Abstract
Deamidation is a prevalent modification of crystallin proteins in the vertebrate lens. The effect of specific sites of deamidation on crystallin stability in vivo is not known. Using mass spectrometry, a previously unreported deamidation in βB1-crystallin was identified at Gln146. Another deamidation was investigated at Asn157. It was determined that whole soluble βB1 contained 13%–17% deamidation at Gln146 and Asn157. Static and quasi-elastic laser light scattering, circular dichroism, and heat aggregation studies were used to explore the structure and associative properties of recombinantly expressed wild-type (wt) βB1 and the deamidated βB1 mutants, Q146E and N157D. Dimer formation occurred for wt βB1, Q146E, and N157D in a concentration-dependent manner, but only Q146E showed formation of higher ordered oligomers at the concentrations studied. Deamidation at Gln146, but not Asn157, led to an increased tendency of βB1 to aggregate upon heating. We conclude that deamidation creates unique effects depending upon where the deamidation is introduced in the crystallin structure.
Keywords: lens crystallins, deamidation, cataracts, oligomer assembly, laser light-scattering, mass spectrometry
The vast majority of cataracts, either congenital or age-related, may result from insolubilization of crystallins, the major proteins in the lens. Post-translational modifications may alter crystallin structure and lead to their insolubilization. Because the turnover of crystallins in the lens is very low, their ability to maintain their native conformation is crucial. The purpose of this study was to determine the effect of a prevalent modification, deamidation, on crystallin structure.
Although deamidation in the lens has been known to occur for years, the extent and the actual sites have only recently been confirmed (Kramps et al. 1978; Voorter et al. 1987; Groenen et al. 1993; Lampi et al. 1998; Lapko et al. 2002; Zhang et al. 2003). Each of the major crystallin polypeptides in the human lens is deamidated during aging and most are deamidated at multiple residues (Lund et al. 1996; Hanson et al. 1998; Lampi et al. 1998; Takemoto and Boyle 1998; Lapko et al. 2002; Zhang et al. 2003). For example, six to eight deamidations have been detected in βB1-crystallin (Lampi et al. 1998; Zhang et al. 2003). This is in contrast to other modifications, such as proteolysis or oxidation, where fewer proteins and residues are susceptible (Lampi et al. 1998). Deamidation is one of the most prevalent modifications in the aging lens, with βB1 being extensively deamidated.
Support for the detrimental role of deamidation in crystallin structure is found in several reports of deamidations associated with the water-insoluble crystallins of adult normal and cataractous lenses (Yang et al. 1994; Lund et al. 1996; Hanson et al. 2000; Takemoto and Boyle 2000; Lapko et al. 2002; Srivastava and Srivastava 2003). A deamidation site in βB1 at Asn157 was reported in the insoluble proteins of adult lenses (Hanson et al. 2000). Deamidated αA and γS-crystallins have also been identified in the insoluble proteins (Lund et al. 1996; Hanson et al. 2000). In γS, the same sites of deamidation were present in the soluble and insoluble proteins from cataractous lenses, but the extent of deamidation was greater in the insoluble proteins (Lapko et al. 2002).
Subunits in the βγ-crystallin family have two domain regions joined by a connecting peptide (Blundell et al. 1981; Bax et al. 1990). The connecting peptide has been proposed to be important in determining the domain–domain interactions (Wright et al. 1998). Each tightly folded domain has two Greek key motifs comprised of four β-pleated sheet strands (Lapatto et al. 1991). These structural features contribute to their high intrinsic stability. Unlike γ-crystallins, β-crystallins interact to form dimers and higher ordered oligomers. Deamidation introduces a negative charge by replacing an amide with a carboxyl group. This modification at regions predicted to be critical to the overall structure might disrupt the stability and the associative properties of the protein. For example, deamidation at Gln204 in βB1 at the intradomain interface markedly altered the stability and associative properties of the protein (Lampi et al. 2001; Kim et al. 2002).
The present study examines two additional sites of deamidation at residues Gln146 and Asn157 in βB1. Based on the recently published structure of βB1-crystallin, Asn157 is predicted to be located in a loop region oriented away from the C-terminal domain and not to affect the tertiary structure if deamidated (Lapatto et al. 1991; Van Montfort et al. 2003). In contrast, the location of Gln146 is predicted to be in the connecting peptide adjacent to a charged residue, Glu147. Provided in this report is evidence that during aging deamidation occurs at residues Asn157 and Gln146 βB1, and that deamidation at residue Gln146 markedly disrupts βB1 structure while deamidation at Asn157 has little effect. Deamidation in the connecting peptide at Gln146 alters both the secondary structure and stability of βB1. These results are significant, because they suggest that some sites of deamidation in β-crystallins are more likely to contribute to cataract formation.
Results
Prevalence of deamidation at Gln146 and Asn157 in adult lenses
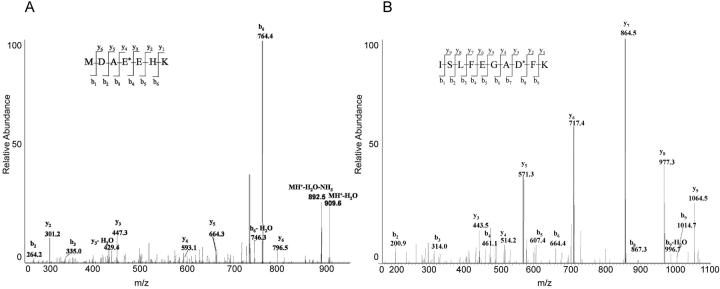
Deamidation was detected in βB1 at residues 146 and 157 in the soluble proteins from normal lenses of 55- and 59-year-old donors. Tryptic digest from these lenses were esterified with either normal (59-year-old) or deuterated (55-year-old) methanol and analyzed by LC-MS. For example in the 55-year-old sample, the singly charged peptide 143–149 of βB1 containing a Glu rather than a Gln at position 146 was detected with an m/z of 927.6, and a doubly charged peptide 150–159 of βB1 containing a deamidation at Asn157 was detected with an m/z of 589.6. These masses correspond to the addition of 17 mass units at each carboxylic acid, in addition to 18 mass unit increases due to the deamidation and subsequent esterification at residues 146 and 157. The tandem (MS/MS) spectra of these peptides from the 55-year-old donor lenses confirmed their identity and sites of deamidation (Fig. 1 ▶).
