google Scholar]
- Baskakov, I.V., Legname, G., Prusiner, S.B., and Cohen, F.E. 2001. Folding of prion protein to its native α-helical conformation is under kinetic control. J. Biol. Chem. 276 19687–19690. [DOI] [PubMed] [Google Scholar]
- Baskakov, I.V., Legname, G., Baldwin, M.A., Prusiner, S.B., and Cohen, F.E. 2002. Pathway complexity of prion protein assembly into amyloid. J. Biol. Chem. 277 21140–21148. [DOI] [PubMed] [Google Scholar]
- Bolen, D.W. and Yang, M. 2000. Effects of guanidine hydrochloride on the proton inventory of proteins: Implications on interpretations of protein stability. Biochemistry 39 15208–15216. [DOI] [PubMed] [Google Scholar]
- Calzolai, L. and Zahn, R. 2003. Influence of pH on NMR structure and stability of the human prion protein globular domain. J. Biol. Chem. 278 35592–35596. [DOI] [PubMed] [Google Scholar]
- Cohen, F.E. and Prusiner, S.B. 1998. Pathologic conformations of prion proteins. Annu. Rev. Biochem. 67 793–819. [DOI] [PubMed] [Google Scholar]
- Hornemann, S. and Glockshuber, R. 1998. A scrapie-like unfolding intermediate of the prion protein domain PrP(121–231) induced by acidic pH. Proc. Natl. Acad. Sci. 95 6010–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosszu, L.L.P., Baxter, A., Jackson, G.S., Power, A., Clarke, A.R., Waltho, J.P., Craven, C.J., and Collinge, J. 1999. Structural mobility of the human prion protein probed by backbone hydrogen exchange. Nat. Struct. Biol. 6 740–743. [DOI] [PubMed] [Google Scholar]
- Kuwata, K., Li, H., Yamada, H., Legname, G., Prusiner, S.B., Akasaka, K., and James, T.L. 2002. Locally disordered conformer of the hamster prion protein: A crucial intermediate of PrPSc? Biochemistry 41 12277–12283. [DOI] [PubMed] [Google Scholar]
- Lu, B.Y. and Chang, J.Y. 2001. Isolation of isoforms of mouse prion protein with PrPSc-like structural properties. Biochemistry 40 13390–13396. [DOI] [PubMed] [Google Scholar]
- McKinley, M.P., Taraboulos, A., Kenaga, L., Serban, D., DeArmond, S.J., Stieber, A., and Prusiner, S.B. 1990. Ultrastructural localization of scrapie prion proteins in secondary lysosomes of infected cultured cells. J. Cell Biol. 111 316a. [PubMed] [Google Scholar]
- Mehlhorn, I., Groth, D., Stöckel, J., Moffat, B., Reilly, D., Yansura, D., Willett, W.S., Baldwin, M., Fletterick, R., Cohen, F.E., et al. 1996. High-level expression and characterization of a purified 142-residue polypeptide of the prion protein. Biochemistry 35 5528–5537. [DOI] [PubMed] [Google Scholar]
- Morillas, M., Vanik, D.L., and Surewicz, W.K. 2001. On the mechanism of α-helix to β-sheet transition in the recombinant prion protein. Biochemistry 40 6982–6987. [DOI] [PubMed] [Google Scholar]
- Nicholson, E.M., Mo, H., Prusiner, S.B., Cohen, F.E., and Marqusee, S. 2002. Differences between the prion protein and its homolog doppel: A partially structured state with implications for scrapie formation. J. Mol. Biol. 316 807–815. [DOI] [PubMed] [Google Scholar]
- Pan, K.-M., Baldwin, M., Nguyen, J., Gasset, M., Serban, A., Groth, D., Mehlhorn, I., Huang, Z., Fletterick, R.J., Cohen, F.E., et al. 1993. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. 90 10962–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner, S.B. 2001. Shattuck lecture—Neurodegenerative diseases and prions. N. Engl. J. Med. 344 1516–1526. [DOI] [PubMed] [Google Scholar]
- Rezaei, H., Choiset, Y., Eghiaian, F., Treguer, E., Mentre, P., Debey, P., Grosclaude, J., and Haertle, T. 2002. Amyloidogenic unfolding intermediates differentiate sheep prion protein variants. J. Mol. Biol. 322 799–814. [DOI] [PubMed] [Google Scholar]
- Safar, J., Roller, P.P., Gajdusek, D.C., and Gibbs Jr., C.J. 1994. Scrapie amyloid (prion) protein has the conformational characteristics of an aggregated molten globule folding intermediate. Biochemistry 33 8375–8383. [DOI] [PubMed] [Google Scholar]
- Santoro, M.M. and Bolen, D.W. 1988. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry 27 8063–8068. [DOI] [PubMed] [Google Scholar]
- Sokolowski, F., Modler, A.J., Masuch, R., Zirwer, D., Baier, M., Lutsch, G., Moss, D.A., Gast, K., and Naumann, D. 2003. Formation of critical oligomers is a key event during conformational transition of recombinant Syrian hamster prion protein. J. Biol. Chem. [Epub ahead of print]. [DOI] [PubMed]
- Swietnicki, W., Petersen, R., Gambetti, P., and Surewicz, W.K. 1997. pH-dependent stability and conformation of the recombinant human prion protein PrP(90–231). J. Biol. Chem. 272 27517–27520. [DOI] [PubMed] [Google Scholar]
- Wildegger, G., Liemann, S., and Glockshuber, R. 1999. Extremely rapid folding of the C-terminal domain of the prion protein without kinetic intermediates. Nat. Struct. Biol. 6 550–553. [DOI] [PubMed] [Google Scholar]
- Yang, M., Liu, D., and Bolen, D.W. 1999. The peculiar nature of the guanidine hydrochloride-induced two-state denaturation of staphylococcal nuclease: A calorimetric study. Biochemistry 38 11216–11222. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Stöckel, J., Mehlhorn, I., Groth, D., Baldwin, M.A., Prusiner, S.B., James, T.L., and Cohen, F.E. 1997. Physical studies of conformational plasticity in a recombinant prion protein. Biochemistry 36 3543–3553. [DOI] [PubMed] [Google Scholar]