Figure 1.
Tandem mass spectra of (A) esterified tryptic peptide 143–149 of βB1-crystallin containing a deamidated Gln146 and (B) esterified tryptic peptide 150–159 of βB1-crystallin containing a deamidated Asn157 from the soluble protein of a 55-year-old human lens. The parent ion of peptide 143–149 was singly charged with m/z = 927.6 (expected = 927.5), and the parent ion of peptide 150–159 was doubly charged with m/z = 589.6 (expected 589.4). The observed y and b fragment ions are indicated in the spectra. The asterisk indicates the position of the deamidated residue.
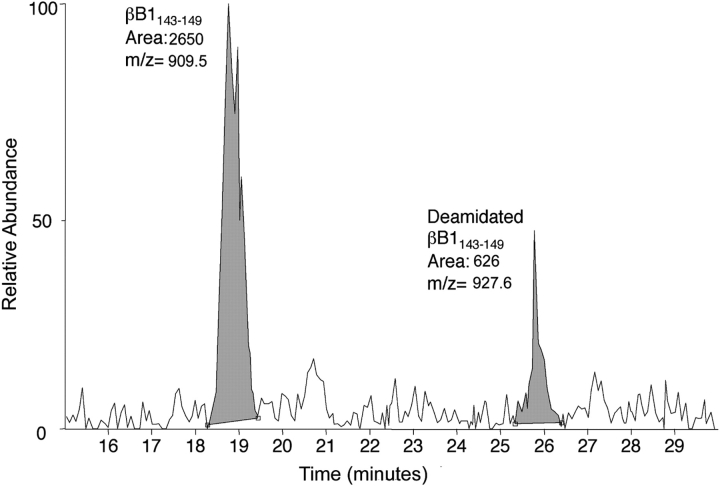
The extent of deamidation at residues 146 and 157 was estimated by tracing the ion intensities for both amidated and deamidated forms of each peptide in the chromatograms, integrating the areas under the peaks, and calculating the percentage of the combined ion currents contributed by the deamidated peptides. The result for one measurement of deamidation in peptide 143–149 for the 55-year-old sample is shown in Figure 2 ▶. The extent of deamidation in the 55-year-old sample (two measurements) was 20% and 16% at residues 146 and 157, respectively, and in the 59-year-old sample (single measurement) 18% and 15% at residues 146 and 157, respectively. However, these values were not corrected for deamidations introduced during the esterification reaction. Over 10 different sites of deamidation in five different recombinantly expressed βB1-crystallins were observed where deamidations should not have been detected. The average extent of deamidation due to derivatization was 2 ± 2%. Therefore, the true extent of deamidation at residues 146 and 157 in normal adult lenses was approximately 17% and 13%, respectively.
Figure 2.
Extracted ion chromatograms for both the singly-charged amidated (m/z range 909.0–910.0, peak at 18.8 min) and singly charged deamidated (m/z range 927.1–928.1, peak at 25.8 min) forms of peptide 143–149 of βB1-crystallin from soluble proteins of 55-year-old lenses. The 19.1% deamidation of peptide 143–149 was calculated by dividing the area under the peak for the deamidated peptide (626) by the total areas of both peaks (3276) × 100.
These values were calculated assuming that the amide and deamidated forms of peptides ionize with similar efficiencies in the mass spectrometer. This assumption was verified by mixing equal amounts of esterified wild-type and deamidated proteins. The ion currents of the nondeamidated and deamidated peptides were then compared. Within the uncertainty of the measurements no difference in ionization efficiencies could be detected.
The effect of deamidation on the structure of human recombinant βB1 proteins
Expression of recombinant human wt βB1 was confirmed as previously described (Lampi et al. 2001). Expression of the βB1 mutants, Q146E and N157D, was also confirmed by derivatization of the tryptic peptides to form methyl esters as described above, detection of ions with masses matching those of the expected deamidated peptides, and confirmation of sequences by MS/MS spectra (data not shown).
Circular dichroism experiments were performed to confirm the folding of the expressed proteins and to investigate structural differences (Fig. 3 ▶). The overall secondary structures of the wt and deamidated βB1s were similar. The spectra in the far-UV range indicated that all three proteins folded with distinct minima and maxima in their ellipticity. wt βB1 (Fig. 3A ▶, blue spectrum) showed a minimum at 205 nm and at 215 nm, as previously reported (Lampi et al. 2001). The spectra of N157D and Q146E (Fig. 3A ▶, red and green spectra) were similar to the spectrum of wt βB1, except for a greater maximum ellipticity for Q146E. The tertiary structures of the three crystallins did not match as well. Differences were noted in the near-UV spectra particularly for Q146E (Fig. 3B ▶).
Figure 3.

(A) Far-UV and (B) near-UV circular dichroism of wt βB1 (blue spectrum), Q146E (green spectrum), and N157D (red spectrum). Protein concentrations ranged from 1.1 to 1.6 mg/mL. All spectra were averaged over 30 scans.
Molecular size and shape of crystallin assemblies
The molar masses of whole soluble crystallins were determined in order to validate the use of multiangle laser light scattering (MALS) to characterize crystallins. The molar masses determined by MALS of whole soluble protein fractions from infant lenses were comparable to values obtained by other methods (Bindels et al. 1981; Chiou et al. 1989). The weight-averaged molar masses (Mw) were 25 ± 2, 29 ± 3, 52 ± 3, 163 ± 5, and 635 ± 19 kD for γ, γS, βLow, βHigh, and α-crystallins (Fig. 4A ▶). As expected, the hydrodynamic radii (RH) increased with increasing molar masses (Fig. 4B ▶). The RH values were 2.0, 2.1, 3.2, 5.8, and 8.7 nm for γ, γS, βLow, βHigh, and α-crystallins. The βLow fraction, comprised of β-dimers, and the βHigh fraction, comprised of higher ordered β-oligomers, provided a reference for the elution of recombinant βB1s below.
Figure 4.
Chromatograms of (A) the molar masses and (B) hydrodynamic radii of whole soluble protein extracted from lenses from a 3-month-old donor. The line tracing represents the signal from the refractive index detector. In (A), the filled diamonds represent the molar masses. In (B), the filled diamonds represent the hydrodynamic radii. A 50-μL sample of 15 mg/mL whole soluble protein was analyzed.
Associative behavior of recombinant βB1s
The effect of deamidation on the βB1 dimer was determined by MALS. Equal amounts (500 μg) of recombinant proteins were analyzed by MALS, and the corresponding chromatograms are shown in Figure 5 ▶. wt βB1 eluted in a single peak with a Mw of 43 ± 3 kD (Fig. 5A ▶, Table 1). Molar masses in different fractions across the peak were consistent, indicating an homogeneous sample. Experiments were repeated at different concentrations. The Mw increased from 31 ± 2 to 52 ± 4 kD with increasing concentrations under the peak from 0.01 to 0.32 mg/mL. Because the mass of recombinant βB1 is 27,890 (Lampi et al. 2001), the Mw at the higher concentrations suggested dimer formation. Furthermore, the elution of wt βB1 was similar to the elution of the βLow fraction, which contains β-crystallin dimers (Figs. 4A ▶). The formation of wt βB1 dimers agrees with previously published reports (Bateman et al. 2001; Lampi et al. 2001).
Figure 5.
Chromatograms of the molar masses for the recombinant proteins (A) wt βB1, (B) N157D, and (C) Q146E. Filled diamonds represent the molar masses determined for each eluting slice. The line represents the refractive index signal; all refractive index tracings are normalized to one another. The concentration of each protein was 10 mg/mL in a 50-μL sample.
Table 1.
Diffusion properties of βB1 and deamidated βB1s
| MW | ± | RH | ± | f | ± | DT | ± | ||||
| Sample description | [ ] Under peak (mg/mL) | Elution (mL) | (kD) | (nm) | (×10−11 kg/sec) | f/f0 — | (×10−11 m2/sec) | ||||
| Wt βB1 | 0.01 | 13.9 | 30 | 1 | 2.0 | 0.2 | 3.4 | 0.4 | 1.0 | 12.3 | 1.5 |
| 0.16 | 12.9 | 43 | 3 | 3.1 | <0.1 | 5.2 | <0.1 | 1.2 | 7.9 | <0.1 | |
| Q146E | 0.04 | 14.0 | 37 | 3 | 2.7 | 0.1 | 4.5 | 0.2 | 1.4 | 9.1 | 0.4 |
| 0.16 | 13.3 | 55 | 3 | 3.4 | <0.1 | 5.7 | <0.1 | 1.4 | 7.2 | <0.1 | |
| N157D | 0.04 | 13.7 | 36 | 2 | 2.6 | 0.1 | 4.4 | 0.1 | 1.3 | 9.4 | 0.3 |
| 0.14 | 13.0 | 42 | 1 | 3.1 | <0.1 | 5.2 | <0.1 | 1.2 | 7.9 | <0.1 | |
The deamidated βB1, N157D eluted in a single peak with a Mw across the width of the peak of 42 ± 3 kD, similar to the elution of wt βB1 (Fig. 5B ▶, Table 1). The Mw of N157D increased from 36 ± 2 to 66 ± 2 kD with increasing βB1 concentration under the peak from 0.04 to 0.51 mg/mL.
In contrast to wt βB1 and N157D, Q146E eluted in several peaks (Fig. 5C ▶): a broad peak similar to the elution of βLow, and a very heterogeneous peak similar to the elution of the βHigh fraction. The Mw of Q146E in the main peak, disregarding the shoulder, was 55 ± 3 kD (Fig. 5C ▶, Table 1). The Q146E protein in the early eluting peak was very heterogeneous with molar masses from early to late eluting fractions from about 500 to 200 kD (Fig. 5C ▶). This peak did not disappear under reducing conditions of 1 mM DTT.
The Mw of the main Q146E peak increased from 36 ± 3 to 55 ± 3 kD with increasing concentration under the peak from 0.04 to 0.16 mg/mL (Table 1). At the higher concentrations prepared for chromatography, Q146E precipitated.
Approximate shape determination of βB1s
To determine if deamidation altered the overall shape of βB1, frictional coefficients were determined by quasi-elastic light scattering. Because it had previously been determined from sedimentation velocity that the wt βB1 dimer was elongated (Lampi et al. 2001), RH were converted to frictional coefficients (f) and compared to frictional coefficients for spheres (f0) of equivalent Mw (Table 1). The f values determined during the chromatography runs above in Figure 5 ▶ were 5.2 ± 0.2, 5.7 ± 0.3, and 5.2 ± 0.2E-11 kg/sec for wt βB1, Q146E, and N157D, respectively (Table 1). The higher f for Q146E was associated with a higher Mw. When the Mw of wt βB1 and N157D matched Q146E more closely, their frictional coefficients also matched more closely. At the lower concentrations f/f0 was close to one for all proteins, indicating they were in a roughly spherical conformation. At higher concentrations, f/f0 was near 1.3, indicating the proteins were elongated and in a conformation similar to βHigh (Chiou et al. 1989).
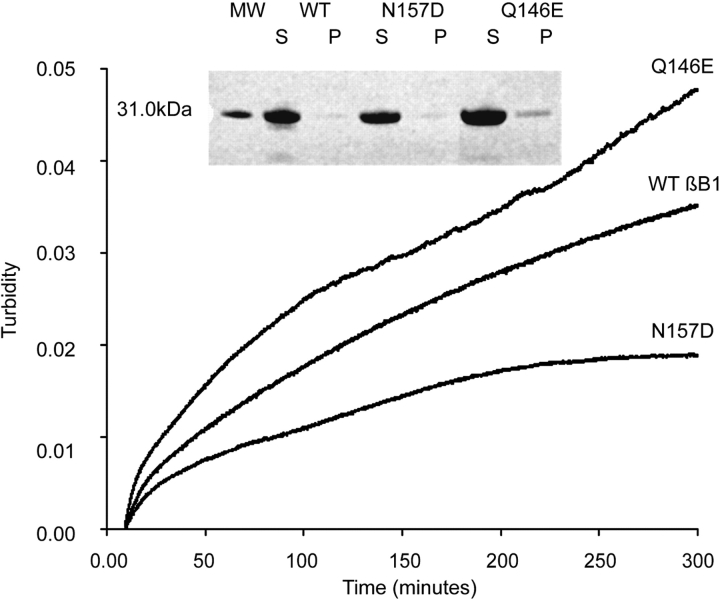
Thermal-precipitation of deamidated βB1s
As a measure of the tendency to aggregate, dilute solutions of βB1 were heated and the increase in turbidity at 405 nm measured. After heating for 300 min, N157D had less turbidity than wt βB1, while Q146E had more turbidity than either wt βB1 or N157D (Fig. 6 ▶). In separate experiments, 1.5-mg/mL samples were heated for 300 min and then analyzed by SDS-PAGE. A very faint band was visualized from the pellet of the Q146E sample only (Fig. 6 ▶, inset). The difference in the measured turbidity between wt βB1 and Q146E was due to a very small amount of precipitate. No precipitate was detected when Q146E was heated with αA-crystallin (data not shown).
Figure 6.
Thermal denaturation/precipitation curves. Thermal denaturation curves of wt βB1 (middle line), N157D (bottom line), and Q146E (top line) were obtained by heating 0.1 mg/mL concentration of proteins at 55°C and measuring change in turbidity at 405 nm. Data shown for wt were the average of two experiments, and are similar to previously reported results (Lampi et al. 2002). Data shown for N157D and Q146E were the average of four to five experiments. Samples were also heated at 1.5 mg/mL and visualized by SDS-PAGE (inset: S, supernatant; P, pellet; MW, molecular weight marker).
For comparison, we have previously reported that deamidation at another residue, Gln204, led to increased precipitation (Lampi et al. 2002). The initial rate of precipitation leading to turbidity for Q204E (0.015 min−1) was 1.7-fold greater than for Q146E (0.009 min−1) and a heavier band of precipitated Q204E was readily detected by SDS-PAGE.
Discussion
Lens crystallins, especially β-crystallins, undergo extensive deamidation during normal aging. Many sites of deamidation have recently been identified (Zhang et al. 2003), but the effect of specific deamidations on β-crystallins in vivo is not known. We report here the effects of two sites of deamidation at Gln146 and Asn157 on recombinantly expressed βB1-crystallin. This investigation found: (1) a new site of deamidation in βB1 at Gln146, (2) βB1 from human lens soluble proteins contained approximately 13%–17% deamidation at Gln146 and Asn157, (3) dimer formation occurred for wt βB1, Q146E, and N157D in a concentration-dependent manner, (4) only Q146E showed formation of higher oligomers, and (5) Q146E, but not N157D, showed an increased tendency to aggregate upon heating. Deamidation at Gln146 located in the connecting peptide of βB1 leads to increased association of the protein. Together with our previously published results on the effects of deamidation at Gln204 in βB1 (Kim et al. 2002; Lampi et al. 2002), we conclude that deamidation creates unique effects depending upon where the deamidation is introduced in the crystallin structure.
Identification and quantification of in vivo sites of deamidation at Gln146 and Asn157
Little quantitative data are available measuring levels of deamidation. In this study, the methods used allowed ready assignment of sites of deamidation. The mass shift of +1 introduced by deamidation can be difficult to assign in MS/MS spectra from ion trap mass spectrometers due to the mass error that is often as great as 0.5 mass units. However, when aided by software for interpretation of MS/MS spectra, the greater mass shift following the conversion of the deamidated residue to an ester allowed unambiguous assignment. Introduction of the ester at the site of deamidation also increased the hydrophobicity of the peptide so that it eluted later during reverse phase chromatography. In addition, the amide and deamidated forms of several different peptides were found to ionize with similar efficiencies in the mass spectrometer. This allowed for direct comparison of the amidated and deamidated peptides and avoided the necessity of assigning percent deamidation by comparison of the expected isotopic distribution to that observed in nonderivatized samples (Hanson et al. 2000).
The approximate 13%–17% deamidation observed in peptides 143–149 and 150–159 of βB1 was consistent with the extent of deamidation observed in other peptides of βB1 during aging (Zhang et al. 2003). Deamidation at multiple sites may disrupt the normal structure of β-crystallins. Although a greater number of lenses must be analyzed to verify the extent of deamidation, the data confirm that residues Gln146 and Asn157 undergo deamidation during aging of human lens and justifies the selection of the deamidated mutants used in these studies.
The effect of deamidation on the association of recombinant βB1
The use of static and quasi-elastic light scattering during chromatography allowed for the simultaneous determination of molecular size and shape of crystallins. All three of the recombinant βB1-crystallins in this study formed dimers in a concentration-dependent manner. Both wt βB1 and N157D eluted in single peaks with molar masses consistent across the width of the peaks, suggesting a single species. At the higher concentrations, the peaks would have been predominantly comprised of dimers. The molar masses at the lower concentrations suggested a monomer–dimer mixture. This was rather surprising, as the crystal structure of βB1 indicates that it has two independent interfaces of interaction that would both have to be disrupted to form a monomer (Van Montfort et al. 2003).
In contrast, Q146E was predominantly a dimer at a concentration that wt βB1 and N157D achieved a monomer–dimer equilibrium. Q146E also formed a higher ordered oligomer that was very polydisperse across the peak. The molar masses across this peak are consistent with oligomers of approximately 6–18 subunits. The subtle tertiary structural changes observed in the near-UV spectra of Q146E most likely contributed to these altered associative properties. This altered structure or the additional charge due to deamidation in the connecting peptide may have led to new hydrophobic or charge–charge interactions between subunits, resulting in higher ordered oligomers. This may also explain the unusual behavior during chromatography. Interestingly, the dimer peak of Q146E eluted after the monomer–dimer peak of wt βB1 and the aggregate peak of 200–500 kD eluted after the βHigh peak of approximately 160 kD. The additional charge and subtle structural changes may have led to column interactions. The three deamidated βB1s we have analyzed thus far, Q146E, N157D, and Q204E, all have different chromatographic behavior. The Q204E dimer eluted ahead of the wt βB1 dimer (Lampi et al. 2001), suggesting that the effects of deamidation are dependent on where the deamidation is introduced.
The effect of deamidation on the shape of recombinant βB1
It is apparent from the frictional coefficients of wt βB1 and N157D that these proteins were in the same conformation, and that this conformation was larger than for the monomer, which generated lower frictional coefficients. Frictional coefficients of Q146E were generally higher as were the molar masses at similar concentrations, suggesting that deamidation at this residue resulted in an overall larger molecule.
The prolate and oblate approximations suggested that the dimer for all three βB1s had a major axis four to six times the length of the minor axis (equations 1 and 2 in Materials and Methods). This shape characterization does not fit previous observations, as the crystal structure of βB1 is a closed dimer that would appear nearly spherical (Van Montfort et al. 2003). To clarify this disagreement, the Kirkwood approximation was used (equation 3 in Materials and Methods). However, although theoretical f values similar to observed f values could be obtained by the Kirkwood approximation, one had to arrange all four domains in an end-to-end fashion, which is implausible based on the structure of βB1.
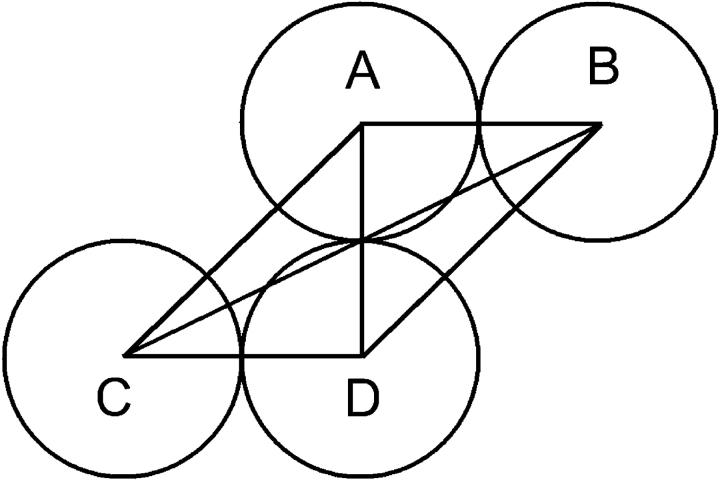
The disagreement can be explained by the terminal, charged extensions of βB1. The published crystal structure has truncated extensions, while those used for these experiments were intact. We have previously shown that the long N-terminal and shorter C-terminal extensions of βB1 are unstructured and have conformational mobility (Lampi et al. 2002). It is therefore plausible that the extensions affect the frictional coefficients significantly, as well as contribute to the overall shape of the aggregate. In support of this hypothesis, a sample of N157D whose SDS-PAGE migration was consistent with truncated extensions had an f of 4.9E-11 kg/sec at a concentration 0.16 mg/mL (data not shown). This is consistent with a Kirkwood approximation using an offset arrangement of spheres (Fig. 7 ▶), which gives an f of 4.8E-11 kg/sec. This conformation is very similar to the published structure of βB1, suggesting that the extensions significantly increase the frictional coefficient. To characterize the shape of intact βB1 in solution a model must be developed that takes the extensions into account.
Figure 7.
Diagram of a Kirkwood approximation of a truncated βB1 dimer using an offset arrangement of spheres (A–D). Each sphere represents a domain. The distance between A and B, C and D, and A and D is 2Rs, where Rs is the radius of the sphere. The distance between B and C is (square root of 5)Rs.
Although the specific shape of the molecule cannot be clearly determined based on these data, Q146E shows a different behavior than wt βB1 and N157D. Both the f and Mw of Q146E increase more rapidly than that of wt and N157D, suggesting an earlier association. f is higher overall for Q146E, suggesting a more extended conformation.
Heat stability of deamidated βB1s
The introduction of a negative charge at Asn157 stabilized βB1 upon heating. This is in contrast to introducing a charge at Gln146, which decreased the heat stability of βB1. Asn157 is located on the exterior of the C-terminal domain, and may increase solvent interaction. This raises the intriguing possibility that deamidation could compensate for age-related loss of the highly charged N-terminal extension of βB1. The structural effects due to deamidation at Gln146 were not able to compensate for the increased solubilizing ability of the additional negative charge.
We have previously reported precipitation of another deamidated βB1, Q204E, during heating (Lampi et al. 2002). Deamidation of βB1 at Gln204 led to a greater tendency to aggregate than did deamidation at Q146E. The precipitation of Q146E during heating was prevented by αA-crystallin (data not shown), while a severalfold excess of αA was required to prevent precipitation of Q204E. Therefore, deamidation at Gln204, located at the intradomain interface of the dimer, may be more detrimental than deamidation at Gln146, located in the connecting peptide between domains.
In conclusion, deamidation at Gln146, but not at Asn157 altered the structure of βB1. This does not preclude deamidation at Asn157 from having an effect on hetero-oligomers, such as those found in the βHigh assembly. Together with our previous studies for deamidation at Gln204, these results suggest that particular sites of deamidation in β-crystallins are more likely to contribute to cataract formation.
Materials and methods
Identification of in vivo sites of deamidation by mass spectrometry
Eyes of organ donors were obtained from the Lions Eye Bank of Oregon and processed as previously described (Lampi et al. 1997). Whole soluble proteins from lenses of 55-year-old and 59-year-old donors were digested by the following method. Samples were reduced and alklyated by dissolving 1 mg of dried protein in 30 μL of 8 M urea, 0.4 M ammonium bicarbonate, followed by addition of 15 μL of 45 mM DTT, incubation at 50°C for 15 min, addition of 15 μL 100 mM iodoacetamide, incubatation at room temperature for 15 min, followed by an additional 15 μL of 45 mM DTT. Twenty micrograms of sequencing-grade, modified trypsin (ProMega) was then added so that the final concentration of urea and ammonium bicarbonate was 2 M and 0.1 M, respectively. Digestion was performed at 37°C overnight, formic acid added to a final concentration of 4%, peptides purified by solid phase extraction using Sep-Pak C18 light cartridges (Waters Corporation), and the samples dried.
Deamidations within digested peptides were then detected by liquid chromatography-mass spectrometry (LC-MS) using an LCQ ion trap mass spectrometer (ThermoFinnigan). Deamidation causes an increase of only one mass unit due to the conversion of an amide to a carboxyl group. To simplify the assignment of deamidation sites in tandem mass spectra, peptides were esterified with either normal (CH3OH) or deuterated (CD3OH) methanol (Aldrich), resulting in a 14 or 17 mass unit increase for each carboxyl group present. The method of Goodlett et al. was used except the esterification reaction was performed for only 5 min (Goodlett et al. 2001). Deamidations between different samples were directly compared by esterifying samples with either regular or deuterated methanol and then analyzing the mixtures.
Portions of esterified peptides (20 μg complex digests or 2 μg digest of recombinant proteins) were then injected onto a 0.5 × 250 mm SB-C18 reversed-phase column (Agilent) and peptides separated by a 15%–35% acetonitrile gradient over 130 min in mobile phase containing 0.2% acetic acid at a 10 μL/min flow rate. Spectra were acquired by alternating MS scans and data-dependent MS/MS scans using dynamic exclusion of ions triggering previous MS/MS scans. Peptides were identified by Sequest analysis of MS/MS spectra (ThermoFinnigan) with static modifications to the masses of Asp, Glu, and peptide C termini of either 14 or 17, depending on whether normal or deuterated methanol was used, respectively. Deamidation sites were identified by performing differential searches of either 15 or 18 mass units to Asn and Gln residues. All identified peptides met these search criteria: Xcorr >1.5 for +1 ions, Xcorr >2.25 for +2 ions, and deltaCN >0.08.
Expression and purification of recombinant proteins
A deamidation at residue 146 in βB1 was introduced by replacing a glutamic acid for a glutamine, and at residue 157 by replacing an aspartic acid for an asparagine. Both mutations were separately introduced into βB1 by PCR-mediated mutagenesis (Quik Change Mutagenesis, Stratagene) using wt βB1 plasmid as the template and internal primers containing the desired mutation. The plasmid containing the human βB1 cDNA (GenBank accession number U35340) in the pCR T7/CT TOPO vector was previously cloned (Lampi et al. 2001). The forward primer to generate βB1Q146E contained the sequence CC ATC AAA ATG GAT GCC GAG GAG CAC AAA ATC TCC. The forward primer to generate βB1N157D contained the sequence C CTG TTT GAA GGG GCC GAC TTC AAG GGC AAC ACC. Reverse primers complementary to the forward primers were also used. Plasmids were sequenced and mutagenesis was confirmed (Nevada Genomics Center). Plasmids were then transformed into BL21Star(DE3) Escherichia coli cells (Invitrogen) for expression as previously described (Lampi et al. 2001).
Both deamidated mutants were purified by cation-exchange chromatography on a 10 × 2.5-cm SP Sepharose Fast Flow column (Amersham Pharmacia Biotech AB) as previously reported for wt βB1 (Lampi et al. 2001). Mass spectrometry was performed to confirm the sites of introduced deamidations as described above. Expression and purification of αA-crystallin was as previously described (Lampi et al. 2002).
Circular dichroism measurements
Circular dichroism spectra were recorded on a JASCO J-500 A spectropolarimeter in the far and near UV range (JASCO). Samples were exhaustively dialyzed into 5 mM NaH2PO4, 5 mM Na2HPO4 (pH 6.8), containing 100 mM NaF, and measured in a 0.005-cm cell for far-UV and in a 1-cm cell for near-UV at 20°C. All spectra were averaged over 30 scans. The far-UV data were subjected to three passes of smoothing and the near-UV data were subjected to four passes of smoothing (Savitzky and Golay 1964). Protein concentration was determined by amino acid analysis.
Associative behavior of expressed proteins
Using a multiangle light scattering (MALS) instrument (miniDAWN, Wyatt Technology), associations of βB1 were observed based on the measured molar masses. The instrument was placed in line with a UV detector and a quasi-elastic light scattering detector (QELS, Wyatt) during size-exclusion chromatography (SEC). SEC was performed with a Superose 12 10/300 GL column (Amersham Biosciences) equilibrated in buffer containing 29 mM Na2HPO4, 29 mM KH2PO4, 100 mM KCl, 0.7 mM EDTA (pH 6.8) with a flow rate of 0.2 mL/min. Samples were concentrated to values between 0.3 and 20 mg/mL, filtered, and 50 μL were applied.
Briefly, MALS directs light from a 685-nm laser through a flow cell such that the intensity of light scattered by the sample is detected simultaneously at several scattering angles. Software provided by the manufacturer calculates the molar mass of a species in a very narrow eluting volume from the intensity of the scattered light according to Rayleigh light scattering principles (Van Holde 1985). The intensity of light scattering by a molecule is proportional to the molar mass, concentration of the solute, and square of the dn/dc of the solute. Measurements were made every 2–5 sec. Concentration was determined from the UV absorbance at 280 nm and an experimentally determined molar extinction coefficient for βB1 of 65,167 M−1cm−1.
The extinction coefficient was determined from the slope of the absorbance at 280 nm versus the concentration of different βB1 solutions times the path length. Known concentrations were based on amino acid analysis. The dn/dc measurement was performed during SEC chromatography using an inline UV detector to measure the concentration in g/mL, and refractive index detector (Opti-lab, Wyatt Technologies) to measure the refractive index, n.
Signals from the UV and MALS detector were normalized using bovine serum albumin. Monodisperse regions under the peak were analyzed. The weight-averaged molar masses (Mw) were reported based on the average protein concentration for the peak area analyzed. The peak area analyzed was approximately 75%. The uncertainty, due to noise in the signal, ranged from 5%–10% with the higher noise at the low concentrations.
Size of expressed proteins
QELS was used to measure the translational diffusional constant (DT). The QELS detector was attached to the 90-degree angle detector on the MALS instrument. For a single species undergoing translational diffusion, the autocorrelation functions are expected to follow a simple exponential function and the data were fit to this model with software provided by the manufacturer. The software reported the radius of hydration assuming a spherical conformation (RH). Frictional coefficient (f) was determined from this value using Stokes law, f = 6πηRH, where η is the viscosity of the solvent. The diffusion coefficient (DT) was determined using the Stokes-Einstein relation, DT = kT/f, where k is the Boltzmann constant and T is the temperature in Kelvin.
βB1 conformations were simulated using several different methods. βB1 was simulated as a prolate and oblate ellipsoid using the following equations where f0 is the theoretical frictional coefficient of βB1 in a spherical conformation and p is the ratio of the major and minor ellipsoid axes (Cantor and Schimmel 1980). By substituting experimentally determined measured f values for fpro and fob, one could determine p, and thus the dimensionality of the molecule if it were in the simulated conformation.
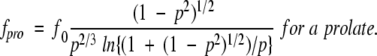 |
(1) |
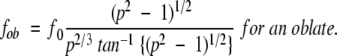 |
(2) |
The Kirkwood theory was also used to simulate conformations of βB1 using a combination of identical spheres that represented the tertiary domains of the molecule. The following equation was used where N is the number of spheres, f1 is the frictional coefficient of an individual sphere, and Rij is the distance between the centers of the spheres. The summation is taken over all pairs of values i and j (Van Holde 1985):
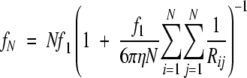 |
(3) |
Size and shape measurements determined for recombinant proteins were then compared to those determined for crystallin assemblies from whole soluble proteins isolated from 3-month-old donor lenses. Due to the complex mixtures of the assemblies, the refractive index and a dn/dc of 0.185, rather than UV absorbance and extinction coefficients, was used to determine concentrations, and DTT was included in the solvent buffer.
Heat-induced aggregation of expressed proteins
The tendency of proteins to aggregate upon heating was measured by the change in turbidity (random light scattering) at 405 nm during heating as previously described (Lampi et al. 2002). Samples at 0.1 mg/mL (3.3 μM) were heated at 55°C in a thermal jacketed cuvette with constant stirring (Cary 4 Bio UV-Visible spectrophotometer, Varian). Samples at room temperature were added to a final volume of 2.5 mL of buffer preheated to 55°C. Due to an initial decrease in absorbance at 405 nm upon mixing, samples were equilibrated with stirring for 10 min. Samples were then heated for an additional 290 min and measurements recorded. Incubations were performed in 10 mM phosphate (pH 6.8), 100 mM NaCl, and 1 mM DTT. Protein at 1.5 mg/mL was similarly heated, centrifuged to separate the pellet and supernatant, and visualized by SDS-PAGE. Measurements were made every 10 sec, and experiments for each sample were repeated three to five times.
Electrophoresis
Electrophoresis was performed using precast 1.0-mm thick, 8 × 8 cm, 10% polyacrylamide Nupage minigels (Invitrogen). Proteins were visualized by staining with Simply Blue Safe Stain (Invitrogen).
Acknowledgments
We thank Christine Slingsby for invaluable discussions of βB1 structure, and April Mixon and Sunyoung Yi for expert technical help. This work was supported in part by National Institutes of Health Grants EY12239 (to K.J.L.), EY07755 (to L.L.D.), and Core Grant EY10572, a Shriners Hospital grant (to H.B.P.), and the Oregon Lions Sight and Hearing Foundation (to K.J.L.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03427504.
References
- Bateman, O.A., Lubsen, N.H., and Slingsby, C. 2001. Association behaviour of human βB1-crystallin and its truncated forms. Exp. Eye Res. 73 321–331. [DOI] [PubMed] [Google Scholar]
- Bax, B., Lapatto, R., Nalini, V., Driessen, H., Lindley, P.F., Mahadevan, D., Blundell, T.L., and Slingsby, C. 1990. X-ray analysis of β B2-crystallin and evolution of oligomeric lens proteins. Nature 347 776–780. [DOI] [PubMed] [Google Scholar]
- Bindels, J.G., Koppers, A., and Hoenders, H.J. 1981. Structural aspects of bovine β-crystallins: Physical characterization including dissociation–association behavior. Exp. Eye Res. 33 333–343. [DOI] [PubMed] [Google Scholar]
- Blundell, T., Lindley, P., Miller, L., Moss, D., Slingsby, C., Tickle, I., Turnell, B., and Wistow, G. 1981. The molecular structure and stability of the eye lens: X-ray analysis of γ-crystallin II. Nature 289 771–777. [DOI] [PubMed] [Google Scholar]
- Cantor, C.R. and Schimmel, P.R. 1980. Biophysical chemistry. W.H. Freeman, San Francisco.
- Chiou, S.-H., Azari, P., Himmel, M.E., Lin, H.-K., and Chang, W.-P. 1989. Physicochemical characterization of β-crystallins from bovine lenses: Hydrodynamic and aggregation properties. J. Protein Chem. 8 19–32. [DOI] [PubMed] [Google Scholar]
- Goodlett, D.R., Keller, A., Watts, J.D., Newitt, R., Yi, E.C., Purvine, S., Eng, J.K., von Haller, P., Aebersold, R., and Kolker, E. 2001. Differential stable isotope labeling of peptides for quantitation and de novo sequence derivation. Rapid Commun. Mass Spectrom. 15 1214–1221. [DOI] [PubMed] [Google Scholar]
- Groenen, P.J., van Dongen, M.J., Voorter, C.E., Bloemendal, H., and de Jong, W.W. 1993. Age-dependent deamidation of α B-crystallin. FEBS Lett. 322 69–72. [DOI] [PubMed] [Google Scholar]
- Hanson, S.R., Smith, D.L., and Smith, J.B. 1998. Deamidation and disulfide bonding in human lens γ-crystallins. Exp. Eye Res. 67 301–312. [DOI] [PubMed] [Google Scholar]
- Hanson, S.R., Hasan, A., Smith, D.L., and Smith, J.B. 2000. The major in vivo modifications of the human water-insoluble lens crystallins are disulfide bonds, deamidation, methionine oxidation and backbone cleavage. Exp. Eye Res. 71 195–207. [DOI] [PubMed] [Google Scholar]
- Kim, Y.H., Kapfer, D.M., Boekhorst, J., Lubsen, N.H., Bachinger, H.P., Shearer, T.R., David, L.L., Feix, J.B., and Lampi, K.J. 2002. Deamidation, but not truncation, decreases the urea stability of a lens structural protein, βB1-crystallin. Biochemistry 41 14076–14084. [DOI] [PubMed] [Google Scholar]
- Kramps, J.A., de Jong, W.W., Wollensak, J., and Hoenders, H.J. 1978. The polypeptide chains of α-crystallin from old human eye lenses. Biochim. Biophys. Acta 533 487–495. [DOI] [PubMed] [Google Scholar]
- Lampi, K.J., Ma, Z., Shih, M., Shearer, T.R., Smith, J.B., Smith, D.L., and David, L.L. 1997. Sequence analysis of βA3, βB3, and βA4 crystallins completes the identification of the major proteins in young human lens. J. Biol. Chem. 272 2268–2275. [DOI] [PubMed] [Google Scholar]
- Lampi, K.J., Ma, Z., Hanson, S.R.A., Azuma, M., Shih, M., Shearer, T.R., Smith, D.L., Smith, J.B., and David, L.L. 1998. Age-related changes in human lens crystallins identified by two-dimensional electrophoresis and mass spectrometry. Exp. Eye Res. 67 31–43. [DOI] [PubMed] [Google Scholar]
- Lampi, K.J., Oxford, J.T., Bachinger, H.P., Shearer, T.R., David, L.L., and Kapfer, D.M. 2001. Deamidation of human βB1 alters the elongated structure of the dimer. Exp. Eye Res. 72 279–288. [DOI] [PubMed] [Google Scholar]
- Lampi, K.J., Kim, Y.H., Bachinger, H.P., Boswell, B.A., Lindner, R.A., Carver, J.A., Shearer, T.R., David, L.L., and Kapfer, D.M. 2002. Decreased heat stability and increased chaperone requirement of modified human βB1-crystallins. Mol. Vision 8 359–366. [PubMed] [Google Scholar]
- Lapatto, R., Nalini, V., Bax, B., Driessen, H., Lindley, P.F., Blundell, T.L., and Slingsby, C. 1991. High resolution structure of an oligomeric eye lens β-crystallin. Loops, arches, linkers and interfaces in β B2 dimer compared to a monomeric γ-crystallin. J. Mol. Biol. 222 1067–1083. [DOI] [PubMed] [Google Scholar]
- Lapko, V.N., Purkiss, A.G., Smith, D.L., and Smith, J.B. 2002. Deamidation in human γ S-crystallin from cataractous lenses is influenced by surface exposure. Biochemistry 41 8638–8648. [DOI] [PubMed] [Google Scholar]
- Lund, A.L., Smith, J.B., and Smith, D.L. 1996. Modifications of the water-insoluble human lens α-crystallins. Exp. Eye Res. 63 661–672. [DOI] [PubMed] [Google Scholar]
- Savitzky, A., and Golay, M.J.E. 1964. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36 1627–1639. [DOI] [PubMed] [Google Scholar]
- Srivastava, O.P. and Srivastava, K. 2003. Existence of deamidated αB-crystallin fragments in normal and cataractous human lenses. Mol. Vision 9 110–118. [PubMed] [Google Scholar]
- Takemoto, L. and Boyle, D. 1998. Deamidation of specific glutamine residues from α-A crystallin during aging of the human lens. Biochemistry 37 13681–13685. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Increased deamidation of asparagine during human senile cataractogenesis. Mol. Vision 6 164–168. [PubMed] [Google Scholar]
- Van Holde, K.E. 1985. Physical biochemistry, 2nd ed., pp. xii, 287. Prentice-Hall, Englewood Cliffs, NJ.
- Van Montfort, R.L., Bateman, O.A., Lubsen, N.H., and Slingsby, C. 2003. Crystal structure of truncated human βB1-crystallin. Protein Sci. 12 2606–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorter, C.E., Roersma, E.S., Bloemendal, H., and de Jong, W.W. 1987. Age-dependent deamidation of chicken α A-crystallin. FEBS Lett. 221 249–252. [DOI] [PubMed] [Google Scholar]
- Wright, G., Basak, A.K., Wieligmann, K., Mayr, E.M., and Slingsby, C. 1998. Circular permutation of βB2-crystallin changes the hierarchy of domain assembly. Protein Sci. 7 1280–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Chamorro, M., Smith, D.L., and Smith, J.B. 1994. Identification of the major components of the high molecular weight crystallins from old human lenses. Curr. Eye Res. 13 415–421. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Smith, D.L., and Smith, J.B. 2003. Human β-crystallins modified by backbone cleavage, deamidation and oxidation are prone to associate. Exp. Eye Res. 77 259–272. [DOI] [PubMed] [Google Scholar]